Abstract
Background
Mounting evidence suggest that there are an association between psoriasis and ulcerative colitis (UC), although the common pathogeneses are not fully understood. Our study aimed to find potential crucial genes in psoriasis and UC through machine learning and integrated bioinformatics.
Methods
The overlapping differentially expressed genes (DEGs) of the datasets GSE13355 and GSE87466 were identified. Then the functional enrichment analysis was performed. The overlapping genes in LASSO, SVM‐RFE and key module in WGCNA were considered as potential crucial genes. The receiver operator characteristic (ROC) curve was used to estimate their diagnostic confidence. The CIBERSORT was conducted to evaluate immune cell infiltration. Finally, the datasets GSE30999 and GSE107499 were retrieved to validate.
Results
112 overlapping DEGs were identified in psoriasis and UC and the functional enrichment analysis revealed they were closely related to the inflammatory and immune response. Eight genes, including S100A9, PI3, KYNU, WNT5A, SERPINB3, CHI3L2, ARNTL2, and SLAMF7, were ultimately identified as potential crucial genes. ROC curves showed they all had high confidence in the test and validation datasets. CIBERSORT analysis indicated there was a correlation between infiltrating immune cells and potential crucial genes.
Conclusion
In our study, we focused on the comprehensive understanding of pathogeneses in psoriasis and UC. The identification of eight potential crucial genes may contribute to not only understanding the common mechanism, but also identifying occult UC in psoriasis patients, even serving as therapeutic targets in the future.
Keywords: bioinformatics, gene expression omnibus, machine learning, potential crucial genes, psoriasis, ulcerative colitis
Abbreviations
- ARNTL2
Aryl Hydrocarbon Receptor Nuclear Translocator‐Like 2
- AUC
area under the ROC curve
- BP
biological process
- CC
cellular component
- CD
Crohn's disease
- CHI3L2
Chitinase 3 Like 2
- CLOCK
circadian locomotor output cycles kaput
- DCs
dendritic cells
- DEGs
differentially expressed genes
- GEO
Gene Expression Omnibus
- GO
Gene Ontology
- IBD
inflammatory bowel disease
- IFN‐γ
Interferon‐γ
- IL‐17
Interleukin‐17
- KCs
keratinocytes
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- KYNU
Kynureninase
- LASSO
least absolute shrinkage and selection operator
- MF
molecular function
- NF
nuclear factor
- NLRP3
nucleotide‐binding and oligomerization domain‐like receptor protein 3
- NOD
nucleotide‐binding and oligomerization domain
- PI3
Peptidase Inhibitor 3
- RAGE
receptor for advanced glycation end products
- ROC
receiver operator characteristic
- ROR2
Receptor Tyrosine Kinase Like Orphan Receptor 2
- S100A9
S100 Calcium Binding Protein A9
- SERPINB3
Serpin Family B Member 3
- SLAMF7
Signaling Lymphocytic Activation Molecule Family 7
- SVM‐RFE
support vector machine‐recursive feature elimination
- TEAD4
TEA domain family member 4
- Th1
T Helper 1
- Tregs
regulatory T cells
- UC
ulcerative colitis
- WGCNA
Weighted Gene Co‐expression Network Analysis
- WNT5A
Wnt Family Member 5A
1. INTRODUCTION
Psoriasis is a chronic inflammatory skin disease with erythema, scaling, and pruritus, which is mediated by genetic, environmental, and immune factors. 1 The incidence of psoriasis has gradually increased in recent years. 2 Psoriasis is considered to be a systemic disease, and some diseases such as atherosclerosis, ulcerative colitis (UC), and diabetes mellitus are associated with psoriasis. 3 , 4 Some studies have shown that patients with psoriasis have a higher risk of suffering from UC. Farzad Alinaghi et al. conducted a meta‐analysis and reported the prevalence of UC in patients with psoriasis reached 0.5% [95%CI, 0.3−0.8%]. 5 And a case‐control study indicated psoriasis was associated with UC by using logistic multivariate model (OR, 1.64; 95% CI, 1.15–2.33). 6
As a chronic inflammatory bowel disease, UC is more common than Crohn's disease (CD), 7 with typical symptoms of abdominal pain, bloody purulent diarrhea, and fecal incontinence. 8 , 9 The onset of UC is mostly attributed to some factors such as genetic factors, diet, and lifestyle as well as mental health, which ultimately lead to the disorder of microbial ecosystem in colon, the interaction of immunocytes and the triggering of inflammatory response. 10 UC is also regarded as a system disease and some patients are affected by extraintestinal manifestations, such as the involvement of skin and joints. 11 A nationwide population‐based matched cohort study in Korea showed that UC patients had an increased risk of psoriasis, with an incidence of 217.68 per 100 000 person‐years. 12 , 13 The common pathogeneses of UC and psoriasis are lifestyle factors, genetic overlap, gut microbial antigens, and shared immune and inflammatory processes. 14 , 15 Many treatments for psoriasis are also suitable for UC, such as infliximab 16 and ustekinumab. 17 Some studies have shown the opinion that inflammatory bowel disease (IBD) and psoriasis are both related to the immune response, which alter the microbiota of gut and skin after environmental triggers for genetically susceptible individuals. 18 Accordingly, genetic susceptibility is regarded as a potential risk. A meta‐analysis of the genome‐wide association scanning showed that there were many IBD loci associated with psoriasis (14 out of 17, 14‐fold). 19
The diagnosis of UC is mainly based on typical clinical symptoms and intestinal endoscopy, but the discomfort caused by the examination makes it difficult for many patients to accept. 20 And the lesions in the early stages of psoriasis are easily confused with other skin diseases. Paying attention to psoriasis patients suffering from gastrointestinal discomfort and UC patients with skin lesions will contribute to the early diagnosis and treatment of comorbidities and improve the prognosis of patients. Therefore, it is of great clinical significance to identify the potential crucial genes shared in psoriasis and UC and explore their common pathogenesis.
In the present study, we obtained gene expression profiles of psoriasis and UC from the Gene Expression Omnibus (GEO) database, and then the potential crucial genes shared in both diseases were identified through weighted gene co‐expression network analysis (WGCNA) and machine learning. Finally, the results were verified in the validation datasets. The end purpose aims at uncovering the new insight into the comorbidity between psoriasis and UC as well as identifying candidate genes for the diagnosis and therapeutic targets.
2. MATERIALS AND METHODS
2.1. Data collection and processing
In order to obtain gene expression profiles and clinical information of diseases, we searched datasets by using the keywords “psoriasis” and “ulcerative colitis” in the GEO database (https://www.ncbi.nlm.nih.gov/geo/). Our filtering criteria include: (1) The organism should be Homo sapiens; (2) The tissue should be from psoriatic skin or colon lesional mucosal; (3) In the datasets, not only a disease group, but also a healthy control group is required; (4) The number of samples for WGCNA should be more than 15. Finally, the datasets GSE13355 and GSE87466 were used for analysis. GSE13355 was based on GPL570 [HG‐U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array, which collected 58 psoriatic lesions and 64 normal healthy controls. 21 , 22 , 23 GSE87466 was based on GPL13158 [HT_HG‐U133_Plus_PM] Affymetrix HT HG‐U133+ PM Array Plate, which collected 91 UC and 21 normal mucosal biopsy samples. 24 The datasets GSE30999 and GSE107499 were used for verifying our results. GSE30999 was based on GPL570 [HG‐U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array, which collected 170 skin biopsy samples from 85 moderate‐to‐severe psoriasis patients. 25 , 26 The dataset GSE107499 was based on GPL15207 [PrimeView] Affymetrix Human Gene Expression Array, which collected 75 UC patients and 44 healthy controls. Initially, data were downloaded using the “GEO query” R package 27 from GEO database. Subsequently, the data underwent normalization using the normalizeBetweenArrays function from the “Limma” R package, 28 aimed at eliminating systematic variations across different arrays, thus ensuring comparability in subsequent analyses. Additionally, annotation processing was conducted, where probes mapping to multiple genes were removed, and in cases where multiple probes map to the same gene, only the probe with the highest signal value was retained.
2.2. Differentially expressed genes screening
The “Limma” R package was used to extract differentially expressed genes (DEGs) in psoriasis and UC datasets. |Log2 fold change|≥1 & p‐value < 0.05 were regarded as the cut‐off standard. The overlapping genes were defined as upregulated or downregulated simultaneously in both datasets. PCA plot and Venn plot were used to visualize the clustering between sample groups and overlapping DEGs, respectively.
2.3. Functional enrichment analysis
Functional enrichment analysis was performed in order to understand the function of the overlapping genes better. The “Org. Hs. eg. Db” R package was applied for ID conversion of molecular lists. The “clusterProfiler” R package 29 was employed for Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. The results of the enrichment analysis were visualized using the “ggplot2” R package. GO enrichment analysis focuses on three categories: biological process (BP), cellular component (CC), and molecular function (MF). KEGG pathway enrichment analysis was used for investigating biological pathways. Functional enrichment analysis was performed by the cluster for investigating gene biological functions and pathways. p‐value < 0.05 was regarded as the cut‐off criterion.
2.4. Weighted gene co‐expression network analysis
The “WGCNA” R package was used to perform the WGCNA analysis. First, the expression matrix was obtained, and the top 5000 genes exhibiting the greatest variation were selected based on the standard deviation of their expression data and included in the subsequent analysis. 30 , 31 Then the fitted index (positive correlation) and the average connectivity (negative correlation) were plotted with the power scatter plot. In order to conduct a scale‐free network, β = 9 was regarded as the optimal soft‐power value. R 2 > 0.8 was used for conducting the topological relationship of a scale‐free network. Topological overlap matrix analysis was used to cluster genes and divide genes into different modules according to their similarity in expression. The minimum module size was set to 30 for creating clusters of highly interconnected genes. The link between module features and clinical features was carried out by Pearson correlation analysis. Finally, gene significance and module significance values larger than 0.8 and 0.8, respectively, were regarded as hub genes in modules. 32
2.5. Machine learning to identify potential crucial genes
Two machine learning algorithms including least absolute shrinkage and selection operator (LASSO) and support vector machine‐recursive feature elimination (SVM‐RFE) together with WGCNA were used to screen out potential crucial genes. LASSO regression model is to compress the variable regression coefficients in the regression model by generating a penalty function (L1 regularization) to prevent overfitting. The “glmnet” R package was used for LASSO. Then cross‐validation graphs were drawn and Nfolds were set as 10. SVM‐RFE is a machine‐learning method that made variable selection fast and automated. The “E1071,” “kernlab,” and “caret” R packages were employed for SVM‐RFE. The point with the smallest cross‐validation error was obtained and then the genes were output.
2.6. Evaluating diagnostic confidence
The receiver operator characteristic (ROC) curve is also called the sensitivity curve performed by the “pROC” package. The basic principle is calculating the corresponding sensitivity and specificity at different cut‐off points in the continuous variable. The area under the ROC curve (AUC) was calculated to show the diagnostic confidence of crucial genes on the test and the validation datasets. The value of AUC was between 0 and 1. The larger the AUC value, the higher the diagnostic confidence.
2.7. Immune infiltration analysis
The CIBERSORT (https:// cibersort.stanford.edu) is an analysis tool that applies the principle of linear support vector regression to deconvolve the expression matrix of human immune cell subtypes. This method provides a set of gene expression values in 22 types of immunocytes based on a pre‐processed reference dataset. 33 In the present study, 1000 permutations were set as the default signature matrix and the “ggplot2” R package was used to visualize. Heatmaps and violin plots were used to show the difference in immune cell infiltration between the disease group and the control group. Then, the correlation between infiltrating immunocytes and potential crucial genes was calculated by the Spearman correlation analysis. p‐values < 0.05 was considered statistically significant. If a gene was associated with three or more infiltrating immune cells, the results were visualized using the lollipop graphs, otherwise, the correlation scatter plot was preferred.
3. RESULTS
3.1. Identification of overlapping genes
Our research process was displayed in the workflow in Figure 1. As shown in the two‐dimensional PCA cluster diagrams, the clustering of samples from psoriasis patients and healthy controls were obvious, as was the case with samples from UC patients and healthy controls (Figure 2A,B). In GSE13355, 484 upregulated and 197 downregulated DEGs were identified (Figure 2C). And in GSE87466, 793 DEGs were up‐regulated and 419 were downregulated (Figure 2D). Then, the upregulated and downregulated DEGs were intersected separately, a total of 112 overlapping DEGs were obtained, including 100 co‐upregulated and 12 co‐downregulated DEGs (Figure 2E,F).
FIGURE 1.
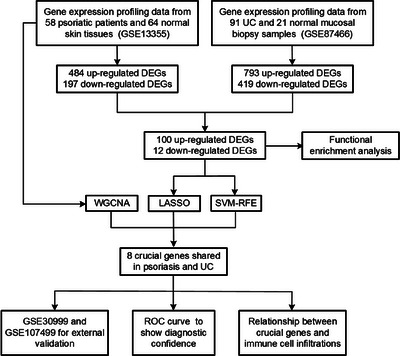
Workflow of process in this study.
FIGURE 2.
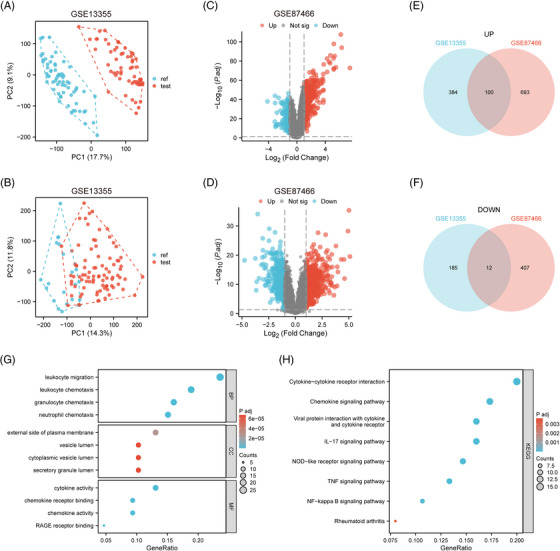
DEGs in GSE13355 and GSE87466. (A) The two‐dimensional PCA cluster diagram in GSE13355. (B) The two‐dimensional PCA cluster diagram in GSE87466. (C) The volcano plot of DEGs in GSE13355. (D) The volcano plot of DEGs in GSE87466. (E) The common up‐regulated DEGs in GSE13355 and GSE87466. (F) The common down‐regulated DEGs in GSE13355 and GSE87466. (G) The GO enrichment analysis of common DEGs shared in GSE13355 and GSE87466. (H) The KEGG pathway enrichment analysis of common DEGs shared in GSE13355 and GSE87466.
3.2. Functional enrichment analysis
GO enrichment analysis in BP revealed 112 overlapping DEGs enriched in positive regulation of leukocyte migration and chemotaxis, granulocyte, and neutrophil chemotaxis. CC was associated with the external side of plasma membrane, cytoplasmic vesicle lumen, and secretory granule lumen. MF was involved in cytokine activity, chemokine receptor binding, chemokine activity, and receptor for advanced glycation end products (RAGE) binding (Figure 2G; Table S1). KEGG pathway enrichment analysis showed that DEGs were enriched in cytokine–cytokine receptor interaction, chemokine signaling pathway, viral protein interaction with cytokine and cytokine receptor, Interleukin‐17 (IL‐17) signaling pathway, nucleotide‐binding and oligomerization domain (NOD) ‐like receptor signaling pathway, tumor necrosis factor (TNF) signaling pathway and nuclear factor (NF) ‐kappa B signaling pathway, which were primarily closely related to inflammatory and immune response. (Figure 2H, Table S2).
3.3. WGCNA construction and key module identification
The cluster dendrogram of genes showed that a total of seven modules were obtained in GSE13355 through WGCNA, with each color representing a module (Figure 3A). The red module was the most positively related with psoriasis (r = 0.97, p = 2e‐79), with 868 hub genes were screened for further analysis (Figure 3B,C).
FIGURE 3.
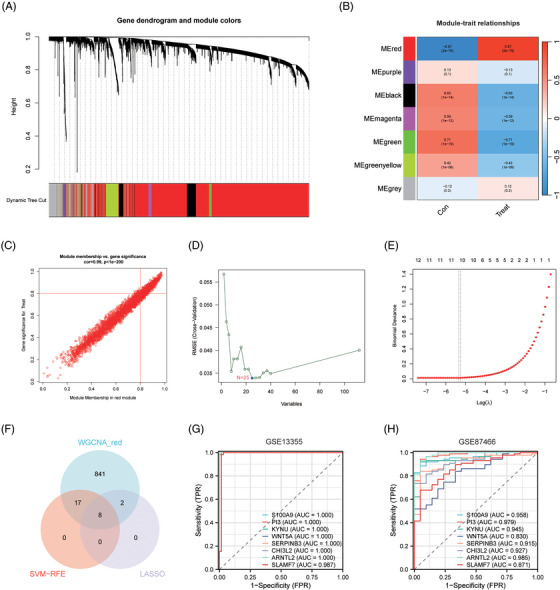
Identification of potential crucial genes by WGCNA, LASSO and SVM‐RFE. (A) The cluster dendrogram of genes in GSE13355. (B) Relationships between modules and clinical traits in GSE13355. (C) The relationship between module membership and gene significance in red module. (D) LASSO algorithm to determinate key genes in DEGs. (E) SVM‐RFE algorithm to screening key genes in DEGs. (F) Venn diagram to show the overlap genes obtained using WGCNA, LASSO, and SVM‐RFE. (G). ROC curve evaluating the diagnostic efficacy of eight crucial genes in GSE13355. (H) ROC curve evaluating the diagnostic efficacy of eight crucial genes in GSE87466.
3.4. Machine learning to screen potential crucial genes
Ten genes were filtered out by the LASSO regression and 25 genes were screened out by the SVM‐RFE algorithm (Figure 3D,E). To identify the potential crucial genes, we obtained the overlapping genes from LASSO, SVM‐RFE, and WGCNA (Figure 3F). And eight genes were identified, which were S100 Calcium Binding Protein A9 (S100A9), Peptidase Inhibitor 3 (PI3), Kynureninase (KYNU), Wnt Family Member 5A (WNT5A), Serpin Family B Member 3 (SERPINB3), Chitinase 3 Like 2 (CHI3L2), Basic Helix‐Loop‐Helix ARNT Like 2 (ARNTL2), and SLAM Family Member 7 (SLAMF7). In GSE13355, the AUC of crucial genes was all identified as 1.00. In GSE87466, the AUC of S100A9 (AUC = 0.958), PI3 (AUC = 0.979), KYNU (AUC = 0.945), SERPINB (AUC = 0.915), CHI3L2 (AUC = 0.927), ARNTL2 (AUC = 0.985), and SLAMF2 (AUC = 0.871) were comparably high (Figure 3G,H). Together, ROC curves showed that the eight genes all had high confidence in the test dataset, which demonstrated their diagnostic ability in discriminating psoriasis and UC patients from controls.
3.5. Correlation analysis of immune cell infiltration and crucial genes
The relative proportions of 22 immune cells in psoriasis samples, UC samples and their respective control samples were presented in the heatmaps (Figures 4A and 5A). The violin plots showed the distribution of immunocytes in psoriasis and UC samples compared with control samples (Figures 4B and 5B). In psoriasis samples, nine types of immune cells were remarkedly different compared with control samples. The proportion of CD4 memory activated T cells, follicular helper T cells, M1 macrophages, activated dendritic cells (DCs), activated mast cells, and neutrophils in psoriasis patients were significantly higher and they were positively correlated. And the proportion of regulatory T cells (Tregs), gamma delta T cells and activated NK cells were reduced and they were negatively correlated. The infiltration of CD4 memory activated T cells, follicular helper T cells, activated NK cells, M0 macrophages, M1 macrophages resting, mast cells, and neutrophils were significantly increased in the UC group. And plasma cells, Tregs, M2 macrophages, activated DCs, and resting mast cells showed the opposite results.
FIGURE 4.
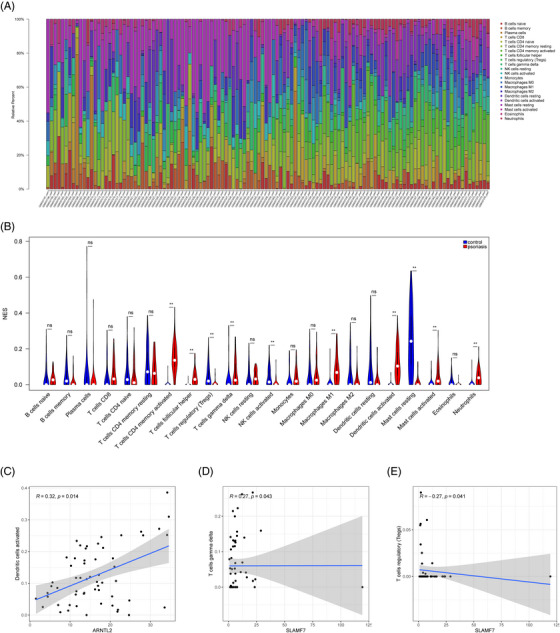
Immune infiltration analysis of crucial genes in psoriasis. (A) The heatmap of immune cell infiltration between psoriasis and controls. (B) The violin plot to show the different distribution of each immune cell between psoriasis and control in GSE13355. (C) Correlation between ARNTL2 and infiltrating immune cells. (D and E) Correlation between SLAMF7 and infiltrating immune cells. ** p < 0.001.
FIGURE 5.
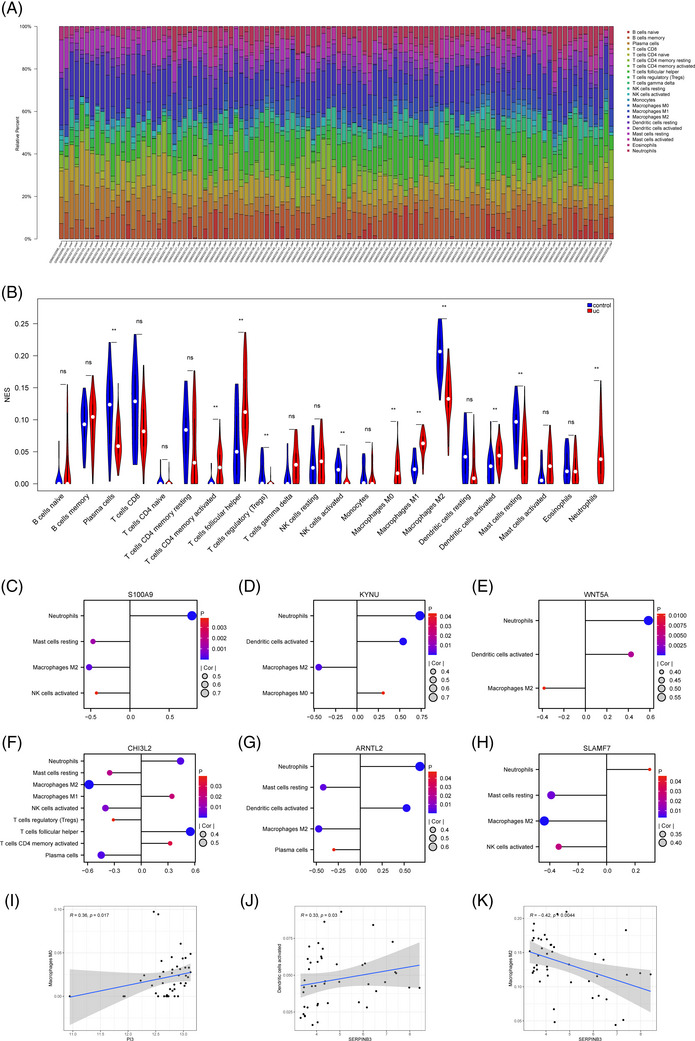
Immune infiltration analysis of crucial genes in UC. (A) The heatmap of immune cell infiltration between UC and controls. (B) The violin plot to show the different distribution of each immune cell between UC and control in GSE87466. (C) Correlation between S100A9 and infiltrating immune cells. (D) Correlation between KYNU and infiltrating immune cells. (E) Correlation between WNT5A and infiltrating immune cells. (F) Correlation between SLMAF7 and infiltrating immune cells. (G) Correlation between ARNTL2 and infiltrating immune cells. (H) Correlation between CHI3L2 and infiltrating immune cells. (I) Correlation between PI3 and infiltrating immune cells. (J and K) Correlation between SERPINB3 and infiltrating immune cells. ** p < 0.001.
In psoriasis samples, ARNTL2 was positively correlated with activated DCs (R = 0.32, p = 0.014). SLAMF7 was positively correlated with gamma delta T cells (R = 0.27, p = 0.043) and negatively with Tregs (R = −0.27, p = 0.041) (Figure 4C–E). And all potential crucial genes showed a close correlation with immune cells in UC samples, particularly CHI3L2 and ARNTL2 (Figure 5C‐K). CHI3L2 was significantly associated with nine types of immune cells, such as neutrophils, M2 macrophages, follicular helper, etc. ARNTL2 had a positive correlation with neutrophils and activated DCs, while had a opposite correlation with resting mast cells, M2 macrophages and plasma cells.
3.6. Validation of crucial genes in the validation datasets
To increase the confidence of our study, the datasets GSE30999 and GSE107499 were retrieved to validate their expression and diagnostic ability in psoriasis and UC. In GSE30999 and GSE107499, the expressions of all crucial genes were significantly upregulated in psoriasis and UC patients (Figure 6A,B). ROC curves showed their high confidence in the validation datasets (Figure 6C,D).
FIGURE 6.
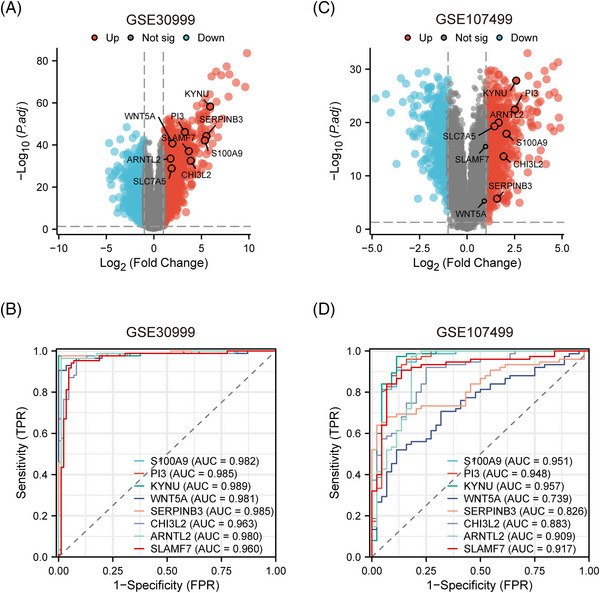
Validation of potential crucial genes. (A) The volcano plot of DEGs in GSE3099. (B) The volcano plot of DEGs in GSE107499. (C) ROC curve evaluating the diagnostic efficacy of eight crucial genes in GSE30999. (D) ROC curve evaluating the diagnostic efficacy of eight crucial genes in GSE107499.
4. DISCUSSION
Currently, bioinformatics has been widely utilized in disease research, particularly for exploring biomarkers and molecular mechanisms. For instance, a study employing a hybrid classifier based on genetic algorithms and SVM has significantly enhanced the predictive accuracy in identifying gene expression signatures for classifying psoriasis. 34 The application of this method highlights the crucial role of bioinformatics in deciphering the complex genetic interplay underlying psoriasis, which offers new insights into the genetic underpinnings of psoriasis. Shokrollah et al. have shed light on potential novel biomarkers for CD patients in the tumorigenesis of colorectal cancer. 35 This is relevant as it suggests a deeper link between IBD and other systemic diseases, offering new insights into their shared mechanisms. As a systemic disease, psoriasis is also related with many diseases and numerous studies have already established the relationship between psoriasis and other diseases like depression, nonalcoholic steatohepatitis, and IBD. 36 , 37 , 38 , 39 , 40 , 41 Our previous work focused on the comorbidity mechanism between psoriasis and nonalcoholic steatohepatitis, and identified several key genes as potential therapeutic targets using bioinformatics methods. 42 This discovery not only aids in understanding the comorbidity mechanisms of these two diseases but also emphasizes the broader applicability of bioinformatics in revealing complex disease relationships. Ding et al. suggested that FOS may play a critical role in both IBD and psoriasis, contributing to a growing body of evidence supporting the genetic commonality between these diseases. 43 Moreover, a two‐sample bidirectional Mendelian randomization study posited a possible causal relationship between IBD and psoriasis. It showed that genetic predisposition to IBD might increase the risk of developing psoriasis, further solidifying their genetic link. 44
Incremental evidence has demonstrated a potential link between psoriasis and UC, yet the mechanism remains unknown. It is critical to identify the potential crucial genes between psoriasis and UC as well as explore the relationship between them to help early diagnosis and treatment. In our study, we got gene expression profiles from GEO database, and 112 DEGs were screened out. The enriched pathways are primarily related to inflammatory and immune response. Subsequently, eight crucial genes shared in the psoriasis and UC datasets were identified via WGCNA, LASSO, and SVM‐RFE, including S100A9, PI3, KYNU, WNT5A, SERPINB3, CHI3L2, ARNTL2, and SLAMF7, which showed high confidence in both training and validation datasets. Therefore, these genes may serve as biomarkers for detecting covert UC in psoriasis patients
GO and KEGG analyses showed the common DEGs were highly related with immune response, indicating immune response played a pivotal role in the initiation of psoriasis and UC. Tang et al. reported that cytokine‐cytokine receptor interaction, viral protein interaction with cytokine and cytokine receptor, IL‐17 signaling pathway and NOD‐like receptor signaling pathway were significantly enriched in psoriasis, 45 which is consistent with our results. Among them, the IL‐17 signaling pathway is considered to be one of the pathways most closely related to the pathogenesis of psoriasis. IL‐23/IL‐17 axis plays an essential role in plaque psoriasis, but also contributes to the progression of UC, and has been regarded as a therapeutic target. 1 , 46 , 47 Transcription factor NFκB is highly expressed in psoriasis and is related to the behavior of keratinocytes (KCs) and immunocytes, indicating the important role of NFκB signaling in psoriasis. 47 NFκB, as the “master switch” for pro‐inflammatory mediators, has also been reported in a variety of diseases including UC. 48 NOD2, a key molecule of the NOD‐like receptor signaling pathway, was observed that upregulated in psoriasis by assaying the transcriptomes of the normal, lesional and non‐lesional psoriatic epidermis. 49 nucleotide‐binding and oligomerization domain‐like receptor protein 3 (NLRP3) is a NOD‐like receptor family, and its activation was significantly related with long‐standing UC. 50
S100A9, PI3, KYNU, and WNT5A have been reported to have a relation with both diseases. S100A9, an intracellular calcium‐binding protein, is mainly produced from KCs and innate immunocytes. 51 S100A9 forming calprotectin heterodimers together with S100A8 bound to receptors on the cell surface such as RAGE, further triggering signaling pathways associated with inflammatory response, cell cycle progression and cell proliferation. 52 It has been reported that S100A9 played a key role in both psoriasis and UC. 53 , 54 Upregulated S100A9 in psoriasis patients could drive chronic inflammation by inducing type 3 immunity. 55 And in the intestinal mucosa of active UC patients, neutrophils could synthesize and secrete S100A9 protein, mediating the inflammatory response. 56 , 57 PI3, as an epithelial host‐defense protein, was significantly elevated in KCs from skin inflammatory lesions, especially psoriasis. 58 , 59 PI3 was proved as Th17‐associate markers, which can accurately identify psoriasis from hidradenitis suppurativa in a cross‐sectional study. 60 KYNU, a tryptophan metabolism enzyme downstream of indoleamine 2,3‐dioxygenase, was highly expressed in psoriatic lesions. 61 It has been demonstrated that KYNU might contribute to the onset of psoriasis by regulating the inflammatory microenvironment in our previous study 62 Wnorowski et al. conducted a meta‐analysis using publicly available transcriptomic datasets as well as found that KYNU was also upregulated in UC patients, and might be recovered after infliximab treatment. 63 WNT5A, as an evolutionary WNT member, can activate both the canonical Wnt/β‐catenin and noncanonical Wnt/Ca2+ pathways. 64 , 65 It was found that WNT5A was highly expressed in psoriasis lesions, yet the specific mechanisms were still unclear. 66 Sato et al. demonstrated that WNT5A‐receptor tyrosine kinase‐like orphan receptor (ROR2) axis can enhance interferon‐γ (IFN‐γ) signaling and promote IL‐12 expression in DCs, inducing T helper 1 (Th1) differentiation in colitiss. 67 However, it is still inconclusive whether WNT5A is pro‐inflammatory or anti‐inflammatory in UC. 68
SERPINB3 can significantly inhibit immunocytes apoptosis and induce abnormal proliferation of autoreactive cells, resulting in autoimmune disorder. 69 In psoriasis, transcriptional factor TEA domain family member 4 (TEAD4) regulated the expression of SERPINB3/4 and promoted the secretion of chemokines, forming crosstalk between KCs and T cells. 70 Among these genes, CHI3L2 and ARNTL2 have not been reported to be associated with psoriasis. As a member of chitinase‐like proteins, CHI3L2 has two kinds of physiological activities, inducing autoimmune response and taking part in tissue remodeling. 71 , 72 TIMER analysis revealed that CHI3L2 was highly associated with tumor immune infiltrating cells in gliomas, such as M1 and M2 macrophages. 71 In order to explore the function of immune cell infiltration, CIBERSORT was conducted for a comprehensive evaluation. CHI3L2 showed a close link with immune infiltrating cells in UC patients. ARNTL2 together with circadian locomotor output cycles kaput (CLOCK) can affect the persistence and period of circadian rhythms. 73 ARNTL2 has been extensively studied in cancers and proved to be associated with poor survival and immune cell infiltration, such as triple‐negative breast cancer, 74 clear cell renal cell carcinoma, 75 and pancreatic cancer. 76 Interestingly, ARNTL2 was identified as a potential biomarker which is related with different clinical manifestations between Uyghur and Han population UC patients. 77 In our study, ARNTL2 was significantly correlated with activated DCs, suggesting that it may participate in the development of UC and psoriasis through mediating immune response, which requires further experiments to verify. SLAMF7, as a self‐ligand immune cell receptor, modulated B cells and adaptive immune response, thus regulating the susceptibility of autoimmunity in multiple sclerosis. 78 SLAMF7 was significantly upregulated in macrophages exposed to IFN‐γ, and the involvement of SLAMF7 drove the inflammatory cascade. 79 In multiple myeloma, SLAMF7 was a marker of inhibitory CD8+ Tregs, and anti‐SLAMF7 antibodies can be used to enhance anti‐tumor immune response. 80 However, there were currently no reports on the functions of SLAMF7 in psoriasis and UC. In the present study, SLAMF7 was positively correlated with gamma delta T cells and negatively with Tregs in psoriasis, and SLAMF7 were significantly related with M2 macrophages, resting mast cells and activated NK cells in UC, which indicated the potential role of SLAMF7 in psoriasis and UC.
In summary, eight crucial genes in psoriasis and UC were ultimately identified and validated using bioinformatics, which may play a significant role in the onset and progression of the two diseases and indicate a common underlying mechanism. These genes are not only promising for further research but could also serve as diagnostic markers, especially for patients experiencing both gastrointestinal and dermatological symptoms. With further experimental validation, these genes have the potential to become new therapeutic targets, offering more effective treatment strategies for patients with psoriasis and UC. Although there exist some limitations in our studies, these results are extremely helpful to understand the relationship between psoriasis and UC as well as identify covert UC in psoriasis patients.
5. CONCLUSION
In our study, we concentrated on the comprehensive understanding of pathogeneses in psoriasis and UC, which may participate in their interaction. Eight genes, including S100A9, PI3, KYNU, WNT5A, SERPINB3, CHI3L2, ARNTL2, and SLAMF7, were ultimately identified as potential crucial genes by machine learning and integrated bioinformatics. These key genes may not only contribute to understanding the common mechanism between psoriasis and UC, but also recognizing occult UC in psoriasis patients, even serving as therapeutic targets in the future.
CONFLICT OF INTEREST STATEMENT
We declare that there are no conflicts of interest in this study.
Supporting information
Supporting information
Supporting information
ACKNOWLEDGMENTS
We thank all the volunteers participated in our study and the researchers uploaded their date onto GEO databases. This work was supported by National Natural Science Foundation of China (grant number 82273541) and Fund of Shaanxi Province (grant number 2021ZDLSF03‐01).
Zhang J, Feng S, Chen M, Zhang W, Zhang X, Wang S, et al. Identification of potential crucial genes shared in psoriasis and ulcerative colitis by machine learning and integrated bioinformatics. Skin Res Technol. 2024;30:e13574. 10.1111/srt.13574
Jing Zhang and Shuo Feng contributed equally to this study.
Contributor Information
Yan Zheng, Email: zenyan66@126.com.
Guorong Wang, Email: 13227701303@qq.com.
DATA AVAILABILITY STATEMENT
In our study, we download these datasets (GSE13355, GSE87466, GSE30999, and GSE107499) in the Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/).
REFERENCES
- 1. Griffiths CEM, Armstrong AW, Gudjonsson JE, Barker J. Psoriasis. Lancet. 2021;397(10281):1301‐1315. [DOI] [PubMed] [Google Scholar]
- 2. Parisi R, Iskandar IYK, Kontopantelis E, et al. National, regional, and worldwide epidemiology of psoriasis: systematic analysis and modelling study. BMJ. 2020;369:m1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Takeshita J, Grewal S, Langan SM, et al. Psoriasis and comorbid diseases: implications for management. J Am Acad Dermatol. 2017;76(3):393‐403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu C, Chen H, Liu Y, et al. Immunity: psoriasis comorbid with atherosclerosis. Front Immunol. 2022;13:1070750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Alinaghi F, Tekin HG, Burisch J, Wu JJ, Thyssen JP, Egeberg A. Global prevalence and bidirectional association between psoriasis and inflammatory bowel disease‐A systematic review and meta‐analysis. J Crohns Colitis. 2020;14(3):351‐360. [DOI] [PubMed] [Google Scholar]
- 6. Cohen AD, Dreiher J, Birkenfeld S. Psoriasis associated with ulcerative colitis and Crohn's disease. J Eur Acad Dermatol Venereol. 2009;23(5):561‐565. [DOI] [PubMed] [Google Scholar]
- 7. Ordas I, Eckmann L, Talamini M, Baumgart DC, Sandborn WJ. Ulcerative colitis Lancet. 2012;380(9853):1606‐1619. [DOI] [PubMed] [Google Scholar]
- 8. Feuerstein JD, Moss AC, Farraye FA. Ulcerative Colitis. Mayo Clin Proc. 2019;94(7):1357‐1373. [DOI] [PubMed] [Google Scholar]
- 9. Segal JP, LeBlanc JF, Hart AL. Ulcerative colitis: an update. Clin Med (Lond). 2021;21(2):135‐139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Eisenstein M. Ulcerative colitis: towards remission. Nature. 2018;563(7730):S33. [DOI] [PubMed] [Google Scholar]
- 11. Rogler G, Singh A, Kavanaugh A, Rubin DT. Extraintestinal manifestations of inflammatory bowel disease: current concepts, treatment, and implications for disease management. Gastroenterology. 2021;161(4):1118‐1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jo UH, Lee JY, Lee H, et al. Various skin manifestations related to inflammatory bowel disease: a nationwide cross‐sectional study on the Korean population. J Dermatol. 2021;48(4):431‐438. [DOI] [PubMed] [Google Scholar]
- 13. Moon JM, Lee JY, Koh SJ, et al. Incidence of psoriasis in patients with inflammatory bowel disease: a nationwide population‐based matched cohort study. Dermatology. 2021;237(3):330‐337. [DOI] [PubMed] [Google Scholar]
- 14. Hedin CRH, Sonkoly E, Eberhardson M, Stahle M. Inflammatory bowel disease and psoriasis: modernizing the multidisciplinary approach. J Intern Med. 2021;290(2):257‐278. [DOI] [PubMed] [Google Scholar]
- 15. Bezzio C, Della Corte C, Vernero M, Di Luna I, Manes G, Saibeni S. Inflammatory bowel disease and immune‐mediated inflammatory diseases: looking at the less frequent associations. Therap Adv Gastroenterol. 2022;15:175628482211153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Syversen SW, Goll GL, Jorgensen KK, et al. Effect of therapeutic drug monitoring vs standard therapy during infliximab induction on disease remission in patients with chronic immune‐mediated inflammatory diseases: a randomized clinical trial. JAMA. 2021;325(17):1744‐1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fiorino G, Allocca M, Correale C, et al. Positioning ustekinumab in moderate‐to‐severe ulcerative colitis: new kid on the block. Expert Opin Biol Ther. 2020;20(4):421‐427. [DOI] [PubMed] [Google Scholar]
- 18. De Francesco MA, Caruso A. The gut microbiome in psoriasis and Crohn's disease: is its perturbation a common denominator for their pathogenesis? Vaccines (Basel). 2022;10(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jostins L, Ripke S, Weersma RK, et al. Host‐microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491(7422):119‐124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li Y, Tang M, Zhang FJ, et al. Screening of ulcerative colitis biomarkers and potential pathways based on weighted gene co‐expression network, machine learning and ceRNA hypothesis. Hereditas. 2022;159(1):42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nair RP, Duffin KC, Helms C, et al. Genome‐wide scan reveals association of psoriasis with IL‐23 and NF‐kappaB pathways. Nat Genet. 2009;41(2):199‐204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Swindell WR, Johnston A, Carbajal S, et al. Genome‐wide expression profiling of five mouse models identifies similarities and differences with human psoriasis. Plos One. 2011;6(4):e18266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ding J, Gudjonsson JE, Liang L, et al. Gene expression in skin and lymphoblastoid cells: refined statistical method reveals extensive overlap in cis‐eQTL signals. Am J Hum Genet. 2010;87(6):779‐789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li K, Strauss R, Ouahed J, et al. Molecular comparison of adult and pediatric ulcerative colitis indicates broad similarity of molecular pathways in disease tissue. J Pediatr Gastroenterol Nutr. 2018;67(1):45‐52. [DOI] [PubMed] [Google Scholar]
- 25. Suarez‐Farinas M, Li K, Fuentes‐Duculan J, Hayden K, Brodmerkel C, Krueger JG. Expanding the psoriasis disease profile: interrogation of the skin and serum of patients with moderate‐to‐severe psoriasis. J Invest Dermatol. 2012;132(11):2552‐2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Correa da Rosa J, Kim J, Tian S, Tomalin LE, Krueger JG, Suarez‐Farinas M. Shrinking the psoriasis assessment gap: early gene‐expression profiling accurately predicts response to long‐term treatment. J Invest Dermatol. 2017;137(2):305‐312. [DOI] [PubMed] [Google Scholar]
- 27. Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23(14):1846‐1847. [DOI] [PubMed] [Google Scholar]
- 28. Smyth GK. Limma: linear models for microarray data. In: Gentalman R, Carey VJ, Huber W, Irizarry RA, Dudoit S, eds. Bioinformatics and Computational Biology Solution Using R and Bioconductor. 2005:397‐420. [Google Scholar]
- 29. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284‐287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang T, Wong G. Gene expression data analysis using Hellinger correlation in weighted gene co‐expression networks (WGCNA). Comput Struct Biotechnol J. 2022;20:3851‐3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang X, Feng H, Li Z, et al. Application of weighted gene co‐expression network analysis to identify key modules and hub genes in oral squamous cell carcinoma tumorigenesis. Onco Targets Ther. 2018;11:6001‐6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang D, Hao W, Niu Q, Xu D, Duan X. Identification of the co‐differentially expressed hub genes involved in the endogenous protective mechanism against ventilator‐induced diaphragm dysfunction. Skelet Muscle. 2022;12(1):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453‐457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tapak L, Afshar S, Afrasiabi M, Ghasemi MK, Alirezaei P. Application of genetic algorithm‐based support vector machine in identification of gene expression signatures for psoriasis classification: a hybrid model. Biomed Res Int. 2021;2021:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shokrollah N, Samadi P, Jalali A, Dalirfardouei R, Afshar S, Pourjafar M. A systems biology approach to identify novel biomarkers in progression from Crohn's disease to colorectal cancer. Asian Pac J Cancer Prev. 2023;24(6):1993‐2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhou Y, Han L, Wang Z, et al. Bioinformatic analysis of the potential common pathogenic mechanisms for psoriasis and metabolic syndrome. Inflammation. 2023;46(4):1381‐1395. [DOI] [PubMed] [Google Scholar]
- 37. Xia X, Yu H, Li Y, Liang Y, Li G, Huang F. Transcriptome analysis identifies biomarkers for the diagnosis and management of psoriasis complicated with depression. Clin Cosmet Investig Dermatol. 2023;16:1287‐1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lapsley CR, Irwin R, McLafferty M, et al. Methylome profiling of young adults with depression supports a link with immune response and psoriasis. Clin Epigenetics. 2020;12(1):85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gau SY, Huang KH, Lee CH, Kuan YH, Tsai TH, Lee CY. Bidirectional association between psoriasis and nonalcoholic fatty liver disease: real‐world evidence from two longitudinal cohort studies. Front Immunol. 2022;13:840106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Koo J, Marangell LB, Nakamura M, et al. Depression and suicidality in psoriasis: review of the literature including the cytokine theory of depression. J Eur Acad Dermatol Venereol. 2017;31(12):1999‐2009. [DOI] [PubMed] [Google Scholar]
- 41. Yousaf A, Raiker R, Davis SM, Gayam S, Zinn Z. Association between psoriasis, psoriatic arthritis and gastrointestinal disease: an exploratory nationwide inpatient sample analysis. Wien Klin Wochenschr. 2021;133(11‐12):586‐593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xian N, Bai R, Guo J, et al. Bioinformatics analysis to reveal the potential comorbidity mechanism in psoriasis and nonalcoholic steatohepatitis. Skin Res Technol. 2023;29(9):e13457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ding RL, Zheng Y, Bu J. Exploration of the biomarkers of comorbidity of psoriasis with inflammatory bowel disease and their association with immune infiltration. Skin Res Technol. 2023;29(12):e13536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li Y, Guo J, Cao Z, Wu J. Causal association between inflammatory bowel disease and psoriasis: a two‐sample bidirectional mendelian randomization study. Front Immunol. 2022;13:916645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tang S, Jiang W, Xu P, et al. Integrated bioinformatic analysis of key biomarkers and signalling pathways in psoriasis. Scott Med J. 2022;67(1):7‐17. [DOI] [PubMed] [Google Scholar]
- 46. Rendon A, Schakel K. Psoriasis pathogenesis and treatment. Int J Mol Sci. 2019;20(6):1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Goldminz AM, Au SC, Kim N, Gottlieb AB, Lizzul PF. NF‐kappaB: an essential transcription factor in psoriasis. J Dermatol Sci. 2013;69(2):89‐94. [DOI] [PubMed] [Google Scholar]
- 48. Ma J, Yin G, Lu Z, et al. Casticin prevents DSS induced ulcerative colitis in mice through inhibitions of NF‐kappaB pathway and ROS signaling. Phytother Res. 2018;32(9):1770‐1783. [DOI] [PubMed] [Google Scholar]
- 49. Tervaniemi MH, Katayama S, Skoog T, et al. NOD‐like receptor signaling and inflammasome‐related pathways are highlighted in psoriatic epidermis. Sci Rep. 2016;6:22745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lazaridis LD, Pistiki A, Giamarellos‐Bourboulis EJ, et al. Activation of NLRP3 inflammasome in inflammatory bowel disease: differences between Crohn's disease and ulcerative colitis. Dig Dis Sci. 2017;62(9):2348‐2356. [DOI] [PubMed] [Google Scholar]
- 51. Mellor LF, Gago‐Lopez N, Bakiri L, et al. Keratinocyte‐derived S100A9 modulates neutrophil infiltration and affects psoriasis‐like skin and joint disease. Ann Rheum Dis. 2022;81(10):1400‐1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shabani F, Farasat A, Mahdavi M, Gheibi N. Calprotectin (S100A8/S100A9): a key protein between inflammation and cancer. Inflamm Res. 2018;67(10):801‐812. [DOI] [PubMed] [Google Scholar]
- 53. Xi L, Garcet S, Ye Z, et al. A shared tissue transcriptome signature and pathways in psoriasis and ulcerative colitis. Sci Rep. 2022;12(1):19740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pekow J, Dougherty U, Huang Y, et al. Gene signature distinguishes patients with chronic ulcerative colitis harboring remote neoplastic lesions. Inflamm Bowel Dis. 2013;19(3):461‐470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Silva de Melo BM, Veras FP, Zwicky P, et al. S100A9 drives the chronification of psoriasiform inflammation by inducing IL‐23/type 3 immunity. J Invest Dermatol. 2023;143(9):1678‐1688. e1678. [DOI] [PubMed] [Google Scholar]
- 56. Zhang Y, Zha Z, Shen W, et al. Anemoside B4 ameliorates TNBS‐induced colitis through S100A9/MAPK/NF‐kappaB signaling pathway. Chin Med. 2021;16(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sinha P, Okoro C, Foell D, Freeze HH, Ostrand‐Rosenberg S, Srikrishna G. Proinflammatory S100 proteins regulate the accumulation of myeloid‐derived suppressor cells. J Immunol. 2008;181(7):4666‐4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pol A, Pfundt R, Zeeuwen P, Molhuizen H, Schalkwijk J. Transcriptional regulation of the elafin gene in human keratinocytes. J Invest Dermatol. 2003;120(2):301‐307. [DOI] [PubMed] [Google Scholar]
- 59. Xu M, Deng J, Xu K, et al. In‐depth serum proteomics reveals biomarkers of psoriasis severity and response to traditional Chinese medicine. Theranostics. 2019;9(9):2475‐2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Navrazhina K, Renert‐Yuval Y, Frew JW, et al. Large‐scale serum analysis identifies unique systemic biomarkers in psoriasis and hidradenitis suppurativa. Br J Dermatol. 2022;186(4):684‐693. [DOI] [PubMed] [Google Scholar]
- 61. Harden JL, Lewis SM, Lish SR, et al. The tryptophan metabolism enzyme L‐kynureninase is a novel inflammatory factor in psoriasis and other inflammatory diseases. J Allergy Clin Immunol. 2016;137(6):1830‐1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wang M, Wang Y, Zhang M, et al. Kynureninase contributes to the pathogenesis of psoriasis through pro‐inflammatory effect. J Cell Physiol. 2022;237(1):1044‐1056. [DOI] [PubMed] [Google Scholar]
- 63. Wnorowski A, Wnorowska S, Kurzepa J, Parada‐Turska J. Alterations in kynurenine and NAD(+) salvage pathways during the successful treatment of inflammatory bowel disease suggest HCAR3 and NNMT as potential drug targets. Int J Mol Sci. 2021;22(24):13497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhou W, Mei J, Gu D, et al. Wnt5a: a promising therapeutic target in ovarian cancer. Pathol Res Pract. 2021;219:153348. [DOI] [PubMed] [Google Scholar]
- 65. Bueno MLP, Saad STO, Roversi FM. WNT5A in tumor development and progression: a comprehensive review. Biomed Pharmacother. 2022;155:113599. [DOI] [PubMed] [Google Scholar]
- 66. Tian F, Mauro TM, Li Z. The pathological role of Wnt5a in psoriasis and psoriatic arthritis. J Cell Mol Med. 2019;23(9):5876‐5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sato A, Kayama H, Shojima K, et al. The Wnt5a‐Ror2 axis promotes the signaling circuit between interleukin‐12 and interferon‐gamma in colitis. Sci Rep. 2015;5:10536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhang M, Xie Z, Zhou G, Wang Y, Zhang S. Conflicting effects of Wnt‐5a in ulcerative colitis: you Wnt some, you lose some. Dig Dis Sci. 2022;67(10):4599‐4601. [DOI] [PubMed] [Google Scholar]
- 69. Vidalino L, Doria A, Quarta S, Zen M, Gatta A, Pontisso P. SERPINB3, apoptosis and autoimmunity. Autoimmun Rev. 2009;9(2):108‐112. [DOI] [PubMed] [Google Scholar]
- 70. Ren C, Liu Q, Ma Y, Wang A, Yang Y, Wang D. TEAD4 transcriptional regulates SERPINB3/4 and affect crosstalk between keratinocytes and T cells in psoriasis. Immunobiology. 2020;225(5):152006. [DOI] [PubMed] [Google Scholar]
- 71. Liu L, Yang Y, Duan H, et al. CHI3L2 is a novel prognostic biomarker and correlated with immune infiltrates in gliomas. Front Oncol. 2021;11:611038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Du H, Masuko‐Hongo K, Nakamura H, et al. The prevalence of autoantibodies against cartilage intermediate layer protein, YKL‐39, osteopontin, and cyclic citrullinated peptide in patients with early‐stage knee osteoarthritis: evidence of a variety of autoimmune processes. Rheumatol Int. 2005;26(1):35‐41. [DOI] [PubMed] [Google Scholar]
- 73. Olkkonen J, Kouri VP, Hynninen J, Konttinen YT, Mandelin J. Differentially expressed in chondrocytes 2 (DEC2) increases the expression of IL‐1beta and Is abundantly present in synovial membrane in rheumatoid arthritis. Plos One. 2015;10(12):e0145279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wang X, Li Y, Fu J, Zhou K, Wang T. ARNTL2 is a prognostic biomarker and correlates with immune cell infiltration in triple‐negative breast cancer. Pharmgenomics Pers Med. 2021;14:1425‐1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wang S, Ma X, Ying Y, et al. Upregulation of ARNTL2 is associated with poor survival and immune infiltration in clear cell renal cell carcinoma. Cancer Cell Int. 2021;21(1):341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Xiao M, Liang X, Yan Z, et al. A DNA‐methylation‐driven genes based prognostic signature reveals immune microenvironment in pancreatic cancer. Front Immunol. 2022;13:803962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lu JJ, Maimaiti M, Liu H, et al. Potential biomarkers associated with differential manifestations of ulcerative colitis (UC) in Uyghur and Han population in China. J Inflamm Res. 2021;14:7431‐7441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. O'Connell P, Blake MK, Godbehere S, Amalfitano A, Aldhamen YA. SLAMF7 modulates B cells and adaptive immunity to regulate susceptibility to CNS autoimmunity. J Neuroinflammation. 2022;19(1):241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Simmons DP, Nguyen HN, Gomez‐Rivas E, et al. SLAMF7 engagement superactivates macrophages in acute and chronic inflammation. Sci Immunol. 2022;7(68):eabf2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Awwad MHS, Mahmoud A, Bruns H, et al. Selective elimination of immunosuppressive T cells in patients with multiple myeloma. Leukemia. 2021;35(9):2602‐2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information
Supporting information
Data Availability Statement
In our study, we download these datasets (GSE13355, GSE87466, GSE30999, and GSE107499) in the Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/).


