Abstract
Staphylococcal enterotoxins are exotoxins produced by Staphylococcus aureus that possess emetic and superantigenic properties. Prior to this research there were six characterized enterotoxins, staphylococcal enterotoxin types A to E and H (referred to as SEA to SEE and SEH). Two new staphylococcal enterotoxin genes have been identified and designated seg and sei (staphylococcal enterotoxin types G and I, respectively). seg and sei consist of 777 and 729 nucleotides, respectively, encoding precursor proteins of 258 (SEG) and 242 (SEI) deduced amino acids. SEG and SEI have typical bacterial signal sequences that are cleaved to form toxins with 233 (SEG) and 218 (SEI, predicted) amino acids, corresponding to mature proteins of 27,043 Da (SEG) and 24,928 Da (SEI). Biological activities for SEG and SEI were determined with recombinant S. aureus strains. SEG and SEI elicited emetic responses in rhesus monkeys upon nasogastric administration and stimulated murine T-cell proliferation with the concomitant production of interleukin 2 (IL-2) and gamma interferon (IFN-γ), as measured by cytokine enzyme-linked immunoassays. SEG and SEI are related to other enterotoxins of S. aureus and to streptococcal pyrogenic exotoxin A (SpeA) and streptococcal superantigen (SSA) of Streptococcus pyogenes. Phylogenetic analysis and comparisons of amino acid and nucleotide sequence identities were performed on related staphylococcal and streptococcal protein toxins to group SEG and SEI among the characterized toxins. SEG is most similar to SpeA, SEB, SEC, and SSA (38 to 42% amino acid identity), while SEI is most similar to SEA, SEE, and SED (26 to 28% amino acid identity). Polyclonal antiserum was generated against purified histidine-tagged SEG and SEI (HisSEG and HisSEI). Immunoblot analysis of the enterotoxins, toxic-shock syndrome toxin 1, and SpeA with antiserum prepared against HisSEG and HisSEI revealed that SEG shares some epitopes with SEC1 while SEI does not.
Staphylococcal enterotoxins (SEs) cause staphylococcal food poisoning and the shock symptoms in some cases of toxic shock syndrome (TSS) (3, 9). SEs are also superantigens which are defined by their unique ability to stimulate virtually all T cells whose T-cell receptor (TCR) bears a particular Vβ element (70). Unlike conventional antigens, superantigens are not processed (18) but bind major histocompatibility complex (MHC) class II molecules outside of the peptide-binding groove and form a trimolecular complex with the TCR (17, 21). Superantigens stimulate the production of cytokines such as interleukin-1 (IL-1), IL-2, gamma interferon (IFN-γ), and tumor necrosis factor alpha (44).
SEs are monomeric proteins produced in a precursor form possessing typical bacterial signal sequences that are cleaved to release the extracellular mature toxins (6), which range in size from 25,200 to 28,300 Da. There are six characterized staphylococcal enterotoxins based on serological groups: staphylococcal enterotoxin types A, B, C, D, E, and H (referred to as SEA, SEB, etc.) (3, 60, 65). Ren et al. (60) first identified the nucleotide sequence and protein product designated SEH. Su et al. (65) purified a new enterotoxin (designated SEH) from Staphylococcus aureus which has the same amino terminal sequence as SEH characterized by Ren et al. The gene, however, has not been cloned, so it is presently unclear if the two toxins are indeed the same. Although SEC is subdivided into three groups (SEC1, SEC2, and SEC3) based upon minor epitopes (3), additional sec variants have been discovered that have >95% deduced amino acid identity among them (43, 67). Overall, SEs share significant nucleotide and amino acid sequence identity (32 to 82% and 21 to 82%, respectively) (2, 6, 8, 11, 12, 15, 27, 33, 60, 67). Within the enterotoxin family, SEA, SEE, and SED fall into one group based upon amino acid identity (52 to 83% amino acid identity), while SEB and the SECs fall into another group (62 to 64% amino acid identity).
Exoproteins of S. aureus and Streptococcus pyogenes form the pyrogenic toxin family based on shared biological properties (6, 9, 31, 67). Members include the SEs and toxic shock syndrome toxin 1 (TSST-1) of S. aureus, streptoccocal pyrogenic exotoxin types A, B, and C (SpeA, SpeB, and SpeC), and streptococcal superantigen (SSA) (6, 31, 57, 67). The pyrogenic toxins stimulate T-cell proliferation, enhance endotoxic shock, suppress immunoglobulin production, and are pyrogenic (reviewed in references 3, 9, and 44). Of these toxins, SpeB and TSST-1 have little, if any, significant amino acid or nucleotide sequence identity with the other toxins (6, 67). The SEs, SpeA, and SSA, however, are similar and share 31 to 98% nucleotide sequence identity and 20 to 98% amino acid sequence identity. In fact, the streptococcal proteins are more similar to some of the SEs than some of the SEs are to each other (6, 31, 57, 67).
Both staphylococcal and streptococcal toxins can cause shock symptoms similar to those caused by TSST-1 (9). The enterotoxins may cause shock symptoms in nonmenstrual cases of TSS where TSST-1 is not produced by the causative isolate (9, 10, 16, 20, 39). Approximately 50% of nonmenstrual TSS cases are caused by S. aureus isolates producing SEs (most often SEB and SEC) (9, 10, 39). However, some nonmenstrual TSS isolates do not produce TSST-1 or any of the characterized enterotoxins (20), suggesting that uncharacterized toxins may be responsible for these cases.
Enzyme-linked immunosorbent assay (ELISA) studies using antisera generated against SEA to SEE reveal that there are enterotoxigenic S. aureus strains which do not produce any of the recognized enterotoxins (4, 35). These strains were isolated from humans, animals, or food, and culture supernatants from these strains cause emesis (vomiting) when administered orally to primates (35). Together, these data demonstrate the need for characterizing new staphylococcal enterotoxins which may be involved in human illness. Here we report the identification and characterization of two new enterotoxins with some unusual genetic and biochemical features, staphylococcal enterotoxin types G and I (SEG and SEI, respectively), from two different enterotoxigenic S. aureus strains.
MATERIALS AND METHODS
Bacterial strains, plasmids, bacteriophage, and growth conditions.
The names and descriptions of all strains used in this study are listed in Table 1. Enterotoxigenic FRI strains (Food Research Institute, University of Wisconsin—Madison) produce an emetic response in nonhuman primates when culture supernatants are orally administered (35). These strains do not express SEA, -B, -C, -D, or -E as tested by ELISA (35).
TABLE 1.
Bacterial strains, plasmids, and phage
| Strain, plasmid, or phagea | Relevant characteristic(s)b | Reference or source |
|---|---|---|
| E. coli | ||
| Strains | ||
| JF626 | Aps, Se | 8 |
| M15(pREP4) | Kmr | Qiagen |
| DH5 | Aps, Se | Gibco BRL |
| DH10B | recA1 deoR mcrA mcrBC mrr hsdRMS | Gibco BRL |
| MJB1308 | Aps, seg, carrying pMJB460 | This work |
| MJB1309 | Aps, sei, carrying pMJB461 | This work |
| Plasmids | ||
| pGEM-7Zf(+) | Apr, contains bla | Promega |
| pGEM-3Zf(+) | Apr | Promega |
| pQE-31, pQE-32 | Apr | Qiagen |
| pMJB38 | Apr, pBR322 containing a 624-bp fragment of sea | 8 |
| pMJB124 | Apr, pGEM7-Zf(+) with a 1.8-kbp insert that contains sec | 14 |
| pMJB460 | Apr, pGEM7-Zf(+) with a 2.5-kbp HindIII seg-containing fragment from FRI572 inserted into the HindIII site (Fig. 2A) | This work |
| pMJB461 | Apr, pGEM7-Zf(+) with a 2.5-kbp HindIII sei-containing fragment from FRI445 inserted into the HindIII site (Fig. 2B) | This work |
| pMJB464 | Apr, pGEM7-Zf(+) digested with SmaI/EcoRI and ligated to a 2.1-kbp seg-containing insert obtained from the digestion of pMJB462 with ClaI (in blunted multiple-cloning site)/EcoRI | This work |
| pMJB465 | Apr, pGEM7-Zf(+) digested with HindIII/SmaI and ligated to a 1.73-kbp HindIII/RsaI sei-containing insert obtained by digesting pMJB461 with HindIII/EcoRI followed by a RsaI partial digest (Fig. 2B) | This work |
| pMJB474 | pQE-32 with a 800-bp insert containing seg, encodes HisSEG (Fig. 2A) | This work |
| pMJB475 | pQE-31 with a 770-bp insert containing sei, encodes HisSEI (Fig. 2B) | This work |
| S. aureus | ||
| Strains | ||
| FRI337 | Sea+ Sed+ | M. S. Bergdollc |
| FRIS6 | Sea+ Seb+ | M. S. Bergdoll |
| FRI578 | See+ | M. S. Bergdoll |
| FRI400 | Sec+ | M. S. Bergdoll |
| FRI569 | Enterotoxigenicd | M. S. Bergdoll |
| FRI393 | ||
| FRI591 | ||
| FRI574 | ||
| FRI772 | ||
| FRI662 | ||
| FRI572 | ||
| FRI445 | ||
| FRI572 | Contains seg | M. S. Bergdoll |
| FRI445 | Contains sei | M. S. Bergdoll |
| RN450 | NCTC 8325 derivative cured of prophages φ11, φ12, and φ13 | R. P. Novick (53) |
| RN4220 | Se | 38 |
| RN7497 | Contains pI524, recipient strain for pRN5548-derived plasmids | S. J. Projane (56) |
| RN8117 | Cmr Cdr, contains pRN5548 and pI524 | S. J. Projan (56) |
| ISP2073 | Spa | 55 |
| MJB894 | Sea+, ISP2073 carrying pMJB193 | Lab strain |
| MJB1315 | Sei+, Spa, ISP2073 carrying pMJB466 | This work |
| MJB1316 | Seg+ (inducible), RN7497 carrying pMJB467 | This work |
| MJB1317 | Seg+, RN7497 carrying pMJB468 | This work |
| MJB1318 | Sei+, Spa, ISP2073 carrying pMJB469 | This work |
| MJB1320 | Sei+ (inducible), RN7497 carrying pMJB471 | This work |
| MJB1321 | Sei+, RN7497 carrying pMJB473 | This work |
| Phage 80α | Generalized transducing phage | 50 |
| Plasmids | ||
| pC194 | Cmr | R. P. Novick (32) |
| pRN5548 | Cmr, blaZ promoter and ribosome-binding site (no start codon) followed by a multiple-cloning site | S. J. Projan (56) |
| pI524 | Cdr, contains β-lactamase control elements | 49 |
| pMJB193 | Apr Cmr, contains 2.5-kbp HindIII fragment containing sea | 7 |
| pMJB462 | pMJB460 + pC194 ligated in the SmaI site of pGEM7Zf(+), contains seg | This work |
| pMJB463 | Cmr, pMJB462 with a translation termination signal introduced into the seg BsmI site by digesting pMJB462 with BsmI, creating blunt ends, and ligating the plasmid to a SMURFT linker (17-mer; Pharmacia Biotech) that contains ochre translation termination signals in all three reading frames | This work |
| pMJB466 | Apr Cmr, pMJB465 + pC194 joined at HindIII site, contains a 1.73-kbp sei-containing insert (Fig. 2B) | This work |
| pMJB467 | Cmr, pRN5548 (digested with PstI/SmaI) ligated to a 1.9-kbp insert containing seg (obtained by isolating the fragment from an NsiI/EcoRV digest of pMJB460) in which seg is transcribed from the inducible S. aureus β-lactamase promoter (Fig. 2A) | This work |
| pMJB468 | Cmr, isogenic to pMJB467 except for a translation termination signal present in seg, pRN5548 with a 1.9-kbp seg-containing insert prepared from pMJB463 as described for pMJB467 | This work |
| pMJB469 | Cmr, pMJB466 with a translation termination signal introduced into sei at the KpnI site (after the first 66 deduced amino acids of SEI) by performing a partial KpnI digest, creating blunt ends, and religating the plasmid | This work |
| pMJB470 | Cmr, pRN5548 with a 1.73-kbp EcoRI/BamHI sei-containing fragment (same genomic fragment as in pMJB465) obtained from pMJB465 | This work |
| pMJB471 | Cmr, pRN5548 with a 1.24-kbp fragment containing sei behind the β-lactamase promoter (plasmid created by digesting pMJB470 with XbaI to remove DNA 5′ of sei) (Fig. 2B) | This work |
| pMJB472 | Cmr, pMJB465 with a translation termination signal introduced into sei at the KpnI site as described for pMJB469 | This work |
| pMJB473 | Cmr, isogenic to pMJB471 except for a translation termination signal in the KpnI site of sei, made by digesting pMJB472 with BamHI/EcoRI and then XbaI, isolating the 1.24-kbp fragment, and ligating it to similarly digested pRN5548 | This work |
| pMJB476 | Cmr, pMJB461 (sei on 2.5-kbp insert) with pC194 inserted into the SmaI site (Fig. 2B) | This work |
Address strain requests to Rodney Welch, Department of Medical Microbiology and Immunology, University of Wisconsin—Madison, Madison, WI 53706.
Ap, ampicillin; Cm, chloramphenicol; Cd, cadmium; Km, kanamycin; blaZ, β-lactamase; Spa, protein A nonproducer; Sea+, Seb+, Sed+, Seg+, and Sei+, producers of staphylococcal enterotoxin types A, B, D, G, and I, respectively; Se, non-enterotoxin producer.
University of Wisconsin—Madison, Food Research Institute, Madison, Wis.
Culture supernatants produce an emetic response when administered orally to primates.
Wyeth-Ayerst Research, Pearl River, N.Y.
S. aureus cultures were grown in 3% N-Z-amine type A (Kraft, Inc., Norwich, N.Y.) and 1% yeast extract (Difco Laboratories, Detroit, Mich.) (3+1) at 37°C with aeration and in Trypticase soy broth (BBL Microbiology Systems) for genomic DNA preparations. Escherichia coli strains were grown in Luria broth at 37°C with aeration (42). Antibiotic concentrations used to maintain plasmids in E. coli were 100 μg of ampicillin/ml, 5 μg of chloramphenicol/ml, and 25 μg of kanamycin/ml; 5 μg of chloramphenicol/ml was used for plasmid maintenance in S. aureus. S. aureus strains containing seg or sei expressed from the β-lactamase promoter were induced by the addition of 10 μg (unless otherwise noted) of 2-(2′-carboxyphenyl)benzoyl-6-aminopenicillanic acid (CBAP; Sigma Chemical Company, St. Louis, Mo.)/ml. E. coli M15 derivatives were grown in 2× YT medium (42) containing 50 μg of carbenicillin/ml and 25 μg of kanamycin/ml at 30°C with aeration.
Chemicals, enzymes, and chromatography resins.
Enzyme reagents were obtained from New England Biolabs, Inc. (Beverly, Mass.), Promega Corp. (Madison, Wis.), and Boehringer Mannheim Biochemicals (Indianapolis, Ind.). Lysostaphin was purchased from Applied Microbiology, Inc. (Brooklyn, N.Y.). [α-32P]dATP and [3H]thymidine were obtained from Amersham Life Sciences (Arlington Heights, Ill.). SEA was purified as previously described (23). SEB, SEC1, SED, SEE, TSST-1, and SpeA were purchased from Toxin Technology (Sarasota, Fla.). Chromatography resins were obtained from the following sources: Ni-nitrilotriacetic acid (NTA) resin was from Qiagen, Inc. (Santa Clarita, Calif.), SP Sepharose Fast Flow and Sephacryl S100 High Resolution were from Pharmacia Biotech (Milwaukee, Wis.), and Amberlite CG-50 was from Sigma Chemical Company.
DNA manipulations.
Genomic DNA was obtained from S. aureus protoplasts as previously described (40). E. coli plasmid DNA was obtained by the alkaline lysis procedure used with the Qiagen kit (Qiagen, Inc.). Staphylococcal plasmid DNA was isolated from cleared lysates and purified by CsCl-ethidium bromide dye-buoyant density centrifugation (54) or by a staphylococcal mini-prep procedure as previously described (68). DNA modification using alkaline phosphatase, phage T4 DNA polymerase, or the Klenow fragment of E. coli DNA polymerase was performed according to Maniatis et al. (42). DNA fragments were isolated from agarose gels by a previously described freeze-squeeze technique (66) with Gene Clean (Bio 101, Inc., La Jolla, Calif.) or the QIAquick Gel Extraction Kit (Qiagen, Inc.).
Southern blot analysis.
Genomic DNA was digested with HindIII, separated on a 1% GTG agarose gel (FMC Bioproducts, Rockland, Maine), and transferred to nitrocellulose filters (Schleicher and Schuell, Keene, N.H.). Southern blot analysis was performed as previously described (except prehybridizing and hybridizing solutions contained 200 μg of sheared salmon sperm DNA/ml) under low-stringency conditions (20% [vol/vol] formamide) (8). The filters were hybridized with a 32P-labeled (42) double-stranded internal structural gene probe from either sea (A-624; a 624-bp HindIII/BamHI fragment from pMJB38 [8]) or sec (C-526; a 562-bp SspI fragment from pMJB124 [14]). The filters were washed at 45°C prior to film exposure.
Extraction of RNA and Northern blot analysis.
RNA was prepared and quantified by A260 readings, and equal amounts of RNA were separated on a 1% agarose–2.2 M formaldehyde gel all as described previously (58). Samples used had A260/A280 ratios between 1.9 and 2.0. RNA was transferred from the gel onto Nytran filters (Schleicher and Schuell) and hybridized to a denatured, 32P-labeled seg probe (SEG-600; a 600-bp BsmI/SpeI internal structural gene probe from pMJB460) as previously described (42). RNA markers were used as size standards (Promega Corp.). Northern analysis was also performed with a 32P-labeled antisense mRNA probe synthesized by in vitro transcription by using the MAXIscript kit (Ambion Inc., Austin, Tex.) according to the manufacturer’s instructions.
DNA sequence analysis.
DNA sequences were obtained for both strands with Sequenase enzyme version 2.0 and the Sequenase kit (United States Biochemical, Cleveland, Ohio) according to the manufacturer’s protocols with either standard or deoxyinosine reagents. The sequencing gels were run with 0.5× Tris-borate-EDTA in the top chamber and 1 M sodium acetate in the lower chamber as previously described (41). The DNA sequence was obtained from pGEM7-based plasmids with SP6 and T7 promoter primers (Promega) in addition to synthetic primers (University of Wisconsin Biotechnology Center, Madison, Wis.) derived from the sequence provided in this paper. DNA sequences were verified by the University of Wisconsin Biotechnology Center using an ABI DNA sequencing apparatus (Applied Biosystems, Inc., Foster City, Calif.). Genetics Computer Group (Madison, Wis.) software and the Lasergene package (DNASTAR, Madison, Wis.) were used to analyze the DNA sequences.
Nucleotide and amino acid sequence analysis.
Nucleotide and amino acid sequences were analyzed using programs from the Lasergene package (DNASTAR). Alignments were performed by the Clustal method of the Megalign program. The amino acid sequence alignment created by the Clustal method was manually changed to alter the gap spacing of the first 25 amino acids of SEI to resemble the SEI alignment with SEA and SEE (to which SEI is most similar). Amino acid sequences compared in a pairwise fashion by the Lipman-Pearson algorithm of the Megalign program had results that were similar to those obtained with Clustal (data not shown).
Plasmid construction and transformation of E. coli and S. aureus.
Plasmids were constructed and introduced into strains as indicated in Table 1. S. aureus RN4220 was transformed by electroporation as described previously (30). Generalized transduction with phage 80α was performed as previously described (62) to move plasmids from RN4220 to S. aureus ISP2073. S. aureus RN7497 was transformed by the protoplast transformation method described by Novick (52). Protoplasts were allowed to regenerate at 30°C for 3 days on DM3 medium (52) containing chloramphenicol.
E. coli DH10B MAX Efficiency cells (Gibco-BRL Life Technologies, Gaithersburg, Md.) were used for the transformation of pMJB460 and pMJB461. Transformants were screened by a colony blot procedure (42) with the C-562 sec probe.
Histidine-tagged enterotoxins.
Amino-terminally histidine-tagged derivatives of mature SEG (HisSEG) and the putative mature form of SEI (HisSEI) were constructed by using the QIAexpressionist system (Qiagen, Inc.). Both proteins were expressed and purified from E. coli M15 (pREP4) according to the manufacturer’s protocol (Qiagen, Inc.).
(i) Production of HisSEG.
E. coli MJB1323(pMJB474) produces an N-terminally histidine-tagged SEG protein that has the sequence HHHHHHGIRMRARYP joined to the Q residue at the N terminus of mature SEG (see Fig. 3). pMJB474 is pQE-32 (encodes a histidine tag) with an 800-bp BsmI (made blunt)/EcoRV seg fragment insert (obtained from pMJB464) ligated to the SmaI site. The extraneous amino acids following the histidine tag result from translation of the multiple-cloning site for the pQE vector.
FIG. 3.
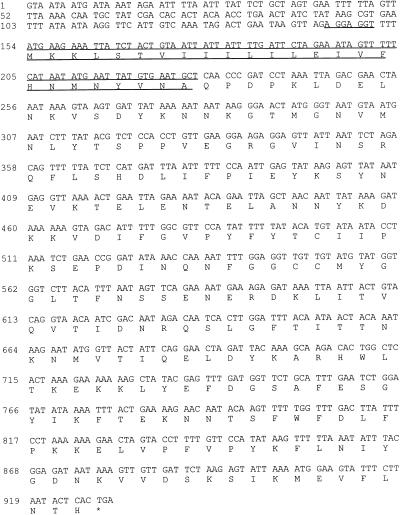
Nucleotide and deduced amino acid sequences for seg. The putative ribosome-binding site is underlined once, and the signal sequence cleaved to form mature SEG is overlined and underlined. The cleavage site for SEG was determined by purifying SEG from S. aureus culture supernatants and performing N-terminal sequence analysis. The asterisk denotes the translation stop codon.
(ii) Production of HisSEI.
E. coli MJB1324(pMJB475) produces an N-terminally histidine-tagged SEI protein with the sequence HHHHHHT joined to the Q residue at the N terminus of mature SEI (predicted from alignment with SEA). pMJB475 was constructed by digesting pQE31 (encodes a histidine tag) with BamHI/SalI and ligating it to a 770-bp BglII/XhoI sei-containing fragment from pMJB465.
Soluble, cytoplasmic HisSEG and HisSEI were purified according to the manufacturer’s protocol utilizing an RNase A and DNase I digest prior to the addition of Ni-NTA resin. The column was washed with 50 mM sodium phosphate (pH 6.0)–300 mM NaCl–10% glycerol–10 mM imidazole (wash buffer) prior to elution with a 40 ml gradient of 10 to 500 mM imidazole in wash buffer, pH 6.0. The His-SE-containing fractions were applied to an S100 size exclusion column equilibrated with buffer containing 50 mM sodium phosphate (pH 7)–300 mM NaCl. The resulting protein preparation contained no other proteins, as determined with a silver-stained polyacrylamide protein electrophoretic gel.
Polyclonal antiserum.
Polyclonal rabbit serum was made against each of the purified histidine-tagged SEs by the Animal Care Unit of the University of Wisconsin—Madison Medical School. New Zealand White rabbits were given an initial intradermal injection of 15 μg of toxin in complete Freund’s adjuvant. Subsequent injections, at 4-week intervals, contained 40, 100, and 150 μg of toxin in incomplete Freund’s adjuvant. Bleeds were performed 2 weeks after each injection.
Western blot analysis.
Culture supernatant samples were prepared by centrifugation of the S. aureus culture and filter sterilization of the supernatants. Purified protein samples were diluted into phosphate-buffered saline. Samples were separated on a denaturing 12% polyacrylamide gel and electrophoretically transferred to nitrocellulose filters (Schleicher and Schuell). Filters were treated with polyclonal rabbit antiserum made against HisSEG or HisSEI. Signals were visualized with the ProtoBlot System AP (Promega Corp.).
Emetic assay.
S. aureus MJB1316, MJB1317, MJB1320, and MJB1321 were grown for 14 to 16 h under the growth and inducing conditions described above. The bacterial cells were removed by centrifugation, and the culture supernatants were concentrated by ultrafiltration through a 10,000-molecular-weight (MW)- cutoff membrane (YM10; Amicon, Beverly, Mass.). Retained proteins were administered in the assay. Concentrated supernatants were filter sterilized by passage through a 0.45-μm-pore-size filter (Gelman Sciences Inc., Ann Arbor, Mich.) and stored on ice. SEG concentrations were quantitated by Western blot analysis (developed with polyclonal HisSEG antiserum) using known concentrations of purified HisSEG as a standard. SEI was quantitated by silver-staining protein gels (46) and comparing serial dilutions of SEI-containing culture supernatants to known concentrations of purified HisSEI. The concentrated equivalents of culture supernatants from isogenic strains possessing SE structural genes containing translation stop signals (described above) served as negative controls. Rhesus monkeys (Macaca mulatta), 2 to 4 kg in size, were given room-temperature samples via nasogastric intubation and were observed for 5 h. SEG-containing culture supernatants were administered at 80 μg/kg of animal weight, and SEI-containing culture supernatants were administered at 150 μg/kg of animal weight. The emetic assays were performed in collaboration with the Wisconsin Regional Primate Research Center, Madison, Wis.
SEG purification and N-terminal amino acid sequence determination.
SEG was purified from MJB1316. All chromatography columns were run at room temperature. The first step in the purification procedure was a permutation of the procedure used to purify TSST-1 (59). MJB1316 was inoculated into 500 ml of 3+1 in 2-liter flasks. The cultures were grown at 37°C with aeration for 7 h, induced with 12 μg of CBAP/ml, and incubated an additional 11 h. Bacterial cells were removed by centrifugation, the culture supernatants were diluted 2.5-fold with distilled, deionized H2O (ddH2O), and the pH was adjusted to 5.3 with HCl. CG-50 resin was prepared as previously described (59) except the pH was adjusted to pH 5.3. For a 1-liter volume of original culture, the swelled equivalent of 12.5 g of CG-50 was added to the diluted culture supernatants and stirred for 80 min at room temperature. After the resin settled, supernatants were removed, and a column (1.25-cm radius) was packed to a bed height of 20 cm (98-ml bed volume). The column was washed with 400 ml of ddH2O at 3.5 ml/min. The column was eluted with 0.5 M sodium phosphate (pH 6.8)–0.2 M NaCl at 2 ml/min. All of the protein eluted in one peak. This bulk protein was dialyzed in 40 mM sodium phosphate, pH 5.4 (loading buffer), clarified, filter sterilized through a 0.45-μm-pore-size filter, and loaded onto an SP Sepharose column with dimensions of 16 mm by 20 cm (Pharmacia Biotech). The column was washed briefly with loading buffer and eluted at a rate of 0.25 ml/min with a pH gradient of pH 5.4 to 7.8 in 40 mM sodium phosphate. One-milliliter fractions were collected and assayed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis for SEG. SEG-containing fractions were pooled, dialyzed in 20 mM sodium phosphate (pH 7)–150 mM NaCl, and applied to a Sephacryl S100 column (16 mm by 60 cm) equilibrated with the same buffer. The flow rate and fraction volume collected were as described for the SP Sepharose column. Protein purity was determined by SDS-PAGE and Coomassie blue staining. Protein concentration was determined by using a protein assay reagent (Bio-Rad Laboratories, Richmond, Calif.) with bovine serum albumin protein standards (Sigma) buffered identically to the assayed proteins.
Amino-terminal sequence analysis was performed for the first 15 amino acid residues of purified SEG by the Macromolecular Structure Facility at Michigan State University (East Lansing, Mich.).
Murine T-cell proliferation assay.
T-cell proliferation assays were performed with splenocytes obtained from 6- to 8-week old BALB/c mice (Harlan Sprague-Dawley, Madison, Wis., or Jackson Laboratory, Bar Harbor, Maine). A total of 106 (150 μl) splenocytes were dispensed into wells of a 96-well tissue culture plate (Falcon; Becton Dickinson and Co., Lincoln Park, N.J.) along with 50 μl of 10−1, 10−2, or 10−3 dilutions of S. aureus culture supernatants made in complete tissue culture medium (RPMI 1640; ICN Biomedicals, Inc., Costa Mesa, Calif.) containing 10% fetal bovine serum (Biocell, Rancho Dominguez, Calif.), 15 mM HEPES, 3 mM glutamine, and 50 μg of gentamicin (Gibco-BRL Life Technologies)/ml. The cells were incubated at 37°C with 5% CO2 for 72 h prior to pulsing for 18 h with 0.5 μCi of [3H]thymidine in 50 μl of complete tissue culture medium. The cells were harvested onto glass fiber filter paper, and the amount of incorporated [3H]thymidine was quantified by liquid scintillation. Three assays were performed and each sample was assayed in quadruplicate. The statistical significance (P ≤ 0.001) was determined by Student’s t test using Minitab (Minitab, Inc.).
IL-2, IL-4, and IFN-γ ELISAs.
Samples of 5 × 106 splenocytes, prepared as described above in complete tissue culture medium, were dispensed into each well of a 24-well tissue culture dish (750 μl). A total of 250 μl of diluted S. aureus culture supernatants (10−1 and 10−2) was added (the ratio of splenocytes to volume of culture supernatant was the same as that used in the murine splenocyte proliferation assay), and the cells were cultured for 48 h prior to collection of the supernatants for cytokine analysis.
Cytokine concentrations were determined by sandwich ELISA. Capture antibodies, biotinylated detection antibodies, and cytokine standards were obtained from Pharmingen (San Diego, Calif.). The ELISA was performed following the manufacturer’s recommendations. Biotinylated antibody was detected by using the Vectastain ABC kit (Vector Laboratories, Burlingame, Calif.), and the absorbance at 405 nm was read on a microplate reader (Bio-Tek Instruments).
Nucleotide sequence accession numbers.
The nucleotide sequence data reported here have been submitted to the GenBank database under accession numbers AF064773 (SEG) and AF064774.
RESULTS
Identification of two S. aureus strains that possess DNA similar to that of a sec probe.
Enterotoxigenic (emesis-causing) S. aureus strains that did not produce SEA to SEE, as previously tested by ELISA (sensitivity of ≥0.625 ng/ml), were isolated from a variety of sources (35). Because the characterized enterotoxins (SEA to SEE and SEH) share nucleotide sequence identity (≥32%), it was considered likely that a new enterotoxin gene would also share nucleotide sequence identity with the characterized toxins. We performed Southern blot analysis to identify new candidate enterotoxin genes from the strains described above.
Two of eight strains (FRI572 and FRI445) contained genomic DNA that hybridized to a sec probe (C-562) under low-stringency conditions (Fig. 1). Genomic DNA from strains FRI445 and FRI572 both contained 2.5-kbp-sized HindIII fragments that hybridized to C-562. C-562 hybridized to genomic DNA from FRI400 (contains sec) and FRIS6 (contains sea and seb). C-562 hybridization to seb is expected as seb and sec share approximately 69% nucleotide sequence identity (6). C-562 did not hybridize to genomic DNA from RN450, which is a nonenterotoxigenic strain (53). No hybridization with genomic DNA from these strains was observed with a sea probe (data not shown).
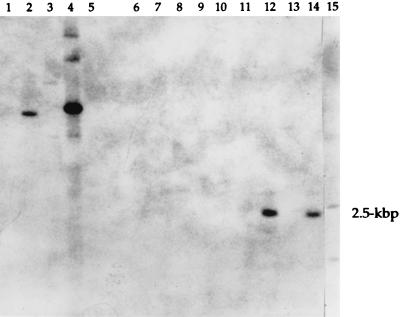
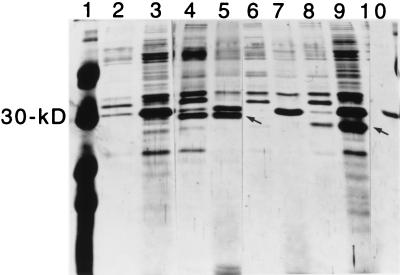
FIG. 1.
Southern hybridization analysis of genomic DNA from enterotoxigenic S. aureus strains using a sec probe. Genomic DNA was digested with HindIII, fractioned by agarose gel electrophoresis, transferred to a nylon membrane, and probed with the 32P-labeled sec probe SEC-524. The lanes contain DNA from S. aureus strains as follows (except lane 10, which contains phage λ DNA): lane 1, FRI337 (produces SEA and SED); lane 2, FRIS6 (produces SEA and SEB); lane 3, FRI578 (produces SEE); lane 4, FRI400 (produces SEC); lane 5, RN450 (non-enterotoxin producer); lane 6, FRI569; lane 7, FRI393; lane 8, FRI591; lane 9, FRI574; lane 10, λ DNA; lane 11, FRI772; lane 12, FRI445; lane 13, FRI569; lane 14, FRI572; lane 15, FRI662. Lanes 6 to 9 and 11 to 15 contain DNA from enterotoxigenic S. aureus strains that do not produce SEA to SEE as determined by ELISA. The 2.5-kbp insert is shown at right.
FRI445 and FRI572 were originally isolated from the nares of residents of Easter Island in the late 1960s (5). Bergdoll and colleagues performed emetic assays with primates and found that FRI572 culture supernatants caused emesis in three of four animals when a concentrated 300-ml equivalent of culture supernatant was administered. FRI445 culture supernatant elicited a response in 5 of 12 animals when a concentrated 50-ml equivalent of supernatant was administered (5, 35). FRI445 was later found to produce 109 ng of SEH/ml as characterized by Su and Wong (64). Therefore, the emetic capabilities of FRI445 culture supernatants cannot be attributed solely to the activity of a putative new enterotoxin. Because it is common for S. aureus strains to produce more than one type of enterotoxin, it is possible that FRI445 and FRI572 each carry multiple enterotoxin genes. Therefore, the new enterotoxins were characterized in recombinant, nonenterotoxin-producing S. aureus strains.
Identification of seg and sei.
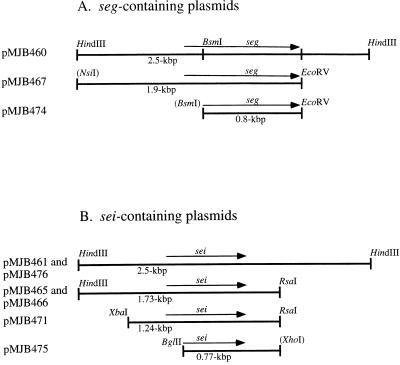
E. coli plasmids pMJB460 and pMJB461, which contain 2.5-kbp genomic DNA fragments from FRI572 and FRI445, respectively, were mapped by Southern blot analysis of restriction endonuclease digests by using C-562 as a probe. The region of the fragment insert that shared similarity with C-562 was localized. Figure 2 depicts diagrams of the seg- and sei-containing E. coli plasmid inserts.
FIG. 2.
Diagrams of cloned genomic DNA plasmid inserts containing seg (A) or sei (B), cloned from S. aureus FRI572 and FRI445, respectively. Subclones shown below the 2.5-kbp insert are drawn to match the regions from which they came. seg and sei are labeled, and the sizes of the inserts are labeled below the fragments. Restriction sites in parentheses were altered in the cloning process or were derived from plasmid multiple-cloning sites and are not available for recleavage. pMJB474 and pMJB475 contain nucleotide sequences for the mature forms of SEG and SEI, respectively.
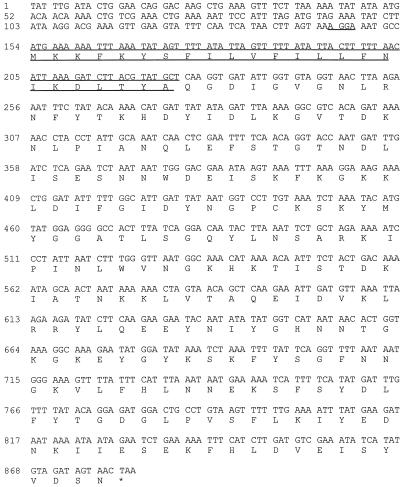
Nucleotide sequence analysis of the fragment cloned from FRI572 revealed a putative enterotoxin gene, designated seg (Fig. 3), consisting of a 774-bp open reading frame (ORF) encoding 258 deduced amino acid residues. Similar analysis of the C-562-hybridizing region of the FRI445 fragment (3′ end) revealed a enterotoxin-like gene containing a frameshift mutation. Directly 5′ of this disrupted gene, however, was found a complete putative enterotoxin gene designated sei (Fig. 4). This gene consisted of a 726-bp ORF and encoded a predicted product of 242 amino acids. An additional enterotoxin-like ORF was discovered upstream of sei. This gene will not be characterized in this paper, and no biological activity could be attributed to a product of this gene in the recombinant S. aureus strains constructed to study sei (see Discussion).
FIG. 4.
Nucleotide and deduced amino acid sequences for sei. The putative ribosome-binding site is underlined once, and the predicted signal sequence cleaved to form mature SEI is overlined and underlined. The asterisk denotes the translation stop codon.
Detection of SEG and SEI in S. aureus strains.
All characterized S. aureus enterotoxins are superantigens; hence, the splenocyte (T-cell) proliferation assay was utilized as a sensitive method for assaying S. aureus transformants for superantigen activity. The S. aureus plasmid pC194 was ligated to E. coli vectors containing either seg or sei on 2.5-kbp fragments (pMJB460 or pMJB461, respectively). These E. coli-S. aureus shuttle vectors [pMJB462 (seg) and pMJB476 (sei)] were introduced into non-enterotoxin-producing S. aureus RN4220, RN450, and ISP2073 for analysis. Culture supernatants from the recombinant S. aureus MJB1310(pMJB462) (seg) did not stimulate splenocyte proliferation, and no unique protein was observed by analysis of the supernatants on silver-stained SDS-PAGE gels (data not shown). Therefore, seg was transcribed from the inducible β-lactamase promoter present in an S. aureus expression vector. Culture supernatants from MJB1316(pMJB467) (Fig. 2A) contained a 27-kDa polypeptide on silver-stained SDS-PAGE gels as would be expected for SEG (Fig. 5). S. aureus MJB1317, in which seg was disrupted by a translation termination signal created after the first 100 bp, served as a negative control and did not produce the 27-kDa protein.
FIG. 5.
Silver-stained polyacrylamide gel of S. aureus 16-h culture supernatants containing SEG and SEI. Samples were analyzed on an SDS–12% PAGE gel. Lane 1 contains protein markers of 46, 30, 21.5, and 14.3 kDa. Samples of S. aureus culture supernatants were from the following strains: lane 2, uninduced RN8117 (host strain for plasmids); lane 3, induced RN8117; lane 4, uninduced MJB1316 (contains seg); lane 5, induced MJB1316 (contains seg); lane 6, uninduced MJB1317 (contains translation stop codon in seg); lane 7, induced MJB1317 (contains translation stop codon in seg); lane 8, uninduced MJB1320 (contains sei); lane 9, induced MJB1320 (contains sei). Purified SEA is in lane 10. Arrows indicate SEG (lane 5) and SEI (lane 9). All samples were run simultaneously, and irrelevant lanes are omitted from the figure.
To determine if SEI was produced from the cloned genomic FRI445 DNA fragment, culture supernatants from the clones were assayed for T-cell proliferation. Culture supernatants from strains containing the 2.5-kbp MJB1326(pMJB476) (Fig. 2B) and the 1.73-kbp MJB1315(pMJB466) (Fig. 2B) sei plasmid inserts stimulated splenocyte proliferation. sei was cloned into an S. aureus expression vector to be transcribed from the β-lactamase promoter [MJB1320(pMJB471)] (Fig. 2B). Upon induction, SDS-PAGE analysis of culture supernatants revealed a distinct 25-kDa protein of the size expected for SEI (Fig. 5). Negative controls for the two SEI-expressing strains described above (MJB1315 and MJB1320) were constructed that were isogenic except for a translation termination signal that was created after the first 200 bp of sei (MJB1318 and MJB1321, respectively). Culture supernatants from these negative controls did not contain the 25-kDa protein.
Northern blot analysis of seg mRNA from FRI572.
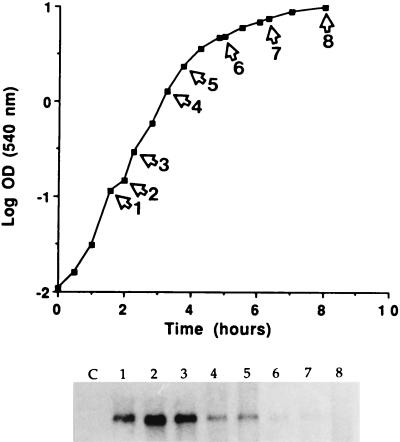
Total cellular RNA from S. aureus FRI572, the seg parent strain, was analyzed at various time points throughout growth for seg mRNA. Cellular RNA from S. aureus RN450, a non-enterotoxin-producing strain, served as a negative control. Northern analysis using SEG-600 (600-bp fragment internal to seg) as a probe identified one 6.7-kb transcript. To verify that this signal corresponded to seg mRNA, and not mRNA transcribed from DNA sequence on the opposite strand, an antisense mRNA probe was used which would bind only to seg mRNA. This seg antisense mRNA probe also hybridized to a 6.7-kb transcript (data not shown). Steady-state seg mRNA accumulated maximally during logarithmic growth, as shown in Fig. 6.
FIG. 6.
Northern hybridization analysis of cellular RNA from S. aureus FRI572 hybridized with the seg probe SEG-600. Samples were taken throughout growth at the labeled time points from cultures grown in shake flasks with medium consisting of 3% N-Z amine and 1% yeast extract. Total cellular RNA was separated on a 1% agarose gel, transferred to a nylon membrane, and hybridized with 32P-labeled SEG-600. Total cellular RNA from non-enterotoxin-producing S. aureus RN450 served as a negative control (lane C). Northern blot analysis determined that FRI572 mRNA is 6.7 kb in length.
Nucleotide and deduced amino acid sequence analysis of seg and sei.
Figures 3 and 4 contain the nucleotide and deduced amino acid sequences for seg and sei, respectively. The 5′-proximal sequences, including initiation codons for seg and sei, were 5′-gttagaGGAGGttttATG-3′ and 5′-tagtaaAGGAaatgccATG-3′, respectively. These ribosome-binding sites (capital letters preceding the ATG initiation codons) are similar to those for E. coli and to other published putative ribosome-binding sites for S. aureus genes (51). There is no obvious putative promoter in the upstream region of seg, which is not surprising due to the fact that SEG is not produced from the 2.5-kbp S. aureus fragment cloned and that seg mRNA from FRI572 is 6.7 kb in length (see above). Inspection of the DNA sequence upstream from sei also did not reveal an obvious promoter sequence. The extracellular form of SEG is predicted to comprise 233 amino acid residues, corresponding to a 27,042-Da protein (based upon the N-terminal amino acid sequence of purified SEG [see below]). The amino acid sequence for the predicted mature form of SEI (determined from amino acid sequence alignment with the characterized enterotoxins) consists of 218 amino acids, corresponding to a 24,928-Da protein.
Both SEG and SEI have typical bacterial signal sequences (45) with positively charged N termini and hydrophobic cores. The precursor form of SEG and the predicted precursor form of SEI contain 25- and 24-amino-acid-residue signal sequences, respectively (Fig. 3 and 4). These sequences, cleaved to produce the mature extracellular forms of SEG and SEI, are similar to those of the characterized enterotoxins (2, 8, 11, 12, 15, 27, 33, 60).
Comparison of nucleotide and derived amino acid sequences of seg and sei with the sequences of other bacterial toxins.
The nucleotide sequences of seg and sei were compared to the nucleotide sequences of sea (8), seb (33), sec1 (11), sec2 (12), sec3 (27), sed (2), see (15), seh (60), speA (69), and ssa (57). Among this group of genes, seg is most closely related to sec3 (46.2% similarity), sec2 (45.5%), sec1 (44.1%), seb (44.1%), ssa (42.8%), and speA (40.4%). The sei gene is related to the characterized toxins, with nucleotide sequence identities ranging from 27.5 to 35.5%. sei is most closely related to sed and sea, with 35.5 and 34.4% nucleotide sequence identities, respectively.
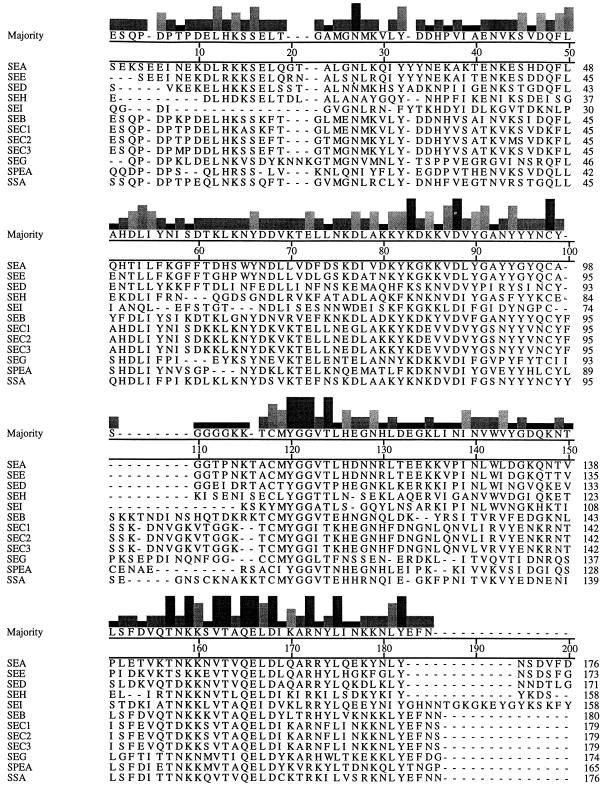
The deduced amino acid sequences of mature SEG and SEI were compared to the deduced amino acid sequences for the mature forms of SEA (8), SEB (33), SEC1 (11), SEC2 (12), SEC3 (27), SED (2), SEE (15), SEH (60), SpeA (69), and SSA (57) by the Clustal method, and the percentages of identity are presented in Table 2. An alignment of all of the enterotoxins with SpeA and SSA appears in Fig. 7. SEG is most closely related to SpeA, SSA, SEB, and the SECs, with 41.6, 40.3, 39.1, and 37.8 to 38.6% amino acid identities, respectively. SEI is most closely related to SEA, SEE, and SED, with 28.4, 27.5, and 26.1% amino acid identities, respectively.
TABLE 2.
Percentages of amino acid sequence identity between the mature forms of staphylococcal and streptococcal toxinsa
| Toxin | % Sequence identity
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SEA | SEB | SEC1 | SEC2 | SEC3 | SED | SEE | SEG | SEH | SEI | SpeA | SSA | |
| SEA | 100 | 27.9 | 23.2 | 24.0 | 24.9 | 52.2 | 81.7 | 20.2 | 30.9 | 28.4 | 25.3 | 27.0 |
| SEB | 100 | 64.0 | 62.8 | 62.8 | 28.1 | 27.4 | 39.1 | 24.9 | 18.8 | 46.2 | 59.4 | |
| SEC1 | 100 | 97.1 | 96.2 | 25.9 | 24.8 | 37.8 | 20.7 | 17.0 | 43.4 | 56.8 | ||
| SEC2 | 100 | 98.3 | 26.3 | 25.2 | 38.6 | 21.2 | 17.0 | 43.0 | 58.1 | |||
| SEC3 | 100 | 26.3 | 25.7 | 38.6 | 22.1 | 17.0 | 43.4 | 58.1 | ||||
| SED | 100 | 54.8 | 18.9 | 30.9 | 26.1 | 29.4 | 25.0 | |||||
| SEE | 100 | 20.9 | 30.9 | 27.5 | 25.8 | 26.1 | ||||||
| SEG | 100 | 24.0 | 20.6 | 41.6 | 40.3 | |||||||
| SEH | 100 | 23.5 | 28.6 | 24.4 | ||||||||
| SEI | 100 | 20.6 | 22.0 | |||||||||
| SpeA | 100 | 43.0 | ||||||||||
| SSA | 100 | |||||||||||
Amino acid sequences were compared by the Clustal method with both gap and gap length penalties of 10.
FIG. 7.
Comparison of amino acid sequences of SEA, SEB, SEC1, SEC2, SEC3, SED, SEE, SEG, SEH, SEI, SpeA (SPEA), and SSA. The consensus sequence is displayed above the individual protein sequences. The histogram above each amino acid residue indicates the degree of sequence conservation at that residue (the height of the histogram increases with the increase in sequence conservation). The alignment was made with the MegAlign program of the Lasergene package (DNASTAR).
SEG and SEI are emetic toxins.
S. aureus MJB1316 and MJB1320, which produce SEG and SEI from the inducible S. aureus β-lactamase promoter, respectively, were used as sources of the toxins for the primate emetic assays. Negative controls were culture supernatants from non-SEG- and non-SEI-producing S. aureus strains (MJB1317 and MJB1321, respectively), which are isogenic to the SEG- and SEI-producing strains (MJB1316 and MJB1320, respectively) except for translation termination signals present in either seg or sei, respectively. Staphylococcal culture supernatants that contained SEG (administered at 80 μg/kg of animal weight), SEI (150 μg/kg of animal weight), or supernatants from the negative-control strains (given as concentrated equivalents to the enterotoxin-containing supernatants) were administered nasogastrically to rhesus monkeys. SEG- and SEI-containing culture supernatants evoked an emetic (vomiting) response in four of six and one of four animals tested, respectively. Animals that received enterotoxin but did not vomit experienced other symptoms of intoxication such as diarrhea or pronounced lethargy. None of the animals that received culture supernatants from negative-control strains produced an emetic response, developed diarrhea, or became lethargic (three animals tested for each control).
SEG and SEI stimulate T-cell proliferation.
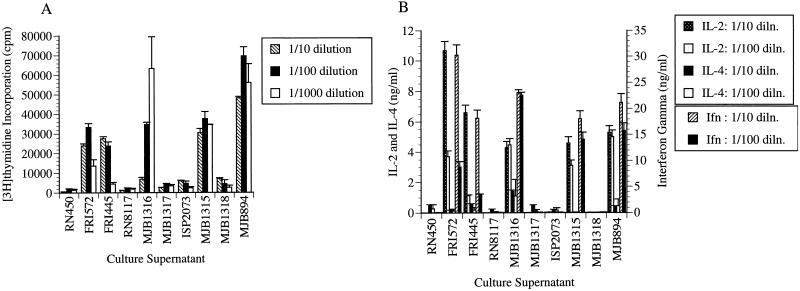
Culture supernatants from SEG- and SEI-producing S. aureus strains and from negative-control strains were tested in a murine splenocyte proliferation assay. Splenocyte proliferation assays were performed in parallel with ELISAs that measured IL-2, IL-4, and IFN-γ concentrations in the splenocyte culture medium after 48 h of stimulation.
Culture supernatants from S. aureus FRI572 (the strain from which seg was cloned) and the SEG-producing S. aureus strain MJB1316 stimulated T-cell proliferation, while supernatants from the non-SEG-producing negative controls MJB1317 (isogenic except for a translation stop codon in seg) and RN8117 (S. aureus host strain containing cloning vector) did not (Fig. 8A). Likewise, culture supernatants from S. aureus FRI445 (the strain from which sei was cloned) and the SEI-producing S. aureus strain MJB1315 stimulated T-cell proliferation, while supernatants from the non-SEI-producing S. aureus negative-control strains MJB1318 (isogenic except for a translation stop codon in sei) and ISP2073 (host strain) did not. T-cell proliferative activities from FRI445 and FRI572 were compared to that of the non-enterotoxin-producing S. aureus strain RN450. SEA-producing S. aureus MJB894 served as a positive control. The differences between enterotoxin-producing strains and negative controls were statistically significant (P ≤ 0.001).
FIG. 8.
(A) Induction of murine splenocyte proliferation by S. aureus supernatants. Dilutions of culture supernatants were incubated with murine splenocytes for 72 h followed by an 18-h pulse with [3H]thymidine. Each sample was tested in triplicate, and the results are reported as the mean counts per minute of a representative experiment. Each sample was tested three times in this assay. Standard deviations are indicated by error bars. The supernatants were from the following strains: RN450 (non-enterotoxin-producing strain), FRI572 (strain from which seg was cloned), FRI445 (strain from which sei was cloned), RN8117 (host strain for seg-containing plasmids), MJB1316 (SEG-producing strain), MJB1317 (negative control; contains a translation stop codon in seg), ISP2073 (host strain for sei-containing plasmids), MJB1315 (SEI-producing strain), MJB1318 (negative control; contains a translation stop codon in sei), and MJB894 (SEA-producing strain). (B) IL-2, IL-4, and IFN-γ production in splenocyte cultures after 48 h of stimulation with dilute culture supernatants as described for panel A. This assay was set up at the same time as the proliferation assay used for the results shown in panel A. Splenocyte culture supernatants were clarified by centrifugation and were analyzed for cytokine production by ELISA. Results are means expressed in nanograms per milliliter compared to recombinant cytokine standards. The standard errors of the means are indicated by error bars.
Culture supernatants from SEG-, SEI-, and SEA-producing S. aureus strains stimulated marked IL-2 and IFN-γ production, as analyzed by ELISA after 48 h of stimulation (Fig. 8B). Little IL-2 or IFN-γ was detected in splenocyte culture supernatants that were stimulated with supernatants from the non-enterotoxin-producing S. aureus negative-control strains (MJB1317, MJB1321, RN450, RN8117, and ISP2073).
Purification of SEG.
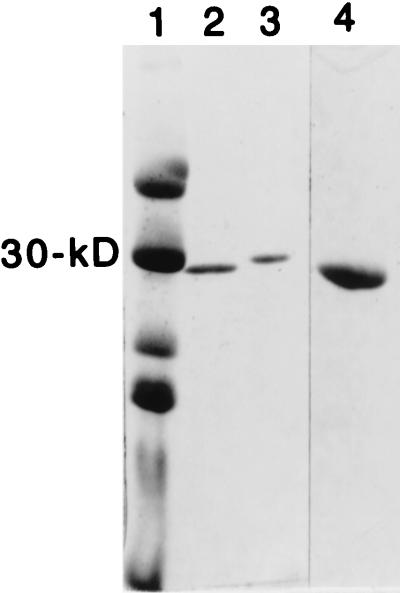
SEG was purified from the culture supernatants of a recombinant S. aureus strain in which seg was transcribed from the inducible β-lactamase promoter (MJB1316). This strain, when induced, produces approximately 5.5 μg of SEG/ml. SEG was chromatographed by using CG-50, SP Sepharose, and Sephacryl S100 columns. A single polypeptide migrating similarly to the 30-kDa marker was evident when 2 μg was analyzed by SDS-PAGE and the gel was stained with Coomassie blue. The predicted size for mature SEG from the derived amino acid sequence is 27,042 Da, corresponding well to the apparent sizes for mature SEG determined from both SDS-PAGE analysis (28,800 Da) (Fig. 9) and from a size exclusion chromatography column (approximately 30 kDa). Amino-terminal sequence analysis of purified SEG revealed QPDPKLDELNKVSDY to be the sequence of the first 15 amino acid residues. This sequence is identical to the predicted amino acid sequence. This verified that SEG was purified and indicated the signal sequence cleavage site (Fig. 3).
FIG. 9.
Coomassie-stained SDS-PAGE gel of purified SEG. Lane 1, molecular mass markers; lane 2, SEG; lane 3, SEA; lane 4, SEG (2 μg).
Purified SEG was tested in the murine T-cell proliferation assay at concentrations of 18.4, 1.84, and 0.184 nM. Maximal proliferation was observed at an SEG concentration of 1.84 nM (data not shown).
Western blot analysis of the characterized toxins.
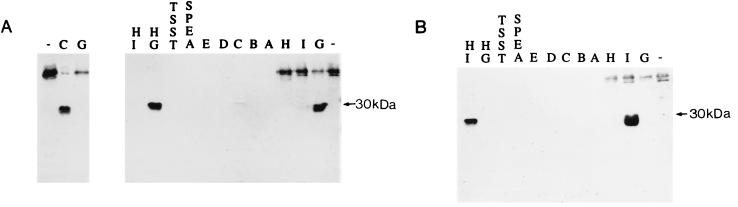
Purified amino-terminal histidine-tagged SEG (HisSEG) and SEI (HisSEI) were used to generate polyclonal rabbit antiserum. Purified HisSEG and HisSEI stimulated murine T-cell proliferation (data not shown), and the antiserum prepared against each reacted with SEG and SEI, respectively. Culture supernatants from S. aureus MJB1316, MJB1320, and FRI569 were utilized as sources of SEG, SEI, and SEH (as characterized by Su and Wong [65]), respectively. Purified toxins were used as the sources of SEA, SEB, SEC1, SED, SEE, TSST-1, and SpeA. MJB1316 and MJB1320 produce approximately 5 μg of SEG and SEI/ml, respectively, and FRI569 produces 230 ng of SEH/ml (64). Purified toxin samples were diluted to a 5-μg/ml concentration in phosphate-buffered saline. Western blot analysis of the denatured proteins detected with antiserum to HisSEG revealed that SEC1 shares some epitopes with SEG (Fig. 10A). Analysis with antiserum prepared against HisSEI revealed that none of the toxins examined have epitopes in common with SEI (Fig. 10B). Ouchterlony immunodiffusion assays (nondenaturing; sensitivity, ≥500 ng/ml) were performed with purified SEA, SEB, SEC3, SED, SEE, HisSEG, and HisSEI (all at 4 μg/ml) against anti-HisSEG or anti-HisSEI (toxins were also examined with anti-HisSEI at a concentration of 20 μg/ml). No lines of identity or spurs were observed under these conditions (data not shown), suggesting that these toxins do not share detectable conformational epitopes with HisSEG or HisSEI.
FIG. 10.
Western blot analyses. The filters were reacted with antiserum prepared against HisSEG (A, right), HisSEI (B), or SEC (A, left). The following samples were analyzed: HisSEI (lanes HI), HisSEG (lanes HG), TSST-1 (lanes TSST), SpeA (lanes SPEA), SEE (lanes E), SED (lanes D), SEC (lanes C), SEB (lanes B), and SEA (lanes A) and culture supernatants from MJB1320 (lanes I, SEI producing), MJB1316 (lanes G, SEG producing), and RN8117 (lanes marked by minus signs, negative-control host strain for seg- and sei-containing plasmids). Signals observed in the region of the gel corresponding to a molecular mass of >30 kDa were presumably due to protein A.
DISCUSSION
Here we describe two new staphylococcal enterotoxins, SEG and SEI, both of which have T-cell proliferative and emetic properties. These toxins are clearly related to the characterized enterotoxins, yet they are distinct in several interesting ways. SEI is a more divergent member of the enterotoxin family than SEG and is approximately as divergent as SEH. Although SEG is as similar to SEB as it is to SEC (approximately 39% amino acid identity to each), immunoblot analysis indicates that SEG shares antigenic epitopes only with SEC (Fig. 10A).
Northern blot analysis of FRI572 RNA revealed that seg mRNA is unusually large (6.7 kb), which suggests that it may be polycistronic. The large transcript is unique to the characterized enterotoxins as each of the known enterotoxin transcripts is just slightly larger than its respective ORF (6, 31). The largest mRNA previously reported is that for sed, which has a transcription start site 266 bp upstream of sed (2). Perhaps the seg transcript includes additional toxin genes or regulatory elements that are cotranscribed with seg. While no other ORFs were present on the 2.5-kbp DNA fragment, there were small stretches of nucleotide sequence which encoded enterotoxin-like sequence segments, indicating that there could be portions of other enterotoxin genes surrounding seg. seg most likely is not the first gene transcribed because there is no apparent production of SEG from the cloned FRI572 DNA fragment (culture supernatants do not possess T-cell stimulatory properties).
Nucleotide sequence analysis of the sei-containing fragment cloned from S. aureus FRI445 revealed that sei is flanked by nucleotide sequences that resemble enterotoxin genes. The downstream potential ORF contains a frameshift mutation, but no putative truncated gene product appears to be produced. Culture supernatants from an S. aureus strain possessing a plasmid containing a subclone covering this area do not stimulate T-cell proliferation. A gene product is not made for the enterotoxin-like gene upstream of sei. T-cell proliferative activity is observed for culture supernatants from S. aureus MJB1315, which contains this putative gene and sei, but not from MJB1318, which is isogenic except for a translation stop codon in sei. Furthermore, TCR Vβ stimulation profiles for culture supernatants from MJB1315 are identical to those from MJB1320, which only expresses SEI (from the β-lactamase promoter) (48).
SEI stimulated T-cell proliferation and elicited an emetic response in one of four animals tested in the primate emetic assay. The animals that did not vomit did develop other symptoms of illness such as diarrhea and prostration. This response is significant because we have never observed an emetic response in animals that received culture supernatants which did not contain toxin (23–26, 30). Although SEI has emetic capabilities, it does contain primary structural differences that may affect its emetic potency: it lacks a disulfide loop due to the absence of a second cysteine residue normally resident in the center of the molecule, and it contains a glycine- and lysine-rich insertion of eight amino acids near the C terminus.
There is evidence that a disulfide-bonded loop is not necessary for emetic activity. Spero and Morlock (63) showed that disruption of the disulfide loop by proteolytic cleavage of amino acid residues within the loop does not completely eliminate the emetic properties of an enterotoxin. Hovde et al. (28) demonstrated that SEC1 mutants containing either or both cysteine residues replaced by a serine residue maintained emetic activity. However, SEC1 variants with cysteine-to-alanine substitutions were not emetic (28). These authors hypothesized that the hydrogen-bonding capabilities of serine residues may contribute to a conformation that supports emetic activity. Bohach et al. (13) suggested that residues conserved in the SEs directly downstream of the disulfide loop may need to be in a proper orientation in order to cause emesis. Although SEI does not have a disulfide-bonded loop, it does have residues following the loop that are conserved in SEA to SEG and SEH.
SEI has primary structural differences which may make it more susceptible to stomach proteases such as trypsin, which cleaves proteins after lysine or arginine residues (SEI contains multiple lysine residues located in the region corresponding to the disulfide loop in other SEs and in the 8-amino-acid insertion of SEI [Fig. 7]). SEI does appear to be less stable than SEA, which may partly account for the greater quantities of toxin required to observe an emetic response. SEI is stable in monkey stomach fluid for >40 min at 37°C but begins to show some degradation at 1 h, whereas SEA is stable for >1 h (data not shown). An SEAV85G mutant used as an unstable enterotoxin control clearly degrades within 20 min (data not shown) (24). Taken together, these data suggest that SEI may be less emetically potent due to primary structural differences that may cause it to be less stable or that do not promote the best conformation (possibly near the loop area of the other enterotoxins) for maximal emetic activity. Although it may be weakly emetic, SEI is fully superantigenic.
SEG and SEI, in nanomolar concentrations, stimulate T-cell proliferation as do other characterized superantigens, presumably by binding MHC class II molecules. Comparisons of the deduced amino acid sequences of SEG and SEI with those of other SEs (Fig. 7) suggest how SEG and SEI may bind MHC class II molecules. SEG possesses residues analogous to F44 and L45 of SEB and to F47 and L48 of SEA (F45 and L46 in SEG), which bind to a site on the α chain of human HLA-DR1 molecules (24, 29, 34, 37). SEI possesses amino acid residues analogous to those in SEA which bind to the human HLA-DR1 β chain through coordination of a zinc atom among SEA H187, H225, D227 (corresponding to H169, H207, and D209 in SEI), and H81 of the HLA-DR1 β chain (19, 26, 29, 37, 61). SEI does not possess N-terminal phenylalanine or leucine residues corresponding to those in SEA and SEB that bind MHC class II molecules, but it has leucine and proline residues in these positions (L29 and P30). Therefore, SEG may bind in a fashion similar to SEB through F45 and L46 to one site on MHC class II molecules, and SEI may bind MHC class II molecules in a fashion similar to SEA through the coordination of a zinc atom. Mutational analysis of SEG and SEI in combination with MHC class II-binding assays is required to explore these possibilities.
The characterization of seg and sei provides additional evidence that the pyrogenic toxin family is quite large. The finding that sei is surrounded by an enterotoxin-like gene on one side and a partial enterotoxin-like gene on the other supports the hypotheses that there are more genes that have yet to be identified and that there may be regions for enterotoxin gene rearrangement in S. aureus. This premise is supported by the fact that the deduced amino acid sequence for the enterotoxin-like gene located 5′ of sei shares 55% deduced amino acid sequence identity with SEI, suggesting that gene rearrangement may have occurred. In fact, the C-terminal two-thirds of the protein is 75% similar to SEI while the N-terminal portion shares only 36% amino acid identity with SEI. We may find many new enterotoxin genes given the fact that many enterotoxin genes are associated with either mobile genetic elements or element-like sequences (reviewed in references 6 and 31). While the genetic locations of seg and sei are unknown, lytic phage could be induced from both FRI572 and FRI445 (strains from which seg and sei were cloned) (47). The high degrees of nucleic acid and deduced amino acid sequence identities among seg and the streptococcal toxin genes ssa and speA support the hypothesis that the toxins of S. aureus and S. pyogenes could have evolved from a common ancestral toxin gene or that exchange of genetic material between the two organisms may have occurred to create this related family of toxins (22, 67).
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant AI-255574. S. Munson was supported by National Institutes of Health Biotechnology Training grant 5T32GM08349.
We thank Steve Projan for his generosity in providing strains and for his protocol and technical advice on protoplast transformations. We also thank Russell Vertein, Doug Cowley, and Kirk Boehm for excellent assistance at the Wisconsin Regional Primate Research Center; Mark Hoffman, Theresa Harris, Cheryl Hertz, Amy Wong, and Merlin Bergdoll for assistance and helpful discussions through the course of this research; and Glenn Chambliss, Martin Voskuil, and M. J. Rosovitz for critical review of the manuscript.
This paper is dedicated to Marsha J. Betley, who spent her career studying the staphylococcal enterotoxins.
REFERENCES
- 1.Arvidson S, Janzon L, Löfdahl S. The role of the δ-lysin gene (hld) in the agr-dependent regulation of exoprotein synthesis in Staphylococcus aureus. In: Novick R, editor. Molecular biology of the staphylococci. New York, N.Y: VCH Publishers, Inc.; 1990. pp. 419–431. [Google Scholar]
- 2.Bayles K, Iandolo J. Genetic and molecular analysis of the gene encoding staphylococcal enterotoxin D. J Bacteriol. 1989;171:4799–4806. doi: 10.1128/jb.171.9.4799-4806.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergdoll M. Enterotoxins. In: Easmon C S F, Adlam C, editors. Staphylococci and staphylococcal infections. New York, N.Y: Academic Press Inc.; 1983. pp. 559–598. [Google Scholar]
- 4.Bergdoll M S. Staphylococcus aureus. In: Doyle M P, editor. Foodborne bacterial pathogens. New York, N.Y: Marcel Dekker; 1989. pp. 463–523. [Google Scholar]
- 5.Bergdoll, M. S. (University of Wisconsin-Madison). Personal communication.
- 6.Betley M J, Borst D W, Regassa L B. Staphylococcal enterotoxins, toxic shock syndrome toxin and streptococcal pyrogenic exotoxins: a comparative study of their molecular biology. Chem Immunol. 1992;55:1–35. [PubMed] [Google Scholar]
- 7.Betley M J, Löfdahl S, Kreiswirth B N, Bergdoll M S, Novick R P. Staphylococcal enterotoxin A gene is associated with a variable genetic element. Proc Natl Acad Sci USA. 1984;81:5179–5183. doi: 10.1073/pnas.81.16.5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Betley M J, Mekalanos J J. Nucleotide sequence of the type A staphylococcal enterotoxin gene. J Bacteriol. 1988;170:34–41. doi: 10.1128/jb.170.1.34-41.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bohach G A, Fast D J, Nelson R D, Schlievert P M. Staphylococcal and streptococcal pyrogenic toxins involved in toxic shock syndrome and related illnesses. Crit Rev Microbiol. 1990;17:251–272. doi: 10.3109/10408419009105728. [DOI] [PubMed] [Google Scholar]
- 10.Bohach G A, Kreiswirth B N, Novick R P, Schlievert P M. Analysis of toxic shock syndrome isolates producing staphylococcal enterotoxins B and C1 with use of Southern hybridization and immunologic assays. Rev Infect Dis. 1989;11:S75–S82. doi: 10.1093/clinids/11.supplement_1.s75. [DOI] [PubMed] [Google Scholar]
- 11.Bohach G A, Schlievert P M. Nucleotide sequence of the staphylococcal enterotoxin C1 gene and relatedness to other pyrogenic toxins. Mol Gen Genet. 1987;209:15–20. doi: 10.1007/BF00329830. [DOI] [PubMed] [Google Scholar]
- 12.Bohach G A, Schlievert P M. Conservation of the biologically active portions of staphylococcal enterotoxins C1 and C2. Infect Immun. 1989;57:2249–2252. doi: 10.1128/iai.57.7.2249-2252.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bohach G A, Stauffacher C V, Ohlendorf D H, Chi Y I, Vath G M, Schlievert P M. The staphylococcal and streptococcal pyrogenic toxin family. Adv Exp Med Biol. 1996;391:131–154. doi: 10.1007/978-1-4613-0361-9_8. [DOI] [PubMed] [Google Scholar]
- 14.Couch J L, Betley M J. Nucleotide sequence of the type C3 staphylococcal enterotoxin gene suggests that intergenic recombination causes antigenic variation. J Bacteriol. 1989;171:4507–4510. doi: 10.1128/jb.171.8.4507-4510.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Couch J L, Soltis M T, Betley M J. Cloning and nucleotide sequence of the type E staphylococcal enterotoxin gene. J Bacteriol. 1988;170:2954–2960. doi: 10.1128/jb.170.7.2954-2960.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crass B A, Bergdoll M S. Involvement of staphylococcal enterotoxins in nonmenstrual toxic shock syndrome. J Clin Microbiol. 1986;23:1138–1139. doi: 10.1128/jcm.23.6.1138-1139.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dellabonna P, Peccoud J, Kappler J, Marrack P, Benoist C, Mathis D. Superantigens interact with MHC class II molecules outside of the antigen groove. Cell. 1990;62:1115–1121. doi: 10.1016/0092-8674(90)90388-u. [DOI] [PubMed] [Google Scholar]
- 18.Fleischer B, Schrezenmeier H. T cell stimulation by staphylococcal enterotoxins. Clonally variable response and requirement for major histocompatibility complex class II molecules on accessory or target cells. J Exp Med. 1988;167:1697–1707. doi: 10.1084/jem.167.5.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fraser J D, Urban R G, Strominger J L, Robinson H. Zinc regulates the function of two superantigens. Proc Natl Acad Sci USA. 1992;89:5507–5511. doi: 10.1073/pnas.89.12.5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garbe P L, Arko R J, Reingold A L, Graves L M, Hayes P S, Hightower A W, Chandler F W, Broome C V. Staphylococcus aureus isolates from patients with nonmenstrual toxic shock syndrome. JAMA. 1985;253:2538–2542. [PubMed] [Google Scholar]
- 21.Gascoigne N R J, Ames K T. Direct binding of secreted T-cell receptor chain to superantigen associated with class II major histocompatibility complex protein. Proc Natl Acad Sci USA. 1991;88:613–616. doi: 10.1073/pnas.88.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goshorn S C, Schlievert P M. Nucleotide sequence of streptococcal pyrogenic exotoxin type C. Infect Immun. 1988;56:2518–2520. doi: 10.1128/iai.56.9.2518-2520.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harris T O, Betley M J. Biological activities of staphylococcal enterotoxin type A mutants with N-terminal substitutions. Infect Immun. 1995;63:2133–2140. doi: 10.1128/iai.63.6.2133-2140.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harris T O, Grossman D, Kappler J W, Marrack P, Rich R R, Betley M J. Lack of complete correlation between emetic and T-cell stimulatory activities of staphylococcal enterotoxins. Infect Immun. 1993;61:3175–3183. doi: 10.1128/iai.61.8.3175-3183.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harris T O, Hufnagle W O, Betley M J. Staphylococcal enterotoxin A internal deletion mutants examined for serological activity and induction of T-cell proliferation. Infect Immun. 1993;61:2059–2068. doi: 10.1128/iai.61.5.2059-2068.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoffman M, Tremaine M, Mansfield J, Betley M. Biochemical and mutational analysis of the histidine residues of staphylococcal enterotoxin A. Infect Immun. 1996;64:885–890. doi: 10.1128/iai.64.3.885-890.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hovde C J, Hackett S P, Bohach G A. Nucleotide sequence of the staphylococcal enterotoxin C3 gene: sequence comparison of all three type C staphylococcal enterotoxins. Mol Gen Genet. 1990;220:329–333. doi: 10.1007/BF00260504. [DOI] [PubMed] [Google Scholar]
- 28.Hovde C, Marr J, Hoffmann M, Sckett S, Chi Y, Crum K, Stevens D, Stauffacher C, Bohach G. Investigation of the role of the disulphide bond in the activity and structure of staphylococcal enterotoxin C1. Mol Microbiol. 1994;13:897–909. doi: 10.1111/j.1365-2958.1994.tb00481.x. [DOI] [PubMed] [Google Scholar]
- 29.Hudson K, Tiedemann R, Urban R, Lowe S, Strominger J, Fraser J. Staphylococcal enterotoxin A has two cooperative binding sites on major histocompatibility complex II. J Exp Med. 1995;182:711–720. doi: 10.1084/jem.182.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hufnagle W O, Tremaine M T, Betley M J. The carboxyl-terminal region of staphylococcal enterotoxin A is required for a fully active molecule. Infect Immun. 1991;59:2126–2134. doi: 10.1128/iai.59.6.2126-2134.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iandolo J J. Genetic analysis of extracellular toxins of Staphylococcus aureus. Annu Rev Microbiol. 1989;43:375–402. doi: 10.1146/annurev.mi.43.100189.002111. [DOI] [PubMed] [Google Scholar]
- 32.Iordanescu S. Recombinant plasmid obtained from two different, compatible staphylococcal plasmids. J Bacteriol. 1975;124:597–601. doi: 10.1128/jb.124.2.597-601.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones C L, Khan S A. Nucleotide sequence of the enterotoxin B gene from Staphylococcus aureus. J Bacteriol. 1986;166:29–33. doi: 10.1128/jb.166.1.29-33.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kappler J W, Herman A, Clements J, Marrack P. Mutations defining functional regions of the superantigen staphylococcal enterotoxin B. J Exp Med. 1992;175:387–396. doi: 10.1084/jem.175.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kokan N P, Bergdoll M S. Detection of low-enterotoxin-producing Staphylococcus aureus strains. Appl Environ Microbiol. 1987;53:2675–2676. doi: 10.1128/aem.53.11.2675-2676.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kornblum J, Kreiswirth B N, Projan S J, Ross H, Novick R P. Agr: a polycistronic locus regulating exoprotein synthesis in Staphylococcus aureus. In: Novick R P, editor. Molecular biology of the staphylococci. New York, N.Y: VCH Publishers, Inc.; 1990. pp. 373–402. [Google Scholar]
- 37.Kozono H, Parker D, White J, Marrack P, Kappler J. Multiple binding sites for bacterial superantigens on soluble class II MHC molecules. Immunity. 1995;3:187–196. doi: 10.1016/1074-7613(95)90088-8. [DOI] [PubMed] [Google Scholar]
- 38.Kreiswirth B N, Löfdahl S, Betley M J, O’Reilly M, Schlievert P M, Bergdoll M S, Novick R P. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983;305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 39.Lee V T P, Chang A H, Chow A W. Detection of staphylococcal enterotoxin B among toxic shock syndrome (TSS)- and non-TSS-associated Staphylococcus aureus isolates. J Infect Dis. 1992;166:911–915. doi: 10.1093/infdis/166.4.911. [DOI] [PubMed] [Google Scholar]
- 40.Lindberg M, Sjöström J-E, Johansson T. Transformation of chromosomal and plasmid characters in Staphylococcus aureus. J Bacteriol. 1972;109:844–847. doi: 10.1128/jb.109.2.844-847.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lurquin P F. Electrolyte gradient gels for DNA sequencing. BioTechniques. 1988;6:942–944. [PubMed] [Google Scholar]
- 42.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 43.Marr J C, Lyon J D, Roberson J R, Lupher M, Davis W C, Bohach G A. Characterization of novel type C staphylococcal enterotoxins: biological and evolutionary implications. Infect Immun. 1993;61:4254–4262. doi: 10.1128/iai.61.10.4254-4262.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marrack P, Kappler J. The staphylococcal enterotoxins and their relatives. Science. 1990;248:705–717. doi: 10.1126/science.2185544. [DOI] [PubMed] [Google Scholar]
- 45.Michaelis S, Beckwith J. Mechanism of incorporation of cell envelope proteins in Escherichia coli. Annu Rev Microbiol. 1982;36:436–465. doi: 10.1146/annurev.mi.36.100182.002251. [DOI] [PubMed] [Google Scholar]
- 46.Morrissey J H. Nonammoniacal silver staining. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1991. p. 10.6.4. [Google Scholar]
- 47.Munson, S. H. 1995. Unpublished data.
- 48.Munson, S. H., R. A. Welch, and J. M. Mansfield. Murine T cell receptor Vβ-specific stimulation by staphylococcal enterotoxin types G and I. Submitted for publication.
- 49.Murphy E, Novick R P. Physical mapping of Staphylococcus aureus penicillinase plasmid pI524: characterization of an invertible region. Mol Gen Genet. 1979;175:19–30. doi: 10.1007/BF00267851. [DOI] [PubMed] [Google Scholar]
- 50.Novick R P. Analysis by transduction of mutations affecting penicillinase formation in Staphylococcus aureus. J Gen Microbiol. 1963;33:121–136. doi: 10.1099/00221287-33-1-121. [DOI] [PubMed] [Google Scholar]
- 51.Novick R P. The staphylococcus as a molecular genetic system. In: Novick R P, editor. Molecular biology of the staphylococci. New York, N.Y: VCH Publishers, Inc.; 1990. pp. 1–37. [Google Scholar]
- 52.Novick R P. Genetic systems in staphylococci. Methods Enzymol. 1991;204:587–636. doi: 10.1016/0076-6879(91)04029-n. [DOI] [PubMed] [Google Scholar]
- 53.Novick R P, Brodsky R. Studies on plasmid replication. I. Plasmid incompatibility and establishment in Staphylococcus aureus. J Mol Biol. 1972;68:285–302. doi: 10.1016/0022-2836(72)90214-8. [DOI] [PubMed] [Google Scholar]
- 54.Novick R P, Murphy E, Gryczan T J, Baron E, Edelman I. Penicillinase plasmids of Staphylococcus aureus: restriction-deletion maps. Plasmid. 1979;2:109–129. doi: 10.1016/0147-619x(79)90010-6. [DOI] [PubMed] [Google Scholar]
- 55.Patel A H, Foster T J, Pattee P A. Physical and genetic mapping of the protein A gene in the chromosome of Staphylococcus aureus 8325-4. J Gen Microbiol. 1989;135:1799–1807. doi: 10.1099/00221287-135-7-1799. [DOI] [PubMed] [Google Scholar]
- 56.Projan, S. J. 1994. Personal communication.
- 57.Reda K B, Kapur V, Mollick J A, Lamphear J G, Musser J M, Rich R R. Molecular characterization and phylogenetic distribution of the streptococcal superantigen gene (ssa) from Steptococcus pyogenes. Infect Immun. 1994;62:1867–1874. doi: 10.1128/iai.62.5.1867-1874.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Regassa L B, Couch J L, Betley M J. Steady-state staphylococcal enterotoxin type C mRNA is affected by a product of the accessory gene regulator (agr) and by glucose. Infect Immun. 1991;59:955–962. doi: 10.1128/iai.59.3.955-962.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reiser R, Rovvins R, Khoe G, Bergdoll M. Purification and some physicochemical properties of toxic-shock toxin. Biochemistry. 1983;22:3907–3912. doi: 10.1021/bi00285a028. [DOI] [PubMed] [Google Scholar]
- 60.Ren K, Bannan J D, Panchli V, Cheung A L, Robbins J C, Fischetti V A, Zabriskie J B. Characterization and biological properties of a new staphylococcal exotoxin. J Exp Med. 1994;180:1675–1683. doi: 10.1084/jem.180.5.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schad E, Zaitseva I, Zaitev V, Dolhsten M, Kalland T, Schlievert P, Ohlendorf D, Svensson L. Crystal structure of the superantigen staphylococcal enterotoxin type A. EMBO J. 1995;14:3292–3301. doi: 10.1002/j.1460-2075.1995.tb07336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schroeder C J, Pattee P A. Transduction analysis of transposon Tn551 insertions in the trp-thy region of the Staphylococcus aureus chromosome. J Bacteriol. 1984;157:533–537. doi: 10.1128/jb.157.2.533-537.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spero L, Morlock B. Biological activities of the peptides of staphylococcal enterotoxin C formed by limited tryptic hydrolysis. J Biol Chem. 1978;253:8787–8791. [PubMed] [Google Scholar]
- 64.Su Y-C, Wong A C L. Detection of staphylococcal enterotoxin H by an enzyme-linked immunosorbent assay. J Food Prot. 1995;59:327–330. doi: 10.4315/0362-028x-59.3.327. [DOI] [PubMed] [Google Scholar]
- 65.Su Y-C, Wong A C L. Identification and purification of a new staphylococcal enterotoxin, H. Appl Environ Microbiol. 1995;61:1438–1443. doi: 10.1128/aem.61.4.1438-1443.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thruing R W J, Sanders J P M, Borst P. A freeze-squeeze method for recovering long DNA from agarose gels. Anal Biochem. 1975;66:213–220. doi: 10.1016/0003-2697(75)90739-3. [DOI] [PubMed] [Google Scholar]
- 67.Van Den Bussche R A, Lyon J D, Bohach G A. Molecular evolution of the staphylococcal and streptococcal pyrogenic toxin gene family. Mol Phylogenet Evol. 1993;2:281–292. doi: 10.1006/mpev.1993.1027. [DOI] [PubMed] [Google Scholar]
- 68.Voskuil M I, Chambliss G H. Rapid isolation and sequencing of purified plasmid DNA from Bacillus subtilis. Appl Environ Microbiol. 1993;59:1138–1142. doi: 10.1128/aem.59.4.1138-1142.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weeks C R, Ferretti J J. Nucleotide sequence of the type A streptococcal exotoxin (erythrogenic toxin) gene from Streptococcus pyogenes bacteriophage T12. Infect Immun. 1986;52:144–150. doi: 10.1128/iai.52.1.144-150.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.White J, Herman A, Pullen A M, Kubo R, Kappler J W, Marrack P. The Vβ-specific superantigen staphylococcal enterotoxin B: stimulation of mature T cells and clonal deletion in neonatal mice. Cell. 1989;56:27–35. doi: 10.1016/0092-8674(89)90980-x. [DOI] [PubMed] [Google Scholar]