Graphical abstract
Keywords: Deep-sea sediment, virome of RNA viruses, Viral community, Virus-encoded gene
Highlights
-
•
Global deep-sea sediment RNA viromes reveal 85,059 viral operational taxonomic units (vOTUs), the largest number of RNA viruses known so far.
-
•
Most vOTUs (98.28%) are unclassified, indicating a vast reservoir of novel RNA viruses in the deep sea.
-
•
A total of 1,463 complete RNA viral genomes are found, expanding our understanding of the RNA viruses in the deep-sea ecosystems.
-
•
Ecosystems rather than geographical locations drive the differentiation of deep-sea RNA viral communities.
-
•
Viral metabolic genes participate in the ecosystem-driven viral community differentiation via mediating the energy metabolism.
Abstract
Introduction
Viruses are the most abundant and diverse life forms on the earth. Both DNA viruses and RNA viruses play important roles in marine ecosystems via regulating biogeochemical cycles.
Objectives
However, the virome of marine RNA viruses has been rarely explored so far. In this study, therefore, the environmental viromes of deep-sea sediment RNA viruses were characterized on a global scale to reveal the global virosphere of deep-sea RNA viruses.
Methods
The viral particles were purified from each of 133 deep-sea sediment samples and then characterized based on metagenomes of RNA viruses.
Results
In this study, we established the global virome dataset of deep-sea RNA viruses purified from 133 sediment samples that were collected from typical deep-sea ecosystems of three oceans. A total of 85,059 viral operational taxonomic units (vOTUs) were identified, of which only 1.72% were hitherto known, indicating that the deep-sea sediment is a repository of novel RNA viruses. These vOTUs were classified into 20 viral families, including prokaryotic (7.09%) and eukaryotic (65.81%) RNA viruses. Furthermore, 1,463 deep-sea RNA viruses with complete genomes were obtained. The differentiation of RNA viral communities was driven by the deep-sea ecosystems as opposed to geographical region. Specifically, the virus-encoded metabolic genes took great effects on the differentiation of RNA viral communities by mediating the energy metabolism in the deep-sea ecosystems.
Conclusions
Therefore, our findings indicate that the deep sea is a vast reservoir of novel RNA viruses for the first time, and the differentiation of RNA viral communities is driven by the deep-sea ecosystems through energy metabolism.
Introduction
Viruses, the highly abundant organisms in oceanic habitats, exhibit considerable diversity and ecological functions [1], [2], [3]. The oceans, covering 70 % of the earth’s surface, are habitats for many kinds of viruses [4]. A total of 195,728 marine viral populations have been previously identified based on the analysis of a global ocean DNA virome dataset [5]. Another study showed that while most marine viruses are novel, they are related to dominant, ecologically relevant microbial hosts [6] and therefore likely influence biogeochemical cycles in marine ecosystems by infecting the hosts [2], [5], [7]. Many virus-encoded genes directly participate in sulfur and nitrogen cycling in the epipelagic zones of the oceans [6], [7]. In the deep-sea hydrothermal vents, the viruses can compensate host metabolism by mediating branched metabolic pathways, thus enabling the hosts to survive in extreme environments [8]. While marine DNA viruses have been well characterized, little is known about the composition and diversity of marine RNA viruses as well as deep-sea viruses.
Although most of the currently characterized marine viruses are double-stranded DNA (dsDNA) viruses [9], RNA viruses constitute over half of the entire marine viral community [10]. In fact, it is estimated that the marine RNA viruses may be as abundant as or even exceed marine DNA viruses [11], [12], [13]. Most marine RNA viruses are positive-sense, single-stranded RNA (+ssRNA) viruses, which are related to the current classification [11]. A few double-stranded RNA (dsRNA) viruses, negative-sense single-stranded RNA (−ssRNA) viruses or retroviruses are known, but very few RNA bacteriophage has been identified in the marine habitats [11]. Almost all of the RNA viruses that constitute the marine virioplankton can infect eukaryotic organisms, especially protists like diatoms, dinoflagellates, raphidophytes, prasinophytes and thraustochytrids [11]. Recently a novel RNA virus was isolated from the deep-sea tubeworm Osedax japonicus, indicating that marine RNA viruses can infect multicellular animals as well [12]. Nevertheless, the RNA viruses of deep-sea sediments are largely unexplored.
The deep sea is one of the largest and the least explored ecosystems on the earth [14], [15]. The extreme conditions of deep-sea habitats, such as high pressure, low temperature, low nutrient levels and the lack of sunlight and hypoxia [16], [17], exert an extraordinary selective pressure on the deep-sea organisms to adapt to the environment [16]. At present, however, the deep-sea viruses that can be imported into deep-sea environments and organisms are insufficiently investigated. In this study, we analyzed the global virome of RNA viruses purified from each of 133 deep-sea sediment samples and found that deep-sea RNA viruses are abundant and highly diverse, and most are hitherto unclassified. In addition, the distribution of these viruses is affected by the environmental factors as opposed to the geographic location.
Materials and methods
Collection of deep-sea sediment samples
The deep-sea sediment samples, derived from oceanic vessel No.1 (Dayang No. 1) geomicrobiology cruises of China, were collected during the 26th, 30th, 34th, 39th, 40th and 45th cruises in the Pacific, Atlantic and Indian Oceans from 2012 to 2018 (Table S1). The samples came from various deep-sea ecosystems including hydrothermal vents, cold seeps, seamounts and ocean basins with depth ranging from 1,100 to 6,105 m. The sampling of sediments was performed using sealable sampling boxes. Upon arrival on the deck, the surface of each sediment sample was removed with a sterile shovel to exclude exogenous contamination. The samples were stored at −80 °C before experiments.
Purification of viral particles from deep-sea sediment samples
A deep-sea sediment (20 g) was resuspended using 10 ml prefiltered (0.015-μm pore size) Milli-Q water and glass beads, and then incubated with shaking for 30 min at 4 °C. Subsequently the mixture was centrifuged at 5,000 × g for 20 min (4 °C) to collect the supernatant. After repeating these steps for six times, all the supernatants were collected and centrifuged at 5,000 × g for 10 min at 4 °C. The supernatant was filtered through a 0.22-μm filter to remove cells. The filtrate was added with PEG6000 at a final concentration of 10 % and then incubated at 4 °C overnight. After ultracentrifugation at 200,000 × g for 2 h, the pellet was collected. The purified viral particles were dissolved in SM buffer (400 mmol/L NaCl, 20 mmol/L MgSO4·7H2O, 50 mmol/L Tris-HCl, pH7.5).
Observation of viral particles with transmission electron microscopy
The purified viral particles were negatively stained by 2 % phosphotungstic acid (pH7.0). Subsequently the samples were examined under JEM-1230 transmission electron microscope (TEM) operating at 120 kV.
RNA extraction, reverse transcription and amplification of deep-sea RNA viruses
To exclude the contamination of exogenous DNA and RNA, the purified deep-sea virions were treated with DNase and RNase at 37℃ for 1 h. Subsequently the mixture was incubated in 20 μL 1 M ethylene diamine tetraacetic acid (EDTA) (pH8.0) at 65℃ for 10 min, followed by the extraction of viral genomic RNAs with the RNA purification kit according to the manufacturer’s protocol (NorgenBiotek Corp, Thorold, Canada). The extracted RNAs were treated with RNase-free DNase at 37℃ for 30 min. Then the reverse transcription was performed with random hexamers using HiScript II 1st strand cDNA synthesis kit (+gDNA wiper) according to the manufacturer’s manual (Vazyme, Nanjing, China), followed by the synthesis of the second-strand cDNA with the second strand cDNA synthesis kit (Beyotime Biotechnology, Shanghai, China). Finally the cDNA was subjected to isothermal amplification using GenomiPhiTM V2 DNA amplification kit (GE Healthcare Life Sciences, Buckinghamshire, UK) with random hexamers. The amplification condition was 95 °C for 3 min, 30 °C for 3 h and 65 °C for 10 min.
Detection of bacterial 16S rRNA gene for the prepared samples
To exclude the bacterial contamination, the viral genomic RNAs extracted from the purified deep-sea virions of each of 133 deep-sea sediments, the first-strand cDNAs, the second-strand cDNAs and the products of isothermal amplification were subjected to PCR using the bacterial 16S rRNA gene-specific primers (515F, 5′-GTGCCAGCMGCCGCGG-3′; 907R, 5′-CCGTCAA TTCMTTTRAGTTT-3′) (M = A/C; R = A/G). The PCR was conducted at 95 °C for 5 min, followed by 30 cycles of 95 °C for 30 s, 55 °C for 30 s, 72 °C for 30 s, and 72 °C for 10 min.
Sequencing of RNA viral metagenome
The products of isothermal amplification from RNA viruses were checked using a 1 % agarose gel, purified using AxyPrep DNA Gel Extraction Kit (Axygen, China) and then quantified with the QuantiFluor-ST fluorescence quantitative system (Promega, CA, USA). After the treatment with an M220 focused ultrasonicator (Covaris Inc., Woburn, MA, USA), DNA was sheared and 400-bp fragments were excised and extracted. The paired-end library was prepared with the TruSeq DNA sample prep kit (Illumina Inc., San Diego, CA, USA). Subsequently the paired-end sequencing (2 × 250 bp) was conducted on an IlluminaHiSeq 2500 system (Illumina Inc., SanDiego, CA, USA). The sequencing was cooperated with Mingke Biotechnology Co., ltd. (Hangzhou, China). After removing the adaptor sequences and duplicate reads, the raw sequence reads were trimmed to get the clean reads (clean data).
Assembly of contigs and identification of viral contigs
To assemble contigs, the clean reads were trimmed with Trimmomatic (version:0.33, default parameters) [18] and then assembled into contigs using metaSPAdes 3.12.0 [19].
To confirm the viral contigs, the assembled contigs were further screened for viral contigs using the commonly used protocols for the identification of viral contigs including VirSorter [20], VirFinder [21], VIBRANT [22] and CAT [23]. Up to date, the known smallest viral genome is 1.7 kb of human hepatitis D virus [24]. Therefore, the contigs of ≥ 1.0 kb in length were selected. The contigs (≥1.0 kb) were screened using the following algorithms: (1) VirSorter categories 1 and 2, (2) VirFinder score ≥ 0.9 and p < 0.05, (3) both VirSorter (categories 1–6) and VirFinder (score ≥ 0.7 and p < 0.05) or (4) all the contigs screened by VIBRANT. The contigs that were not selected by the VirSorter, VirFinder and VIBRANT algorithms were further screened using the CAT algorithm [23]. In this algorithm, the contigs with < 40 % of the genomes classified as bacteria, archaea or eukaryotes were considered as viral contigs [5]. All the viral contigs identified by Virsorter, VirFinder, VIBRANT and CAT algorithms were pooled together and the duplications were removed. These algorithms identify viral contigs by searching the viral RefSeq, the Kyoto encyclopedia of genes and genomes (KEGG), Pfam or/and viral orthologous groups (VOG) databases. In different algorithms, the databases as well as the algorithms are different. Thus, the viral contigs obtained from different algorithms were pooled as described before [5]. To get more viral contigs, the contigs (≥1.0 kb) that were not identified by VirSorter, VirFinder, VIBRANT and CAT were searched for RNA-dependent RNA polymerase (RdRp) and reverse transcriptase (or RNA-dependent DNA polymerase) based on viral RefSeq database (release 201) [25], NCBI non-redundant protein sequence database and IMG/VR database. The contigs contained at least one of RdRp and reverse transcriptase (or RNA-dependent DNA polymerase) were considered to be viral contigs. The viral contigs represented the viral genomes.
To evaluate the completeness of viral genomes, the sequences of viral contigs were subjected to CheckV v7.0 analysis [26].
Identification of viral operational taxonomic units
Nucmer pipeline of MUMmer 4.0 [27] was used to classify the viral contigs into viral operational taxonomic units (vOTUs). The viral contigs were classified into vOTUs if the coverage between viral contigs was larger than 80 % of the shortest contig, and this coverage shared ≥ 95 % mummer-based average nucleotide identity according to the strategy described previously [5].
Establishment of the known RNA virus genomic sequences database
To facilitate the identification of deep-sea viral taxonomy, all of the genomic sequences of the known RNA viruses, which contained the viral taxonomic information or/and the viral origins (such as environments), were collected from the public databases to establish the known RNA virus genomic sequences database. The public databases included NCBI (National Center for Biotechnology Information) viral RefSeq database (https://www.ncbi.nlm.nih.gov/refseq/), NCBI non-redundant protein sequence database (https://ftp.ncbi.nlm.nih.gov/blast/db/FASTA/), GenBank (https://www.ncbi.nlm.nih.gov/genbank/) and IMG/VR database (https://img.jgi.doe.gov/cgi-bin/vr/main.cgi). These public databases contained the information of viral taxonomy or/and viral origins. The established known RNA virus genomic sequences database was used to perform taxonomy annotation of vOTUs.
Identification of viral taxonomy
To identify viral taxonomy, Prodigal was utilized to predict open reading frames (ORFs) of each Votu [28]. Based on protein sequences of the predicted ORFs, viral taxonomy of vOTUs were identified using blastp (ftp://ftp.ncbi.nlm.nih.gov/blast/executables/magicblast/LATEST) as described previously [5]. The vOTUs were aligned with our self-built database (the known RNA virus genomic sequences database). In the meanwhile, the RNA-dependent RNA polymerase (RdRp) and reverse transcriptase of vOTUs were also aligned with our self-built database. Although MG-RAST (metagenomics RAST) is an online platform for metagenomic analysis, it is challenging to identify the taxonomy of RNA viruses solely based on metagenomics. In our self-built database, the viral taxonomic information or/and the viral origin information (such as environments) was available. The vOTUs would be classified into a known virus if greater than 50 % ORFs were aligned to that kind of virus and the blastp bit score was ≥ 50 according to the standard viral taxonomy method [5].
Analysis of RNA viruses with circular genomes
To obtain the RNA viruses with circular genomes, the sequences of vOTUs of deep-sea RNA viruses were loaded into the Cenote-Taker software (https://github.com/mtisza1/Cenote-Taker) [29]. Based on the overlapping ends, the viruses with circular genomes were identified.
Calculation of the relative abundance of vOTUs and viral ORFs
To reveal the relative abundance of vOTUs, the vOTUs of every sample were mapped to the reads using the software bowtie2 v2.4.4 [30]. After removal of the reads with low-quality mapping by BamM v1.7.3 (https://github.com/Ecogenomics/BamM) with parameters --percentage_id 0.95 --percentage_aln 0.75, the reads per kilobase per million mapped reads (RPKM) values were obtained based on the analysis using CoverM v0.3.1 (https://github.com/wwood/CoverM) (--percentage_id 0.95 --percentage_aln 0.75 -rpkm) [31].
To evaluate the relative abundance of viral ORFs of each sample, bowtie2 v2.4.4 was used to map the reads of each sample to viral ORFs [30]. After removal of the reads with low-quality mappings by BamM v1.7.3 (https://github.com/Ecogenomics/BamM) (--percentage_id 0.95 --percentage_aln 0.75), the reads per kilobase per million mapped reads (RPKM) values were generated using CoverM v0.3.1 (https://github.com/wwood/CoverM) (--percentage_id 0.95 --percentage_aln 0.75 -rpkm) [31].
Diversity analysis of deep-sea RNA viruses
To characterize the diversity of vOTUs across three oceans (the Pacific, Atlantic and Indian Oceans) or four ecosystems (hydrothermal vent, cold seep, seamount and ocean basin), α-diversity indices (Shannon, Simpson, Chao and Ace) were calculated using vegan in R. Boxplots were prepared using GraphPad Prism 8.0 (https://www.graphpad.com/) to show the changes of four indices in different oceans or ecosystems. At the same time, the β diversity of deep-sea RNA viruses was characterized using principal coordinate analysis (PCoA) with vegan package in R [32].
Principal co-ordinate analysis (PCoA) of deep-sea RNA viruses
PCoA was carried out to reveal the diversity of deep-sea RNA viruses using vegan package in R [31]. Bray-Curtis dissimilarity matrices were generated from both the subsampled and total reads of global deep-sea RNAs viruses by vegdist (method = bray) after a cube root transformation by function nthroot (n = 3) [5].
Identification of the proteins encoded by vOTUs of deep-sea RNA viruses
To identify the proteins encoded by deep-sea RNA viruses, the ORFs predicted from the deep-sea vOTUs in this study were annotated by (1) blast hit analysis against the Kyoto encyclopedia of genes and genomes (KEGG) database [33], (2) blast against the UniProt Reference Clusters database [34], (3) searching for matches against the InterPro protein signature database using InterProScan [35] and (4) HMM searches against Pfams [36]. After removal of duplications of the annotated viral proteins using 4 algorithms, the unique viral proteins were clustered by CD-HIT at 60 % identity, 80 % coverage and “-g 1 -n 4 -d 0” [37]. The resulting protein clusters were compared with the Refseq Virus database and IMG/VR v3 by blastp in DIAMOND with an e-value threshold of 1e−5, identity of 30 % and coverage of 50 % [25].
Phylogenetic analysis of deep-sea RNA viruses with circular viral genomes
To reveal the relationship between the classified deep-sea RNA viruses with circular viral genomes and the known RNA viruses, the phylogenetic analysis was carried out using MEGA (version 7.0.26) [38]. The amino acid sequences of gag protein, protease and reverse transcriptase of deep-sea RNA viruses with circular viral genomes were aligned with the homologous sequences in NCBI nr database (https://www.ncbi.nlm.nih.gov/protein/?term=) through the MUSCLE algorithm (v3.8). The phylogenetic trees were produced by neighbor-joining method [39]. The confidence coefficient was tested by bootstrap test (500 replicates).
Results
Global virome of deep-sea RNA viruses
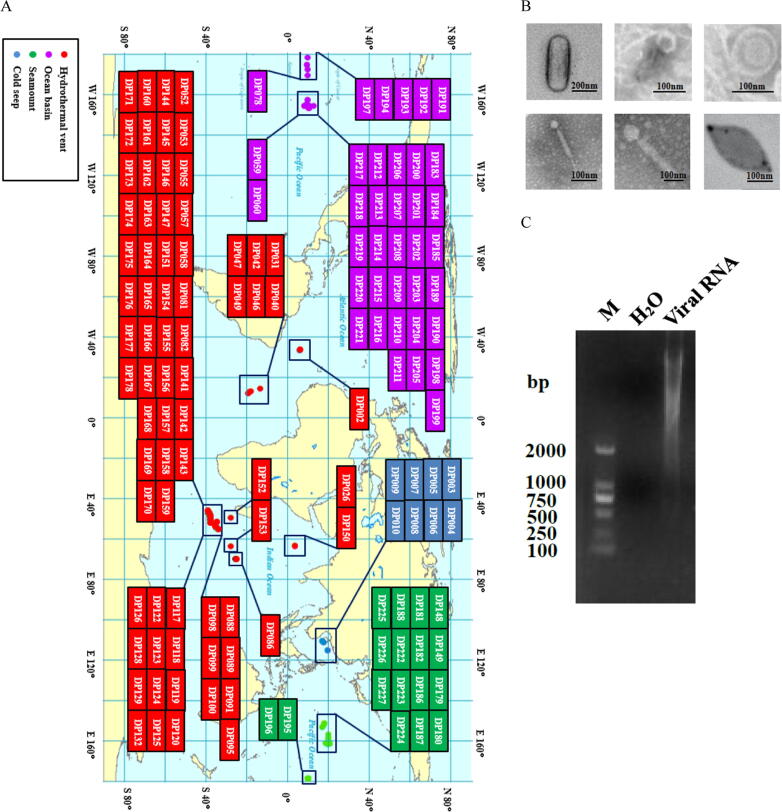
A total of 133 deep-sea sediment samples were collected during six cruises of the geomicrobiology cruise of China, which traveled more than 3,200,000 km from 2012 to 2018 (Fig. 1A and Table S1). The samples were retrieved from four deep-sea ecosystems, including hydrothermal vents (71 samples), cold seeps (8 samples), seamounts (17 samples) and ocean basins (37 samples), from the Pacific, Atlantic and Indian Oceans (Fig. 1A and Table S1). The depths of sampling stations ranged from 1,100 to 6,105 m below surface, and the average depth was 3,545.3 m (Table S1). The environmental types of sampling stations represented the typical deep-sea ecosystems on the planet.
Fig. 1.
Global virome of deep-sea RNA viruses. (A) The distribution of global deep-sea sediment stations. The spots represent the locations of deep-sea stations. (B) The representative images of the purified viral particles from 133 deep-sea sediments. The viruses were examined using transmission electron microscopy. Scale bar, 100 nm or 200 nm. (C) The representative amplified products of viral RNAs from deep-sea sediments. The viruses purified from each of 133 sediments were subjected to RNA extraction, reverse transcription and isothermal amplification of cDNA. Distilled water was used as a control. M, DNA marker.
To characterize the global virome of deep-sea RNA viruses, the viruses from each of 133 sediments were purified, followed by RNA extraction, reverse transcription and isothermal amplification of cDNA. Transmission electron microscopic analysis indicated that the purified viral particles were obtained (Fig. 1B). The size of the amplified DNAs from the viral RNAs ranged from a few hundred base pairs (bp) to several kilo bp (kb) (Fig. 1C), suggesting that the purified viral particles had excluded contaminating extracellular vesicles since these nanoparticles usually contain small RNAs [40]. At the same time, when the RNAs extracted from the purified viral particles of each of 133 sediments were subjected to isothermal amplification with random hexamers, no DNA was amplified for all samples (Fig S1), indicating that the extracted RNAs had no DNA contamination, which excluded the presence of bacteria, archaea and eukaryotes. Furthermore, no bacterial 16S rRNA sequences were detected in the extracted viral RNAs or corresponding cDNA samples from each of 133 sediments (Fig S2), confirming their original sources of the deep-sea viral genomes. These data indicated that the extracted viral genomic RNAs could be used for sequencing the global virome of deep-sea RNA viruses.
To minimize sequencing errors, we established 400-bp fragment libraries of viral metagenomic DNAs obtained from the genomic RNAs, which contained the longest reads for next-generation sequencing or high-throughput sequencing analysis. A total of 2,889,656,790 completely sequenced reads and 673.721 Gb clean data were generated from all 133 samples, with each sample yielding more than 3.5 Gb clean data, from which the adapter sequences and duplicate reads were removed (Table S1). The level of reads duplication in all samples was<10 %. Further analyses were performed on this dataset of deep-sea RNA viruses.
Extreme diversity of deep-sea RNA viruses
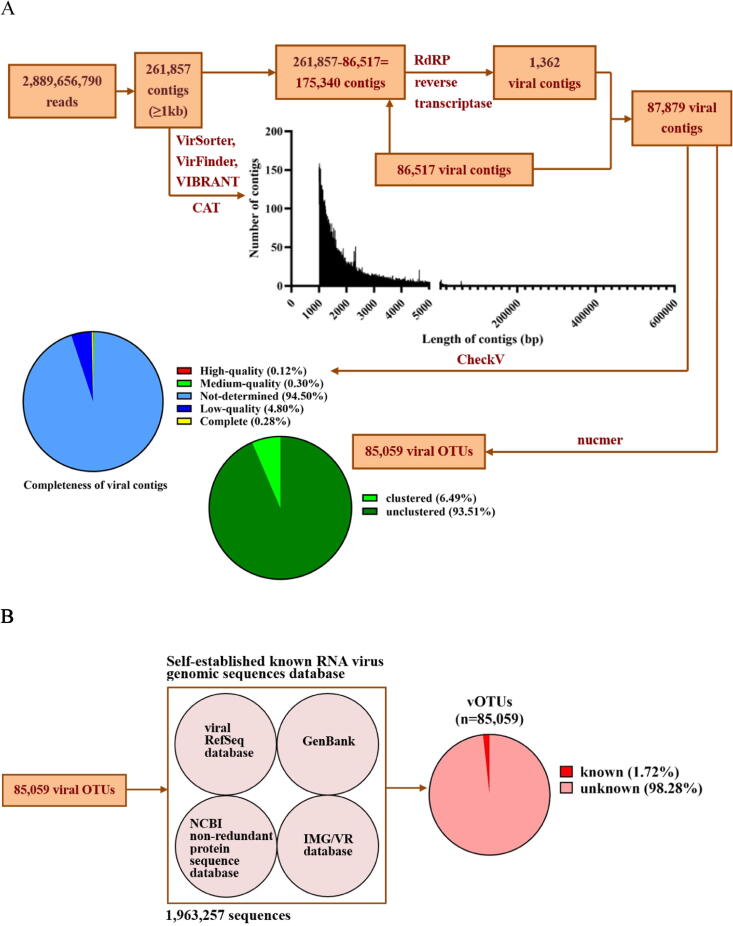
To identify the community of global deep-sea RNA viruses, the sequenced reads of the deep-sea RNA viruses were assembled into 261,857 contigs (≥1.0 kb) (Fig. 2A). The coverage for contigs was more than 10 times. To confirm the viral contigs, the 261,857 assembled contigs were further screened for viral contigs. A total of 86,517 contigs were assigned to viral contigs by VirSorter, VirFinder, VIBRANT and CAT analyses (Fig. 2A and Table S2). To identify any more viral contigs that might have been missed in the first-round search, a second-round search for RNA-dependent RNA polymerase (RdRp) and reverse transcriptase (or RNA-dependent DNA polymerase), the signature proteins of RNA viruses, was carried out. Another 1,362 contigs were found (Fig. 2A and Table S2), which represented 1,362 RNA virotypes. All 87,879 viral contigs have been deposited into the National Omics Data Encyclopedia database (accession number OEP002537). To facilitate the viral taxonomy, the 87,879 viral contigs were further clustered. Based on nucmer analysis, all viral contigs were classified into 85,059 viral operational taxonomic units (vOTUs) (Fig. 2A and Table S2), i.e. 85,059 deep-sea RNA viruses. Most of vOTUs (96 %) were unique in 133 deep-sea sediment samples. These data indicated the extreme diversity of deep-sea RNA viruses.
Fig. 2.
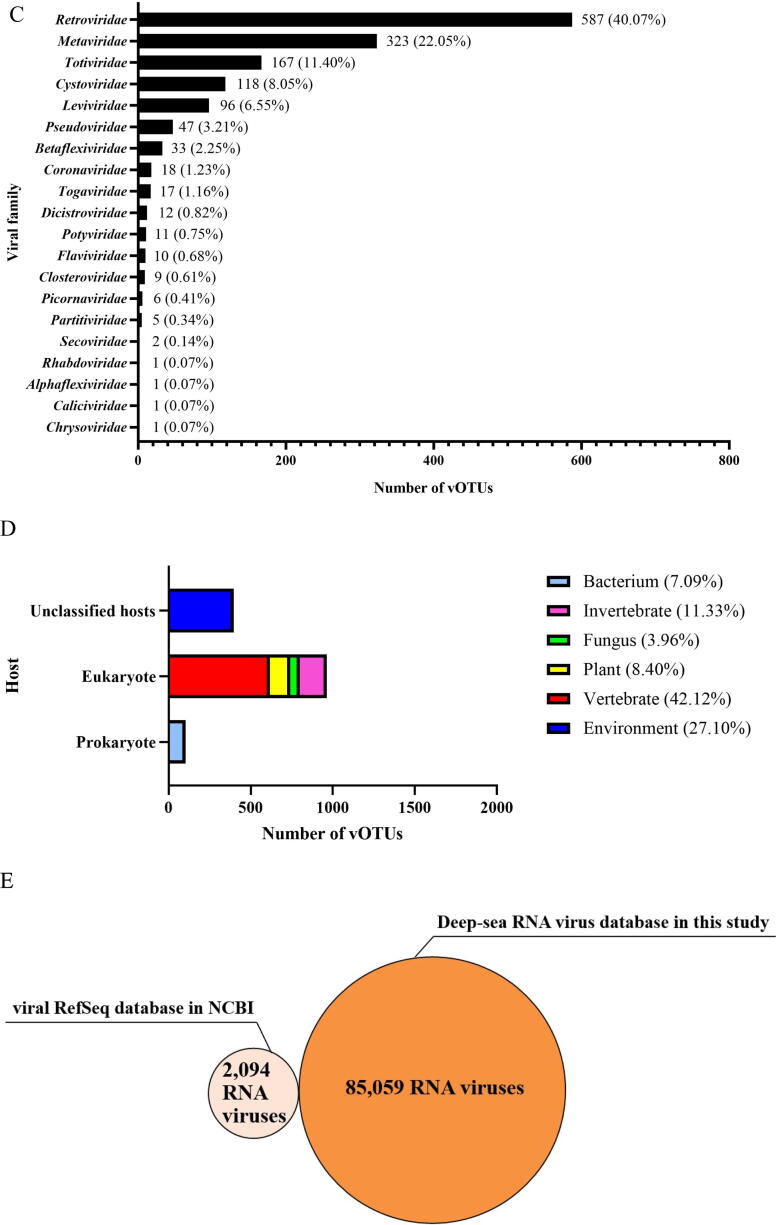
Extreme diversity of deep-sea RNA viruses. (A) Identification of viral operational taxonomic units (vOTUs). After assembly of total reads, 261,857 contigs (≥1kb) were obtained. The subsequent analyses by VirSorter, VirFinder, VIBRANT, CAT, RNA-dependent RNA polymerase (RdRp) and reverse transcriptase (or RNA-dependent DNA polymerase) analyses generated 87,879 viral contigs, the completeness of which was determined by CheckV. Based on the analysis using nucmer pipeline of MUMmer 4.0, the viral contigs were clustered into 85,059 vOTUs. (B) The relative proportion of unknown or known vOTU in the entire dataset. (C) The families of classified vOTUs and their abundance. (D) The proportion of hosts of the vOTUs matching the known RNA viruses. (E) The number of deep-sea RNA virus database in our study and viral RefSeq database in NCBI.
Since most viral metagenomic sequences cannot be annotated and classified [41], it is challenging to identify the taxonomy of RNA viruses solely on the basis of metagenomics. Therefore, we established a dataset of 1,963,257 genomic sequences of known RNA viruses from public databases that contained the viral taxonomic and/or viral origin information (such as environments). Only 1.72 % vOTUs matched the known sequences and the majority of vOTUs were unknown (Fig. 2B and Table S3), indicating that the deep-sea sediment was a repository of novel RNA viruses.
The vOTUs that matched the known viral sequences were taxonomically assigned to 20 families of RNA viruses (Fig. 2C and Table S3). Retroviridae was the most abundant family that accounted for 40.07 % of all known vOTUs, followed by Metaviridae (22.05 %), Totiviridae (11.40 %), Cystoviridae (8.05 %), Leviviridae (6.55 %), Pseudoviridae (3.21 %), Betaflexiviridae (2.25 %), Coronaviridae (1.23 %) and Togaviridae (1.16 %) (Fig. 2C). The remaining 11 families with abundance<1 % accounted for 4.03 % of all known vOTUs (Fig. 2C). These results indicated that deep-sea sediments harbored diverse RNA viruses.
Depending on their hosts, 7.09 % of the classified vOTUs were prokaryotic, 65.81 % were eukaryotic, and 27.1 % could not be assigned to any host (Fig. 2D and Table S3). No archaeal RNA virus was detected, and the vertebrate-infecting viruses accounted for 42.12 % of the known eukaryotic viruses (Fig. 2D and Table S3). These data revealed that the eukaryotic viruses accounted a large proportion of the classified RNA viruses in the deep-sea sediments.
Taken together, we established a dataset of 85,059 deep-sea RNA viruses, which is considerably larger than the viral RefSeq database of NCBI (National Center for Biotechnology Information) that includes only 2,094 RNA viruses (Fig. 2E). Our findings indicate a highly diverse community of RNA viruses the in deep-sea sediments, expanding our understanding of RNA viruses on the earth.
RNA viruses with complete circular genomes in deep sea
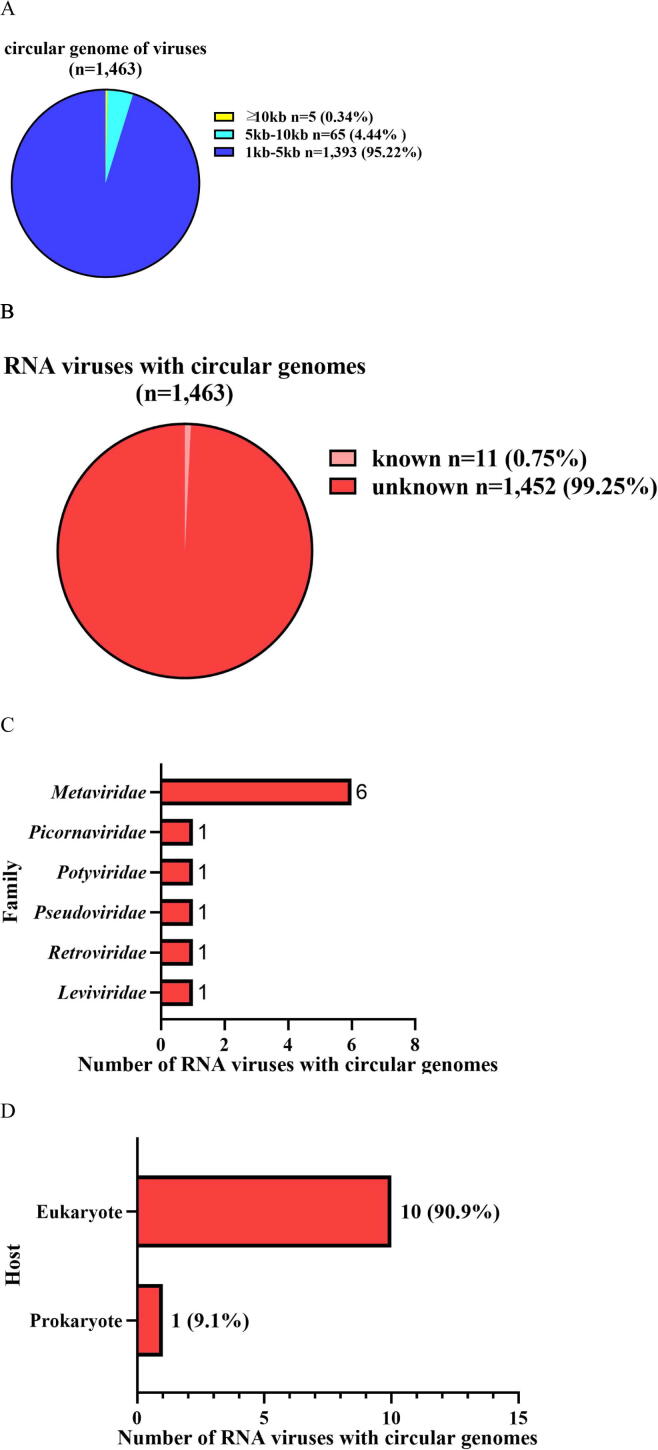
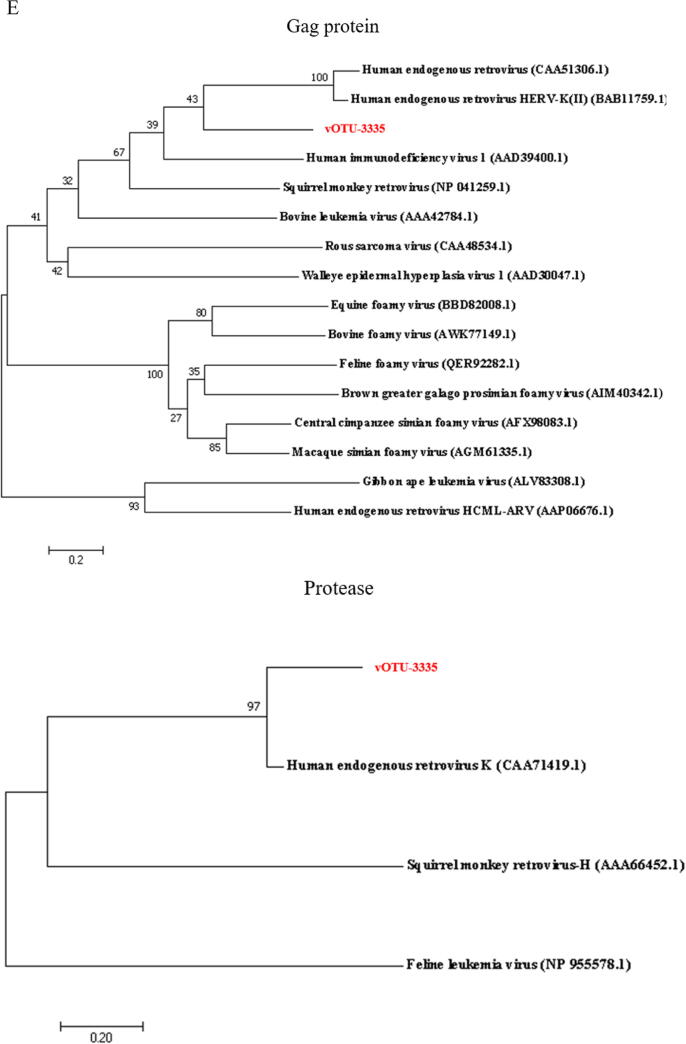
In an attempt to obtain the RNA viruses with complete genomes, the sequences of vOTUs were further analyzed to identify circular viral genomes (CVGs) based on overlapping ends. A total of 1,463 CVGs were obtained which ranged from 1.044 kb to 18.595 kb, representing 1,463 deep-sea RNA viruses with complete genomes (Fig. 3A and Table S4). Only 0.75 % of the CVGs matched the genomic sequences of known viruses, and the remaining were indicative of novel viruses (Fig. 3B and Table S4). The CVGs were classified into 6 families (Fig. 3C and Table S4), of which Metaviridae was the most abundant viral family, followed by Picornaviridae, Potyviridae, Pseudoviridae, Retroviridae and Leviviridae. Based on their hosts, 10 CVGs belonged to eukaryotic viruses and 1 CVG to prokaryotic viruses (Fig. 3D and Table S4).
Fig. 3.
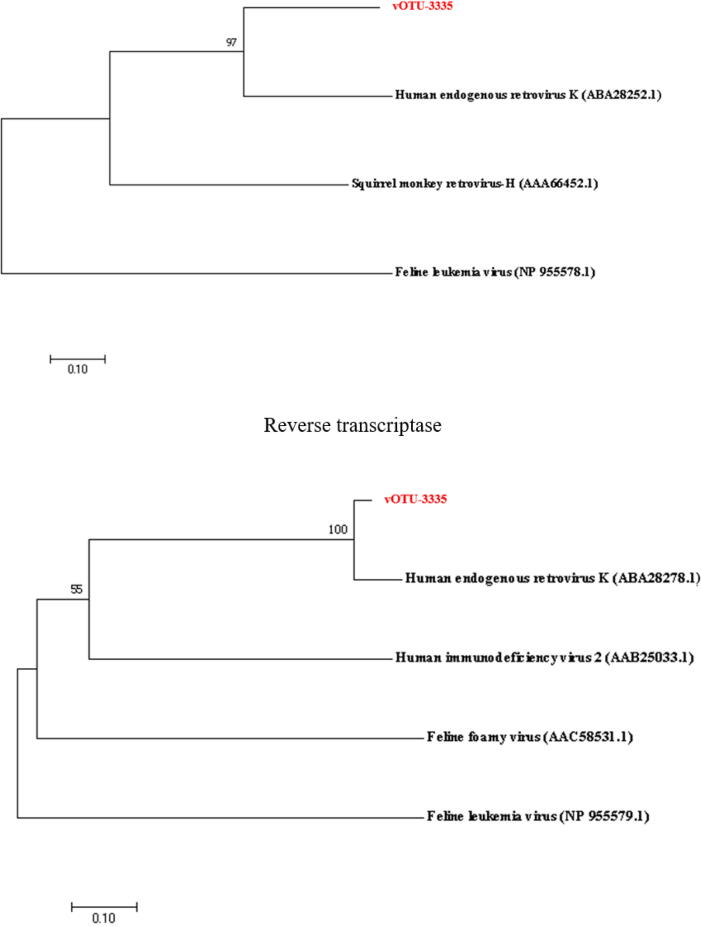
RNA viruses with complete circular genomes in deep sea. (A) The length and proportion of RNA viruses with circular viral genomes (CVGs). (B) The known and unknown RNA viruses with CVGs. (C) The number of the known RNA viruses with complete CVGs at the family level. (D) The hosts of RNA viruses with circular genomes at the host domain level. (E) The phylogenetic analysis of the classified deep-sea CVG (vOTU-3335) belonging to Retroviridae. The gag protein, protease and reverse transcriptase encoded by deep-sea RNA virus vOTU-3335 and human endogenous retrovirus of Retroviridae were included in the phylogenetic analysis. The numbers on the branches represent the percentage of confidence in the bootstrap test (500 replicates).
To explore the evolutionary relationship between deep-sea viruses and the currently known RNA viruses, further characterization was performed on the deep-sea CVGs belonging to Retroviridae, a well-known family of RNA viruses. The deep-sea RNA virus (vOTU-3335) of Retroviridae encoded 4 viral proteins, which were homologous to the gag protein, 2 proteases and reverse transcriptase of human endogenous retrovirus of Retroviridae, respectively. Phylogenetic analysis of these 4 viral proteins showed that the classified deep-sea CVGs belonging to Retroviridae (vOTU-3335) formed an independent branch of the phylogenetic tree except for reverse transcriptase (Fig. 3E), suggesting that the classified deep-sea CVGs were different from the known RNA viruses.
Collectively, our findings reveal a large repository of novel RNA viruses with complete genomes.
Roles of deep-sea ecosystems in the differentiation of deep-sea RNA viruses
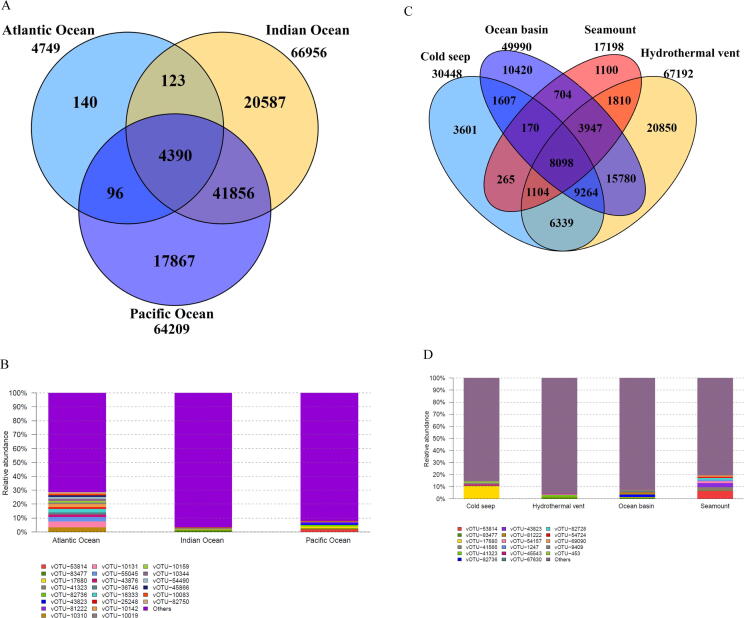
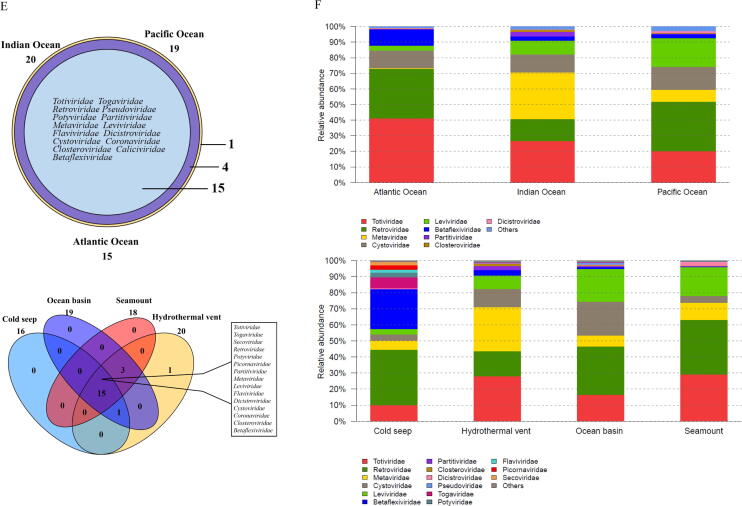
To explore possible effects of deep-sea ecosystems on the differentiation of deep-sea RNA viruses, we analyzed the distribution patterns of RNA viruses in deep-sea ecosystems across three oceans (the Atlantic, Indian and Pacific Oceans). Most RNA viruses were unique to each ocean except Atlantic Ocean, and 4,390 vOTUs were the core RNA viruses that existed in all three oceans (Fig. 4A and Table S5). However, the abundance of the core RNA viruses was less than < 1 % (Fig. 4B). In addition, while we detected the majority of vOTUs was unique to each ecosystem, 8,098 vOTUs were common to all ecosystems (hydrothermal vent, cold seep, seamount and ocean basin) (Fig. 4C and Table S6), representing the core RNA viruses in different deep-sea ecosystems. The relative abundance of these core RNA viruses was also<1 % (Fig. 4D). These data demonstrate that the vast majority of deep-sea RNA viruses are ecosystem-specific, implicating that it is the characteristic features of these ecosystems that drive the differentiation of the resident viral communities.
Fig. 4.
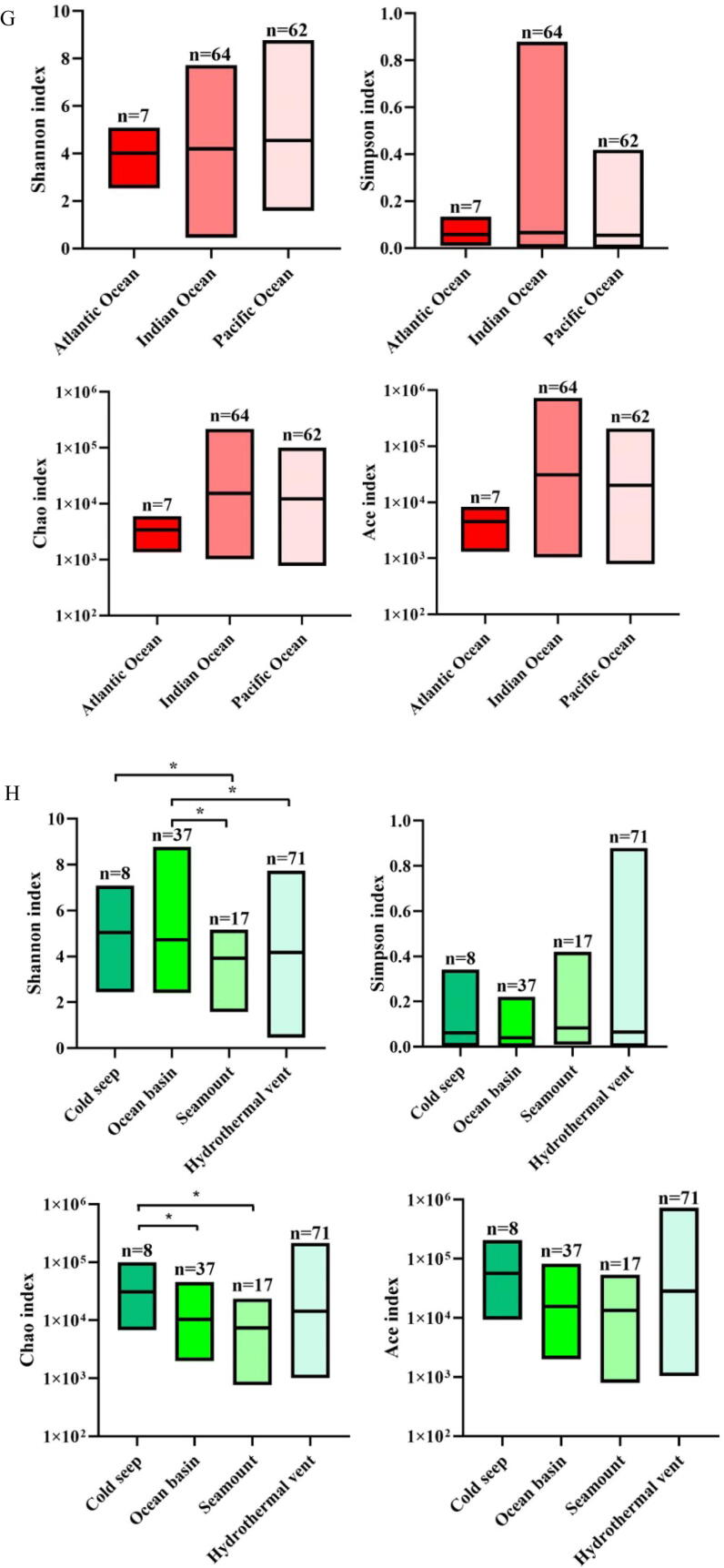
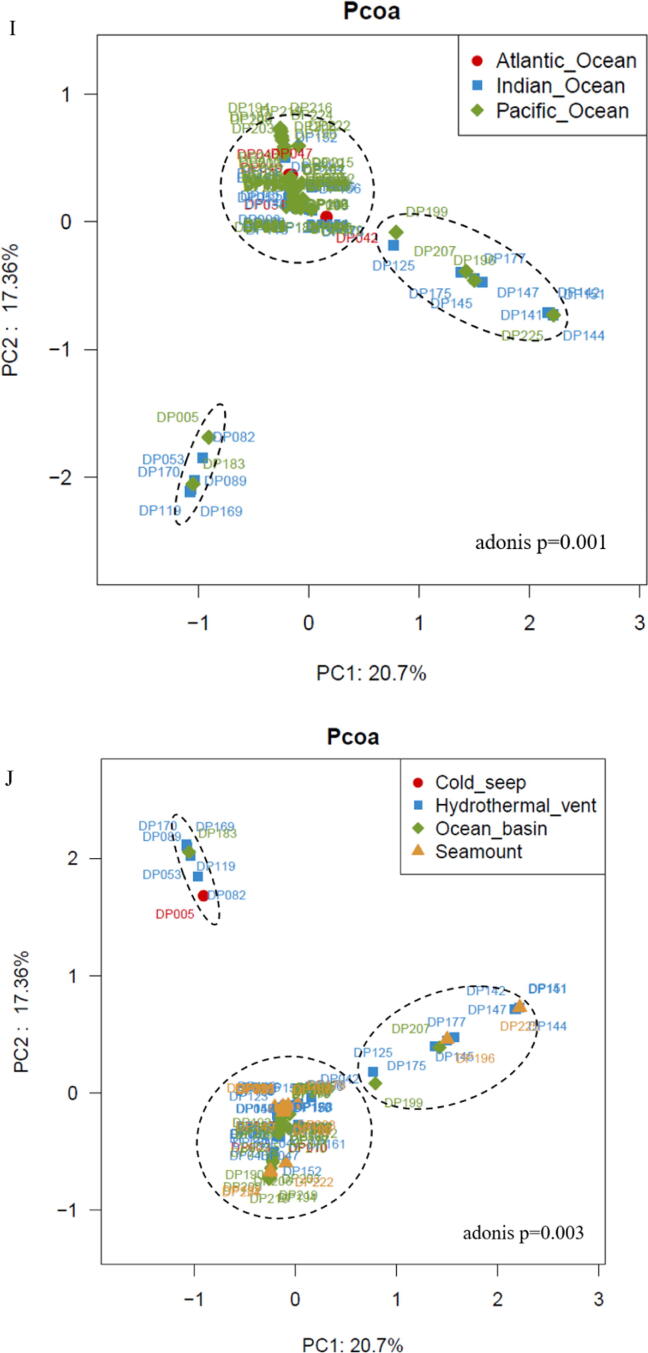
Roles of deep-sea ecosystems in the differentiation of deep-sea RNA viruses. (A) The number of vOTUs in three oceans. The core RNAs viruses existing in all oceans consisted of 4,390 vOTUs. (B) The relative abundance of vOTUs in three oceans. “Others” indicate the vOTUs with relative abundance<1 %. (C) The number of vOTUs in four deep-sea ecosystems. (D) The relative abundance of vOTUs in hydrothermal vents, cold seeps, seamounts and ocean basins. “Others” represent the vOTUs with relative abundance<1 %. (E) The distribution of the known viral families of RNA viruses in three oceans (Atlantic Ocean, Indian Ocean and Pacific Ocean) or four deep-sea ecosystems (hydrothermal vent, cold seep, seamount and ocean basin). Fifteen viral families were the core families of three oceans or four ecosystems. The number of the known viral families is indicated. (F) The relative abundance of the known viral families in Atlantic Ocean, Indian Ocean and Pacific Ocean or in hydrothermal vents, cold seeps, seamounts and ocean basins. “Others” show the viral families with relative abundance<1 %. (G) Boxplots indicating the diversity index of communities of deep-sea RNA viruses in three oceans. The top, medium and bottom part of each box correspond to the highest, median and lowest diversity index of single viral community. The letter “n” represents the number of samples in each ocean. (H) The diversity index of RNA viruses of four deep-sea ecosystems. The letter “n” represented the number of samples in each ecological ecosystem. The statistical significance between diversities was indicated with asterisks (*, p < 0.05). (I) Principal co-ordinates analysis (PCoA) of the vOTUs from Atlantic Ocean, Indian Ocean and Pacific Ocean (adonis p = 0.001). (J) PCoA of the vOTUS from hydrothermal vents, cold seeps, seamounts and ocean basins (adonis p = 0.001).
Fifteen of the 20 known families of deep-sea RNA viruses formed the core viral families of all oceans and ecosystems (Fig. 4E). Among the core viral families, Totiviridae, Retroviridae, Cystoviridae and Leviviridae were the dominant families globally. However, the relative abundances of these dominant families varied significantly among the three oceans as well as the four ecosystems (Fig. 4F). The most dominant family in the Atlantic Ocean, Indian Ocean and Pacific Ocean was Totiviridae (41.18 %), Totiviridae (26.74 %) and Retroviridae (31.69 %), respectively, and in the cold seeps, hydrothermal vents, ocean basins and seamounts was Retroviridae (34.41 %), Totiviridae (28.04 %), Retroviridae (30.22 %) and Retroviridae (33.94 %), respectively. Although Coronaviridae was the core family in the different oceans and ecosystems, its abundance was < 1 % in the deep-sea sediments. These results are indicative of low inter-family diversities of global deep-sea RNA viruses, suggesting that the deep-sea RNA virus communities are shaped by environmental factors.
The results demonstrated that the Pacific Ocean had the most diverse RNA viruses, whereas the lowest diversity was observed in the Atlantic Ocean (Fig. 4G). The viral diversity index analyses showed significant differences across the four ecosystems (p < 0.05) (Fig. 4H and Table S7). Among the different ecosystems, the cold seeps and seamounts had the highest and lowest diversity of RNA viruses, respectively (Fig. 4H). Thus, the viral communities of deep-sea sediments are highly diverse and spatially different across the global scale.
Principal co-ordinates analysis (PCoA) was performed to further explore the global distribution and diversity of deep-sea RNA viruses, and the results showed that the vOTUs could be clustered into 3 groups (Fig. 4I and 4 J). All vOTUs of the Atlantic Ocean and most vOTUs of the Pacific Ocean and Indian Ocean were clustered into one group, while the remaining vOTUs of the Pacific and Indian Oceans were aggregated into two groups (Fig. 4I), suggesting that the deep-sea viral communities were similar in the three oceanic regions. Except for a few sediment samples collected from cold seeps, ocean basins, seamounts and hydrothermal vents, the vOTUs of almost all samples from each ecosystem were clustered (Fig. 4J), indicating that the unique features of these ecosystems (hydrothermal vents, cold seeps, seamounts or ocean basins) rather than the geographical location influence the differentiation of the deep-sea RNA virus communities.
Collectively, these findings reveal that the differentiation of deep-sea RNA viral communities is driven by the deep-sea ecosystems.
Influence of viral genes on the community differentiation of deep-sea RNA viruses
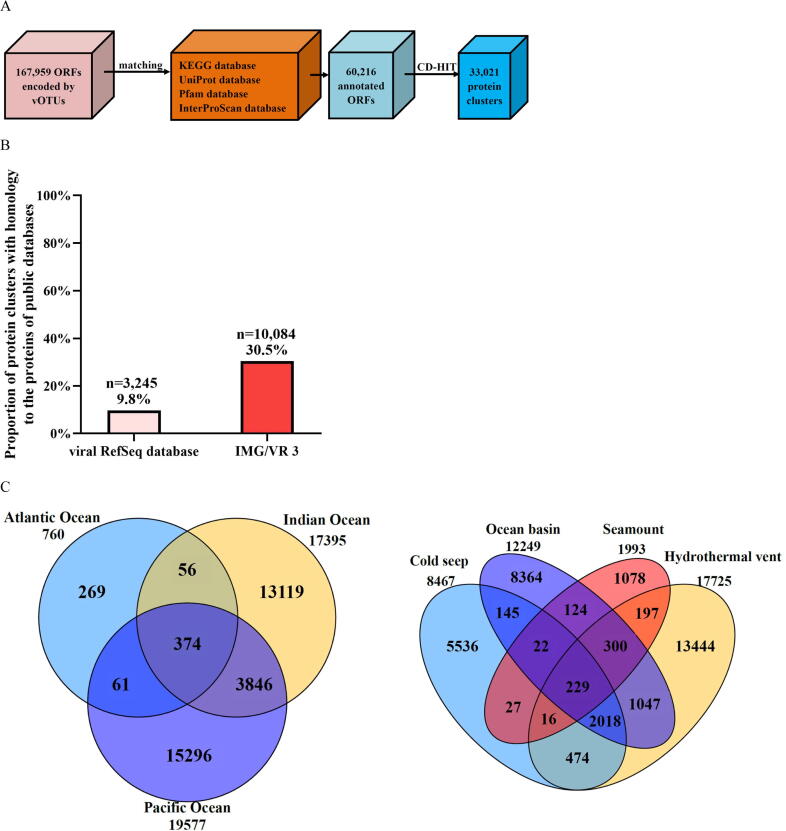
To determine the mechanism that the deep-sea ecosystems drove the differentiation of deep-sea RNA viruses, the roles of viral genes encoded by 85,059 vOTUs in the differentiation of communities of deep-sea RNA viruses were investigated. Based on the predicted open reading frames (ORFs), 167,959 putative RNA viral genes were identified, generating 60,216 annotated ORFs (Fig. 5A and Table S8). After clustering by CD-HIT, 33,021 protein clusters of deep-sea vOTUs were obtained (Fig. 5A). Only 9.8 % (n = 3,245) protein clusters matched the proteins in NCBI Viral RefSeq database, while 30.5 % (n = 10,084) protein clusters were homologous to the known viral proteins in IMG/VR v3 database (Fig. 5B). Thus, most proteins encoded by deep-sea RNA viruses were novel, further underscoring our hypothesis that the deep sea is a reservoir of novel RNA viruses. , only 229 protein clusters were shared by the four deep-sea ecosystems, and 374 protein clusters were the core clusters for the three oceans (Fig. 5C). Most protein clusters were unique to the local ecosystem, indicating that the deep-sea ecosystem influenced the differentiation of viral communities, which was consistent with the results of vOTUs.
Fig. 5.
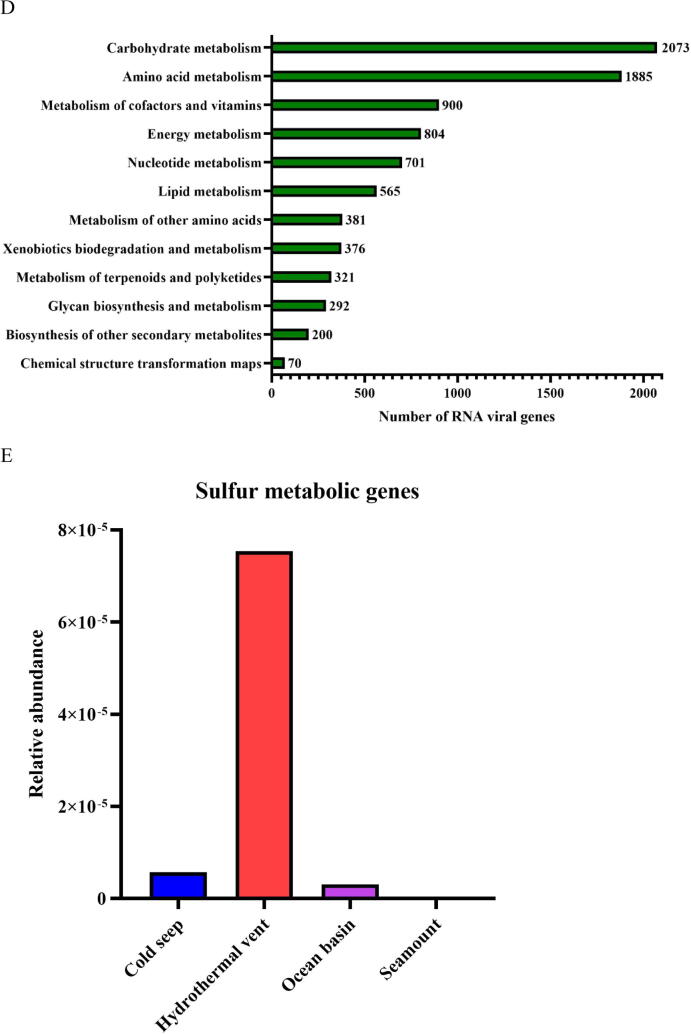
Influence of viral genes on the community differentiation of deep-sea RNA viruses. (A) Diagram showing the identification of protein clusters from vOTUs of global deep-sea RNA viruses. vOTUs, viral operational taxonomic units; ORFs, open reading frames. (B) Proportion of protein clusters of deep-sea vOTUs homologous to the known viral proteins in NCBI Viral RefSeq and IMG/VR v3 databases. (C) The distribution of protein clusters of deep-sea RNA viruses in three oceans and four ecosystems. The numbers indicate the quantity of protein clusters of RNA viruses. (D) The metabolic pathways mediated by putative viral genes of deep-sea RNA viruses based on the KEGG pathway database. (E) The relative abundance of sulfur metabolism genes encoded by deep-sea RNA viruses in different ecosystems. The genes encoded sulfate/thiosulfate transport system ATP-binding protein, adenylylsulfate kinase, sulfate adenylyltransferase subunit 2, phosphoadenosine phosphosulfate reductase, sulfite reductase (NADPH) hemoprotein beta-component, sulfate adenylyltransferase subunit 1, 5′-bisphosphate nucleotidase, sulfate/thiosulfate transport system permease protein, sulfate/thiosulfate transport system permease protein and sulfur-oxidizing protein.
To further explore the roles of deep-sea RNA viruses in host metabolism, we identified the virus-encoded proteins that potentially regulate metabolic pathways. Of the 17,140 viral proteins annotated in the KEGG pathway database, 49.99 % (n = 8,568) were classified into the “Metabolism” category. These putative viral metabolism-related genes were further divided into 12 subgroups, of which “Carbohydrate metabolism” was most enriched (2,037 genes), followed by “Amino acid metabolism” (1,885 genes), “Metabolism of cofactors and vitamins” (900 genes) and “Energy metabolism” (804 genes) (Fig. 5D). These suggested that the virus-encoded genes might affect the metabolic pathways in their hosts.
Given the crucial roles of sulfur metabolism in deep-sea ecosystems [5], [42], we analyzed the type and abundance of virus-encoded genes that mediate sulfur metabolism in four deep-sea ecosystems. A total of 10 viral genes were identified that were involved in assimilatory sulfate reduction and thiosulfate oxidation, which are the most important processes in sulfur metabolism [43], [44], [45]. The 10 genes encoded sulfate/thiosulfate transport system ATP-binding protein, adenylylsulfate kinase, sulfate adenylyltransferase subunit 2, phosphoadenosine phosphosulfate reductase, sulfite reductase (NADPH) hemoprotein beta-component, sulfate adenylyltransferase subunit 1, 5′-bisphosphate nucleotidase, sulfate/thiosulfate transport system permease protein, sulfate/thiosulfate transport system permease protein and sulfur-oxidizing protein. The relative abundance of these 10 genes was significantly higher in hydrothermal vents compared to the other deep-sea ecosystems (Fig. 5E), which is consistent with the high sulfur concentration in hydrothermal vent ecosystem, indicating that sulfur metabolism, the most important energy metabolism in deep-sea hydrothermal vents [5], [42], is the cornerstone of this ecosystem.
Taken together, the differentiation of deep-sea communities of RNA viruses is influenced by the deep-sea ecosystem, which might be driven by the virus-mediated energy metabolism.
Discussion
Marine viruses regulate biogeochemical cycles in the oceans by infecting hosts, thus possessing a significant impact on marine ecosystems [2]. Most studies conducted on marine viruses have been focused on DNA viruses [13]. Although the majority of the known RNA viruses with clear taxonomy are able to infect humans [46], [47], little is known regarding the global diversity and distribution of RNA viruses in deep sea. In the present investigation, we identified 85,059 RNA viruses from global deep-sea sediments collected from three oceans and four ecosystems, which is considerably greater than the 2,094 RNA viruses currently included in the viral RefSeq database of NCBI, and is indicative of the extremely diverse deep-sea viral communities. In this study, the results showed that there was no DNA contamination for the RNAs extracted from all purified virions (Fig S1) and no bacterial contamination for the extracted viral RNAs and the corresponding cDNAs (Fig S2). At the same time, based on the alignment analysis using blastn and blastp, no vOTU of all the attained vOTUs were identical or very similar to any of the known eukaryotic, archeal or prokaryotic genomes and no ORF encoded by the vOTUs shared identical or very similar amino acid sequences to the known proteins. These data indicated that the vOTUs obtained in this study originated from deep-sea sediments. Furthermore, the viruses isolated from ocean sediments better represent the relatively stable deep-sea communities compared to those isolated from ocean waters since the latter can be carried around by oceans currents [48]. Although many viruses in sea waters have been identified [5], [48], [49], they may not be truly representative of the biogeographical patterns of marine viral communities due to the influence of ocean currents. To this end, the viruses isolated from deep-sea sediments better reflected the biogeographical features of deep-sea viral communities. In our study, only 1.72 % of the deep-sea RNA viruses were homologous to the known RNA viruses, and the majority of the genes encoded by the deep-sea RNA viruses were unknown, indicating that the deep sea is a reservoir of novel RNA viruses. The huge unclassified vOTUs, accounting for 98.28 % in all vOTUs revealed in this investigation, might represent important sources for further datamining, such as RNA-targeting CRISPR (clustered regularly interspaced short palindromic repeats) systems and ribozymes. The ORFs encoded by these unclassified vOTUs could also be characterized to explore the evolution of RNA viruses. These issues merit to be investigated in the future. Furthermore, very few RNA bacteriophages have been reported so far [50], [51]. We identified 104 RNA bacteriophages belonging to the Cystoviridae, Leviviridae and Pseudoviridae families. Our findings provide new information on RNA bacteriophages, the very few viruses. However, no archaeal RNA viruses were detected in this study. Therefore, our study presents the novel insights into the global virosphere of deep-sea RNA viruses for the first time.
Studies show that the viral diversity and abundance differ significantly among the marine ecosystems [52]. The virions isolated from the extreme thermal environments have little sequence homology with the published viruses despite morphological similarities [53]. Multi-zone viral populations are predominant in the Antarctic and bathypelagic regions, whereas zone-specific regional viral populations dominate in temperate and tropical epipelagic regions and Arctic regions [5]. However, the relationship between the deep-sea ecosystems and the deep-sea sediment viruses has not been explored so far. We analyzed the RNA viromes of typical global deep-sea ecosystems including hydrothermal vents, cold seeps, seamounts and ocean basins. The deep-sea sediment samples of hydrothermal vents were collected from the Atlantic and Indian Oceans (Fig. 1A and Table S1). We found that the differentiation of deep-sea RNA virus communities is driven by the environmental factors rather than the geographical locations. Recent studies have shown that marine viruses encode metabolic genes that can modify host metabolism during virus infection [6], [8]. In this study, we identified 8,568 RNA virus-encoded genes involved in carbohydrate, amino acid and energy metabolisms. The relative abundance of virus-encoded sulfur metabolism genes was highest in hydrothermal vent fields, which is consistent with the higher sulfur concentration and active sulfur-based chemosynthesis in these regions, indicating that the virus-mediated energy metabolism plays essential roles in driving the differentiation of RNA viral communities in different deep-sea ecosystems. Our findings provide new insights into deep-sea RNA virus communities and their roles in the local as well as global marine ecosystems.
Data Accessibility.
The data we obtained from next sequencing were uploaded on National Omics Data Encyclopedia database (accession number OEP002537).
CRediT authorship contribution statement
Xinyi Zhang: Investigation, Formal analysis, Data curation, Writing – original draft, Writing – review & editing. Haitao Wan: Investigation. Min Jin: Resources. Liquan Huang: Conceptualization. Xiaobo Zhang: Conceptualization, Supervision, Funding acquisition, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was financially supported by China Ocean Mineral Resources R & D Association (DY135-B-04) and the National Key Research and Development Program of China (2018YFC0310703). The bioinformation analysis of this study was supported by Mingke Biotechnology Co., Ltd. (Hangzhou, China).
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2023.04.003.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Breitbart M. Marine viruses: truth or dare. Ann Rev Mar Sci. 2012;4:425–448. doi: 10.1146/annurev-marine-120709-142805. [DOI] [PubMed] [Google Scholar]
- 2.Chow C.E., Suttle C.A. Biogeography of viruses in the sea. Annu Rev Virol. 2015;2:41–66. doi: 10.1146/annurev-virology-031413-085540. [DOI] [PubMed] [Google Scholar]
- 3.Güemes A.G.C., Youle M., Cantú V.A., Felts B., Nulton J. Rohwer, Viruses as winners in the game of life. Review of Virology. 2016;3:197–214. doi: 10.1146/annurev-virology-100114-054952. [DOI] [PubMed] [Google Scholar]
- 4.Zhang X. Virus Infection and Tumorigenesis-Hints From Marine Hosts’ Stress Responses. 2019; Springer.
- 5.Gregory A.C., Zayed A.A., Conceição-Neto N., et al. Marine DNA viral macro- and microdiversity from pole to pole. Cell. 2019;177(5):1109–1123. doi: 10.1016/j.cell.2019.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roux S., Brum J.R., Dutilh B.E., et al. Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature. 2016;537(7622):689–693. doi: 10.1038/nature19366. [DOI] [PubMed] [Google Scholar]
- 7.York A. Marine microbiology: algal virus boosts nitrogen uptake in the ocean. Nat Rev Microbiol. 2017;15:573. doi: 10.1038/nrmicro.2017.113. [DOI] [PubMed] [Google Scholar]
- 8.He T., Li H., Zhang X. Deep-sea hydrothermal vent viruses compensate for microbial metabolism in virus-host interactions. MBio. 2017;8:e00893–e917. doi: 10.1128/mBio.00893-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brum J.R., Ignacio-Espinoza J.C., Roux S., et al. Ocean plankton. Patterns and ecological drivers of ocean viral communities. Science. 2015;348(6237):1261498. doi: 10.1126/science.1261498. [DOI] [PubMed] [Google Scholar]
- 10.Steward G.F., Culley A.I., Mueller J.A., Wood-Charlson E.M., Belcaid M., Poisson G. Are we missing half of the viruses in the ocean? ISME J. 2013;7(3):672–679. doi: 10.1038/ismej.2012.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Culley A.I., Mueller J.A., Belcaid M., Wood-Charlson E.M., Poisson G., Steward G.F. The characterization of RNA viruses in tropical seawater using targeted PCR and metagenomics. MBio. 2014;5(3):e01210–e1304. doi: 10.1128/mBio.01210-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Urayama S.I., Takaki Y., Nishi S., Yoshida-Takashima Y., Deguchi S., Takai K., et al. Unveiling the RNA virosphere associated with marine microorganisms. Mol Ecol Resour. 2018;18(6):1444–1455. doi: 10.1111/1755-0998.12936. [DOI] [PubMed] [Google Scholar]
- 13.Vlok M., Lang A.S., Suttle C.A. Marine RNA virus quasispecies are distributed throughout the oceans. mSphere. 2019;4:e00157–e219. doi: 10.1128/mSphereDirect.00157-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sebastián M., Auguet J.C., Restrepo-Ortiz C.X., Sala M.M., Marrasé C., Gasol J.M. Deep ocean prokaryotic communities are remarkably malleable when facing long-term starvation. Environ Microbiol. 2018;20(2):713–723. doi: 10.1111/1462-2920.14002. [DOI] [PubMed] [Google Scholar]
- 15.Tortorella E., Tedesco P., Esposito F., January G.G., Fani R., Jaspars M., et al. Antibiotics from deep-sea microorganisms: current discoveries and perspectives. Mar Drugs. 2018;16(10):355. doi: 10.3390/md16100355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Danovaro R., Corinaldesi C., Dell'Anno A., Snelgrove P.V.R. The deep-sea under global change. Curr Biol. 2017;27(11):461–465. doi: 10.1016/j.cub.2017.02.046. [DOI] [PubMed] [Google Scholar]
- 17.Pilkington L.I. A chemometric analysis of deep-sea natural products. Molecules. 2019;24(21):3942. doi: 10.3390/molecules24213942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nurk S.D., Meleshko A.K., Pevzner P.A. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 2017;27:824–834. doi: 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roux S., Enault F., Hurwitz B.L., Sullivan M.B. VirSorter: mining viral signal from microbial genomic data. PeerJ. 2015;3:e985. doi: 10.7717/peerj.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren J., Ahlgren N.A., Lu Y.Y., Fuhrman J.A., Sun F. VirFinder: a novel k-mer based tool for identifying viral sequences from assembled metagenomic data. Microbiome. 2017;5:69. doi: 10.1186/s40168-017-0283-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kieft K., Zhou Z. Anantharaman K VIBRANT: automated recovery, annotation and curation of microbial viruses, and evaluation of viral community function from genomic sequences. Microbiome. 2020;8:90. doi: 10.1186/s40168-020-00867-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.von Meijenfeldt F.A.B., Arkhipova K., Cambuy D.D., Coutinho F.H., Dutilh B.E. Robust taxonomic classification of uncharted microbial sequences and bins with CAT and BAT. Genome Biol. 2019;20:217. doi: 10.1186/s13059-019-1817-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang C.R., Lo S.J. Evolution and diversity of the human hepatitis d virus genome. Adv Bioinforma. 2010;2010 doi: 10.1155/2010/323654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buchfink B., Xie C., Huson D. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 26.Nayfach S., Camargo A.P., Schulz F., Eloe-Fadrosh E., Roux S., Kyrpides N.C. CheckV assesses the quality and completeness of metagenome-assembled viral genomes. Nat Biotechnol. 2021;39:578–585. doi: 10.1038/s41587-020-00774-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marcais G., Delcher A.L., Phillippy A.M., Coston R., Salzberg S.L., Zimin A. MUMmer4: a fast and versatile genome alignment system. PLoS Comput Biol. 2018;14:e1005944. doi: 10.1371/journal.pcbi.1005944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hyatt D., Hyatt D., Chen G.L., LoCascio P.F., Land M.L., Larimer F.W., et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinf. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tisza M.J., Pastrana D.V., Welch N.L., et al. Discovery of several thousand highly diverse circular DNA viruses. Elife. 2020;9:e51971. doi: 10.7554/eLife.51971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Z., Pan D., Wei G., et al. Deep sea sediments associated with cold seeps are a subsurface reservoir of viral diversity. ISME J. 2021;15:2366–2378. doi: 10.1038/s41396-021-00932-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dixon P. VEGAN, a package of R functions for community ecology. J Veg Sci. 2003;14:927–930. [Google Scholar]
- 33.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suzek B.E., Wang Y., Huang H., McGarvey P.B., Wu C.H. UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics. 2015;31:926–932. doi: 10.1093/bioinformatics/btu739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zdobnov E.M., Apweiler R. InterProScan–an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 36.Punta M., Coggill P.C., Eberhardt R.Y., L. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li W., Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 38.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Price M.N., Dehal P.S., Arkin A.P. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garcia-Martin R., Wang G., Brandão B.B., Zanotto T.M., Shah S., Patel S.K., et al. MicroRNA sequence codes for small extracellular vesicle release and cellular retention. Nature. 2022;601:446–451. doi: 10.1038/s41586-021-04234-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quince C., Walker A.W., Simpson J.T., Loman N.J., Segata N. Shotgun metagenomics, from sampling to analysis. Nat Biotechnol. 2017;35:833–844. doi: 10.1038/nbt.3935. [DOI] [PubMed] [Google Scholar]
- 42.Anantharaman K., Breier J.A., Dick G.J. Metagenomic resolution of microbial functions in deep-sea hydrothermal plumes across the Eastern Lau Spreading Center. ISME J. 2016;10:225–239. doi: 10.1038/ismej.2015.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cech G.M., Szalewska-Pałasz A., Potrykus K., Kloska A. Virus-host interaction gets curiouser and curiouser. PART II: functional transcriptomics of the E. coli DksA-deficient cell upon phage P1vir infection. Int J Mol Sci. 2021;22(11):6159. doi: 10.3390/ijms22116159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kieft K., Breister A.M., Huss P., et al. Virus-associated organosulfur metabolism in human and environmental systems. Cell Rep. 2021;36(5) doi: 10.1016/j.celrep.2021.109471. [DOI] [PubMed] [Google Scholar]
- 45.McKay L.J., Nigro O.D., Dlakić M., Luttrell K.M., Rusch.DB, Fields MW, Inskeep WP Sulfur cycling and host-virus interactions in Aquificales-dominated biofilms from Yellowstone's hottest ecosystems. ISME J. 2021 doi: 10.1038/s41396-021-01132-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolf Y.I., Kazlauskas D., Iranzo J., Lucía-Sanz A., Kuhn J.H., Krupovic M., et al. Origins and evolution of the global RNA virome. MBio. 2018;9(6):e02329–e10418. doi: 10.1128/mBio.02329-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luo M., Terrell J.R., Mcmanus S.A. Nucleocapsid structure of negative strand RNA virus. Viruses. 2020;12:835. doi: 10.3390/v12080835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suttle C.A. Viruses in the sea. Nature. 2005;437:356–361. doi: 10.1038/nature04160. [DOI] [PubMed] [Google Scholar]
- 49.Zayed A.A., Wainaina J.M., Dominguez-Huerta G., et al. Cryptic and abundant marine viruses at the evolutionary origins of Earth’s RNA virome. Science. 2022;376:156–162. doi: 10.1126/science.abm5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krishnamurthy S.R., Janowski A.B., Zhao G., Barouch D., Wang D. Hyperexpansion of RNA bacteriophage diversity. PLoS Biol. 2016;14:e1002409. doi: 10.1371/journal.pbio.1002409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harb L, Chamakura K, Khara P, Christie PJ, Young R, Zeng L. ssRNA phage penetration triggers detachment of the F-pilus. Proc Natl Acad Sci USA 2020; 117(41): 25751–25758. [DOI] [PMC free article] [PubMed]
- 52.Suttle C.A. Marine viruses-major players in the global ecosystem. Nat Rev Microbiol. 2007;5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 53.Snyder J.C., Young M.J. Advances in understanding archaea-virus interactions in controlled and natural environments. Curr Opin Microbiol. 2011;14:497–503. doi: 10.1016/j.mib.2011.07.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.