Abstract
Hereditary transthyretin (TTR) amyloidosis (ATTRv amyloidosis) is autosomal dominant and caused by mutation of TTR gene. Heterozygous ATTR Tyr114Cys (p.Tyr134Cys) amyloidosis is a lethal disease with a life expectancy of about 10 years after onset of the disease. However, the molecular pathogenesis of ATTR Tyr114Cys amyloidosis is still largely unknown. In this study, we took advantage of disease-specific induced pluripotent stem (iPS) cells and generated & characterized the heterozygous ATTR Tyr114Cys amyloidosis-specific iPS cells (Y114C iPS cells), to determine whether Y114C iPS cells could be useful for elucidating the pathogenesis of ATTR Tyr114Cys amyloidosis. We successfully differentiated heterozygous Y114C iPS cells into hepatocyte like cells (HLCs) mainly producing TTR protein. On day 27 after differentiation, the expression of hepatocyte maker albumin was detected, and TTR expression was significantly increased in HLCs differentiated from Y114C iPS cells. LC–MS/MS analysis showed that both WT TTR & ATTR Y114C protein were indeed expressed in the HLCs differentiated from Y114C iPS cells. Notably, the number of detected peptides derived from ATTR Y114C protein was lower than that of WT TTR protein, indeed indicating the clinical phenotype of ATTR Tyr114Cys amyloidosis. Taken together, we first reported the heterozygous Y114C iPS cells generated from patient with ATTR Tyr114Cys amyloidosis, and suggested that Y114C iPS cells could be a potential pathological tool, which may contribute to elucidating the molecular pathogenesis of heterozygous ATTR Tyr114Cys amyloidosis.
Keywords: Hereditary transthyretin amyloidosis, Transthyretin, Tyr114Cys, iPS cells
1. Introduction
Hereditary transthyretin amyloidosis (ATTRv amyloidosis) a life-threatening, autosomal-dominant systemic amyloidosis. ATTRv amyloidosis is caused by variant Transthyretin (TTR) mutation [[1], [2], [3]]. Patients with ATTRv amyloidosis develop sensorimotor polyneuropathy, autonomic dysfunction, cardiac failure, and other symptoms. Those critical symptoms lead to death typically within 10 years from disease onset [1]. TTR is one of the major amyloidogenic proteins, causes two kinds of systemic amyloidosis, hereditary ATTRv amyloidosis and aging-related wild-type ATTR (ATTRwt) amyloidosis [4]. TTR is mainly synthesized in the liver [5], but also in the choroid plexuses of the brain [6]. Although TTR usually forms tetramer in blood, TTR tetramer comprised of amyloidogenic TTR (ATTR) caused by TTR gene mutation. ATTR dissociates to monomer more easily, and then the monomer misfolds and forms insoluble amyloid fibrils [7]. The fibrils deposit and injure various organs, which causes various symptoms such as peripheral neuropathy, heart failure and central nervous system (CNS) symptoms [1,8,9]. As of today, more than 150 different point mutations have been identified in patients with ATTRv amyloidosis [10]. It has been reported that ATTRv amyloidosis exhibits several different phenotypes. Among them, the molecular pathogenesis underlying most common neuropathy-dominant type of ATTR Val30Met (p.Val50Met) amyloidosis has been intensively explored. Therefore, effective disease-modifying therapies such as TTR tetramer stabilizers and TTR gene silencing therapies have been developed for ATTR Val30Met amyloidosis [11,12]. In contrast to the potentially curable ATTR Val30Met amyloidosis, the specific pathogenesis of patients with various point mutations, such as heterozygous ATTR Tyr114Cys (p.Tyr134Cys) amyloidosis caused by substitution of tyrosine to cysteine at codon 114, has yet to be determined.
Patients with ATTR Tyr114Cys amyloidosis exhibit initial symptoms in their thirties with a life expectancy of about 10 years after onset of the disease [13]. Leptomeningeal-dominant type of ATTR Tyr114Cys amyloidosis exhibits various CNS symptoms, such as cerebral hemorrhage, transient ischemic attack, fluctuating consciousness, extrapyramidal signs and rapid progressive dementia that presents with cerebral amyloid angiopathy (CAA) [14]. Although those symptoms pathogenesis are thought to be caused by its amyloid deposition in cerebral blood vessels, the pathogenesis of ATTR Tyr114Cys amyloidosis is still largely unknown. It is well-documented that TTR is predominantly synthesized in the liver, and that the systemic symptoms of ATTR Val30Met amyloidosis patients are mainly caused by ATTR V30M in serum. In contrast, ATTR Tyr114Cys amyloidosis patients show symptoms mainly in the CNS and rarely systemic peripheral neurological symptoms. Moreover, unlike most common ATTR Val30Met amyloidosis, highly amyloidogenic ATTR Y114C in the blood shows low concentration [15]. Thus, although TTR is known to be predominantly synthesized in the liver, the kinetics and involvement of ATTR Y114C produced by liver in the pathogenesis of ATTR Tyr114Cys amyloidosis has yet to be determined. Despite the urgent need to elucidate the molecular pathogenesis of heterozygous ATTR Tyr114Cys amyloidosis, as of this moment, there is no specific disease model represents the relevant processes in patients with ATTR Tyr114Cys amyloidosis available.
Induced pluripotent stem (iPS) cells have an unlimited replicative ability and the potential to differentiate into most cell types in organisms [[16], [17], [18]]. iPS cells have pluripotency that can differentiate into cells such as nerves, myocardium, intestines, liver, and bones [[19], [20], [21], [22], [23]]. iPS cells can be produced by introducing reprogramming factors using viral vectors and mRNA mainly into somatic cells of skin, blood, and urine [16,24]. In particular, disease-specific iPS cells, generated from patients with hereditary diseases, maintain the patient's genetic information and can be useful pathological models by taking advantage of the pluripotency of iPS cells. For example, disease-specific iPS cells of amyotrophic lateral sclerosis (ALS), an inherited neurodegenerative disease, can be differentiated into neurons that cause the disease, thereby reproducing neuronal degeneration. In addition, this pathological model is screened therapeutic drug candidates for ALS [20,25]. Therefore, disease-specific iPS cells are very useful tools for elucidating the pathogenesis of hereditary diseases because they exhibit potential to differentiate into various disease-responsible cells with maintaining genetic contexts. From the viewpoint of pathological analysis, we previously established the heterozygous ATTR Val30Met amyloidosis-specific iPS cells (V30M iPS cells) [26]. The results showed that V30M iPS cells successfully differentiated into hepatocytes indeed expressing both wild type (WT) TTR & ATTR V30M differently, suggesting that ATTR V30M produced by liver may be responsible for systemic symptoms of ATTR Val30Met amyloidosis [26]. Thus, V30M iPS cells could be very useful for elucidating the pathogenesis of ATTR Val30Met amyloidosis.
In this study, we generated and characterized the heterozygous ATTR Tyr114Cys amyloidosis-specific iPS cells (Y114C iPS cells), to determine whether Y114C iPS cells could be useful for elucidating the pathogenesis of ATTR Tyr114Cys amyloidosis.
2. Materials and methods
2.1. Generation of ATTRv amyloidosis-specific iPS cells
ATTRv amyloidosis-specific iPS cells were generated as described in our previous study [26]. Skin biopsy samples were obtained from 2 Japanese patients with ATTR Tyr114Cys amyloidosis in Kumamoto University Hospital. The patients exhibited the typical CNS symptoms of ATTR Tyr114Cys amyloidosis, such as, cerebral amyloid angiopathy and cerebral hemorrhage, and also, systemic symptoms, such as, gastrointestinal dysfunction (constipation and diarrhea). Skin fibroblasts from ATTR Tyr114Cys amyloidosis were maintained in Dulbecco's Modified Eagle Medium (DMEM; Thermo Fisher Scientific, Carlsbad, CA, USA) supplemented with 10 % fetal bovine serum (FBS; Thermo Fisher Scientific). Induction of human iPS cells was performed as described previously [26]. Briefly, Y114C iPS cells were induced from skin fibroblasts of ATTR Tyr114Cys amyloidosis patients with conventional sendai virus (SeV) vectors carrying octamer-binding transcription factor 3/4 (OCT3/4), sex determining region Y-box 2 (SOX2), kruppel like factor 4 (KLF4), and a temperature-sensitive vector, which has c-MYC. This study was approved by the Research Ethics Committee of Kumamoto University Hospital (approved number: No. 981), and conducted after obtaining written informed consent.
2.2. Differentiation into hepatocyte like cells
ATTRv amyloidosis-specific iPS cells (Y114C and V30 M iPS cells) and 201B7 cells (WT iPS cells) as control [16] were differentiated into hepatocyte like cells (HLCs) using feeder free method modified from reported protocols (Fig. 1A) [26]. The following modifications: these iPS cells were cultured overnight with StemFit AK02 N (Reprocell, Kanagawa, Japan) supplemented with 10 μM Y27632 (FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan) and were then dissociated by using TrypLE Select (Thermo Fisher Scientific). Those cells were plated at 500,000 cells per well in 6-well plates (Corning, corning, NY, USA) that had been previously coated with SynthemaxII-SC Substrate (Corning). Two days later, the cells were cultured from day 0 (D0) to day 3 (D3) in DMEM supplemented with 100 ng/ml Activin A (R&D systems, Minneapolis, MN, USA), 2 % B27 Supplement, minus insulin (Thermo Fisher Scientific), 1 % nonessential amino acids (NEAA; Thermo Fisher Scientific), 1 % l-glutamine (L-Glu; Thermo Fisher Scientific), 1 % penicillin and streptomycin (P/S; nacalai tesque, Kyoto, Japan), and 0.1 mM 2-mercaptoethanol (2-Me; Merck, Darmstadt, Germany). On D3, the cells were replaced into 24-well plates (Corning) that had been previously coated with SynthemaxII-SC Substarate, using DMEM supplemented with 10 % FBS (Thermo Fisher Scientific), 2 % Insulin, Transferrin, Selenium Solution (Thermo Fisher Scientific), 1 % NEAA (Thermo Fisher Scientific), 1 % L-Glu (Thermo Fisher Scientific), 1 % P/S (nacalai tesque), and 0.1 mM 2-Me (Merk), 10 μM Y27632 (FUJIFILM Wako Pure Chemical Corporation). For differentiation from D4 to D5, KnockOut DMEM/F-12 (Thermo Fisher Scientific) supplemented with 10 ng/ml Bone morphogenetic protein (BMP4; protintech, Rosemont, IL, USA), 10 ng/ml Fibroblast growth factor 10 (FGF10; PeproTech, Cranbury, NJ, USA) and 2 % B27 Supplement (Thermo Fisher Scientific), 1 % NEAA (Thermo Fisher Scientific), 1 % L-Glu (Thermo Fisher Scientific), 1 % P/S (nacalai tesque), and 0.1 mM 2-Me (Merk). For differentiation from D5 to D17, Hepatocyte Culture Medium BulletKit (HCM; Lonza, Basel, Switzerland) except for gentamicin supplemented with, 20 ng/ml Hepatocyte growth factor (HGF; PeproTech), 10 ng/ml Oncostatin M (OsM; Merck). For differentiation from D17 to D27, HCM except for gentamicin supplemented with, 20 ng/ml HGF, 10 ng/ml OsM, 1 % Matrigel (Corning), 10 mM nicotinamide (Merck). Medium was replaced 4 ml/well every day (D0-3), 1 ml/well every day (D3-5) and 0.5 ml/well every 2 days (D5-27).
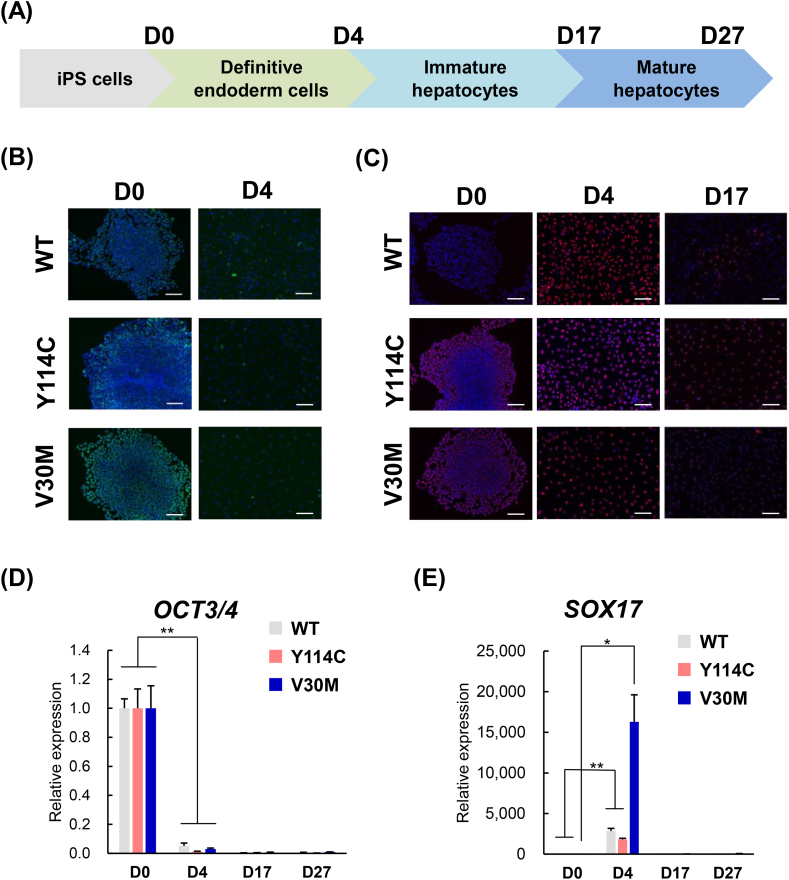
Fig. 1.
Differentiation of Y114C iPS cells into definitive endoderm cells.
(A) Schematic experimental procedure of differentiation into hepatocyte like cells. iPS cells (Y114C, V30M, WT) were differentiated on D0 to D3 in DMEM supplemented with 100 ng/ml Activin and 2 % B27 Supplement, minus insulin. From D4 to D5, kockout DMEM supplemented with 10 ng/ml BMP4, 10 ng/ml FGF10, and 2 % B27 Supplement was used. From D5 to D17, HCM except for gentamicin supplemented with 10 ng/ml OsM and 20 ng/ml HGF was used. From D17 to 27, HCM except for gentamicin supplemented with 10 ng/ml OsM, 20 ng/ml HGF, nicotinamide and Matrigel was used. (B) WT, V30M and Y114C iPS cells differentiated on D0 and D4 were stained for pluripotency marker OCT3/4 (green), (C) WT, V30M and Y114C iPS cells differentiated on D0, D4 and D17 were stained for endoderm marker SOX17 (red), and counterstained with DAPI (blue). (D, E) OCT3/4 and SOX17 mRNA expression levels in WT, V30M and Y114C iPS cells were determined D0, D4, D17 and D27 by real-time RT-PCR. SOX17 mRNA expression on D0 and D27 in WT iPS cells were not detected. Values are expressed as the means ± S.D. of 4 independent experiments. *p < 0.05 and **p < 0.01 vs. D0 and D4 in WT, V30M and Y114C iPS cell via paired t-test. Scale bar, 100 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
2.3. Real-time PCR analysis
Total RNA extraction from cells was performed by phenol chloroform extraction method using TRIzol Reagent (Thermo Fisher Scientific). Total RNA was reverse-transcribed into cDNA for 15 min at 37 °C by the PrimeScript RT reagent (Takara Bio Inc, Shiga, Japan). For quantification of mRNA, each PCR assay was performed using LightCycler System with SYBR Premix DimerEraser (Takara Bio Inc). Real-time PCR conditions were as follows: denaturation for 15 s at 95 °C and annealing and extension for 60 s at 60 °C, for up to 40 cycles. Target messenger RNA (mRNA) levels were expressed as arbitrary units and were determined by using the standard curve method by LightCycler 480 (Roche, Basel, Switzerland). Human 18S ribosomal RNA was used as an internal control. The following primers were used OCT3/4: forward 5′-CGAAAGAGAAAGCGAACCAG-3′, reverse 5′-ACACTCGGACCACATCCTTC-3′; NANOG: forward 5′-CAGAAGGCCTCAGCACCTAC-3′, reverse 5′-ACTGGATGTTCTGGGTCTGG-3′; SOX2: forward 5′-AACCCCAAGATGCACAACTC-3′, reverse 5′-CGGGGCCGGTATTTATAATC-3′; sex determining region Y-box 17 (SOX17): forward 5′-AGCAGAATCCAGACCTGCAC-3′, reverse 5′-TTGTAGTTGGGGTGGTCCTG-3′; forkhead box A2 (FOXA2): forward 5′-GAGGGCTACTCCTCCGTGA-3′; reverse 5′-CACGTACGACGACATGTTCA-3′ alpha-fetoprotein (AFP): forward 5′-CTACCTGCCTTTCTGGAAGAACTTTG-3′, reverse 5′-GATCGATGCTGGAGTGGGCTTT-3′; albumin (ALB): forward 5′-GTGAAACACAAGCCCAAGGCAACA-3′, reverse 5′-TCCTCGGCAAAGCAGGTCTC-3′; TTR: forward 5′-CA CATTCTTGGCAGGATGGCTTC-3′, reverse 5′-CTCCCAGGTGTCATCAGCAG-3′ and 18S: forward 5′-CGGCTACCACATCCAAGGAA-3′, reverse 5′- GCTGGAATTACCGCGGCT-3′.
2.4. Immunocytochemistry
For whole-mount immunocytochemical analysis, iPS cells cultures were fixed in 4 % paraformaldehyde (FUJIFILM Wako Pure Chemical Corporation) for 30 min, followed by permeabilization with 0.1 % Triton-X (nakalai tesque) in phosphate-buffered saline (PBS; Thermo Fisher Scientific) for 10 min at room temperature, washed once with 0.1 % Tween-20 (MP, Irvine, CA, USA) in PBS (PBST) then incubated with 20 % Blocking One (nakalai tesque) in PBS-T in a humidified chamber for 1 h at room temperature. The cells were incubated with diluted (1:200) antibody in 20 % Blocking One in PBS-T. After washing 3 times with PBS-T, the cells incubated with diluted (1:1000) secondary antibody in 20 % Blocking One for 2 h at room temperature in the dark. After washing off the secondary antibody in PBS-T, cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). The following antibodies were used as first antibodies: Mouse anti-OCT3/4 antibody (Santa Cruz Biotechnology, Dallas, TX, USA), goat anti-SOX17 antibody (R&D systems), Goat anti-human Albumin antibody (Fortis Life Sciences, Montgomery, TX, USA). Secondary antibodies used were Alexa 488-conjugated and Alexa 568-conjugated antibodies (Thermo Fisher Scientific).
2.5. Western blotting
Culture supernatants added sample buffer was incubated for 5 min at 95 °C, which apply into sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to Poly Vinyli dene Fluoride (PVDF) membranes (GE Healthcare, Chicago, IL, USA). Membranes were blocked with 5 % nonfat dried milk (Cell Signaling Technology, Danvers, MA, USA) overnight at 4 °C and PBS-T and were then incubated 1 h at room temperature with diluted (1:1000) polyclonal rabbit anti-human TTR antibody (Agilent Technologies, Santa Clara, CA, USA) in PBS-T. After the membranes were washed, they were incubated in diluted (1:1000) polyclonal Goat anti-Rabbit immunoglobulins/HRP (Agilent Technologies) for 1 h at room temperature. After this process, specific protein bands were detected with ECL select (Cytiva, Tokyo, Japan) by an enhanced chemiluminescence system ImageQuant Las4000mini (Cytiva).
2.6. Liquid chromatography–tandem mass spectrometry (LC-MS/MS)
The supernatant sampled D27 was incubated for 2 h at 4 °C with 5 μg of polyclonal rabbit anti-human TTR antibody (Agilent Technologies). PureProteome Protein G Magnetic Beads (Merck) were added to the reaction to capture the immune complexes and were agitated for 2 h at 4 °C. After the immune complexes were washed three times with PBST, 60 μl of sample buffer was added and incubation proceeded for 5 min at 95 °C. Sample buffers (40 μl), which eluted TTR protein from the beads, were fractionated via SDS-PAGE. Silver staining of gel was performed with ProteoSilver™ Plus Silver Stain Kit (Merk) according to the manufacturer's protocol. High intensity bands near 15 kDa from each lane were cut and diced in the tube and destained by the manufacturer's protocol. Peptides derived from TTR were eluted from gel bands as previously reported [27]. After gel pieces were washed and dried in a DNA SpeedVac System (DNA120OP-115; Thermo Fisher Scientific), gel pieces were reduced with 10 mM dithiothreitol solution for 1 h at 56 °C. Then, gel pieces were washed and alkylated with 55 mM iodoacetamide under agitation in the dark for 45 min at room temperature and the supernatant was removed. Gel pieces were washed, dried and digested within 50 ml of 50 mg/ml trypsin solution in the tube (Promega, Madison, WI, USA) overnight at 37 °C. The eluted peptides mixture was dried and dissolved in 40 μL of MS-grade water containing 0.1 % trifluoroacetic acid and 2 % acetonitrile and analyzed by a LC-MS/MS system (LTQ Velos Pro, Thermo Fisher Scientific; Advance Splitless Nano-Capillary LC dual solvent delivery system, Bruker-Michrom, Auburn, CA, USA; HTS-xt PAL autosampler, CTC Analytics, Zwingen, Switzerland; XYZ nanoelectrospray ionization source, AMR, Tokyo, Japan). Five-micro litter peptide solution was applied into a L-trap column (0.3 × 5 mm, 5 mm, Chemical Evaluation Research Institute, Tokyo, Japan) and separated by a capillary reversed-phase C18 column (0.1 × 150 mm, 3 mm, Chemical Evaluation Research Institute) with 5–40 % solution B gradient (solvent A: MS-grade water containing 0.1 % formic acid; solvent B: 100 % acetonitrile) in 40 min. All MS/MS spectra were searched against for Homo sapiens entries in the Swiss-Prot database with added ATTR Y114C amino acid sequence using SEQUEST (Proteome Discoverer software, Thermo Fisher Scientific). False discovery rate <0.05 was considered to search the results. Precursor and fragment mass tolerances were 2 Da and 0.8 Da, respectively. Dynamic modification of oxidation of methionine and static modification of carbamidomethyl on cysteine were considered. The maximum of 2 missed cleavages with trypsin was allowed [27]. Detection of ATTR V30 M amino acid sequence described previously [26].
2.7. Statistical analysis
Data are represented as the mean ± standard deviation (S.D.). The paired t-test were used to assess the differences between the two groups. The p value of <0.05 was considered to be significant. *p < 0.05 and **p < 0.01.
3. Results
3.1. Differentiation of Y114C iPS cells into definitive endoderm cells
Because amyloidogenic TTR proteins are mainly synthesized in the liver [5], we first determined whether ATTRv Tyr114Cys amyloidosis-specific iPS cells (Y114C iPS cells) indeed differentiated into hepatocyte like cells (HLCs). For cell differentiation, iPS cells (WT, V30M and Y114C) were cultured with serial changes of media as shown in Fig. 1A. To test whether Y114C iPS cells can differentiate into definitive endoderm cells, we analyzed the expression of the pluripotency marker OCT3/4, SOX2 and NANOG, and also the endoderm marker SOX17 and FOXA2 (Fig. 1B–E and Supplemental Fig. 1). Immunocytochemical analysis clearly showed that, as a consequence of Y114C iPS cell differentiation (D4), the OCT3/4 was markedly decreased at protein level (Fig. 1B), while the endoderm marker SOX17 expression was increased (Fig. 1C), consistent with WT and V30M iPS cell differentiation. Moreover, similar to the results observed in differentiated WT and V30M iPS cells, real-time RT-PCR analysis also showed that, the decrease in expression of the pluripotency marker OCT3/4 (also, NANOG and SOX2) at mRNA level was accompanied by Y114C iPS cells differentiation (Fig. 1D), while the endoderm marker SOX17 expression (also, FOXA2) was detected on D4 (Fig. 1E and Supplemental Fig. 1B). These results indicated Y114C iPS cells successfully differentiated into definitive endoderm cells.
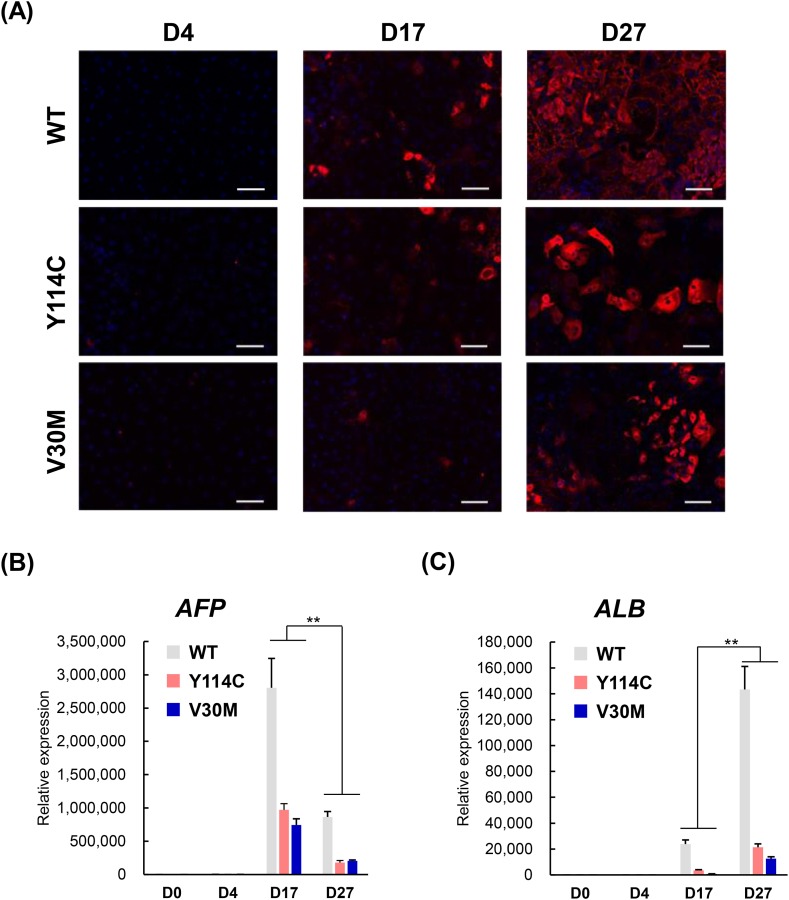
3.2. Differentiation of definitive endoderm cells into hepatocyte like cells
Next, we investigated whether the definitive endoderm cells could be differentiated into HLCs. We analyzed the expression of endoderm marker SOX17 and hepatocyte marker albumin (ALB) by immunocytochemical analysis on D4, D17 and D27 of differentiated Y114C iPS cells (Fig. 1, Fig. 2A). Consistent with WT and V30M iPS cells differentiation, as a consequence of Y114C iPS cells differentiation (D17, D27), SOX17 was decreased at protein level (Fig. 1C), whereas the hepatocyte marker ALB expression was significantly increased (Fig. 2A). Moreover, the hepatoblast marker α-fetoprotein (AFP) expression at mRNA level was observed on D17 (Fig. 2B), and then ALB expression was markedly increased on D27 (Fig. 2C), also similar to the results observed in WT and V30M iPS cell differentiation. Taken together, those results clearly showed that Y114C iPS cells could be indeed differentiated into HLCs.
Fig. 2.
Differentiation of Definitive Endoderm cells into hepatocyte like cells.
(A) WT, V30M and Y114C iPS cells differentiated on D4, D17 and D27 were stained for hepatocyte marker ALB and counterstained with DAPI (blue). (B, C) Hepatoblast marker AFP and ALB mRNA expression levels in WT, V30M, and Y114C iPS cells were determined D0, D4, D17 and D27 by real-time RT-PCR. Values are expressed as the means ± S.D. of 3–4 independent experiments. **p < 0.01 vs. D17 and D27 in WT, V30M, and Y114C iPS cells via paired t-test. Scale bar, 100 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
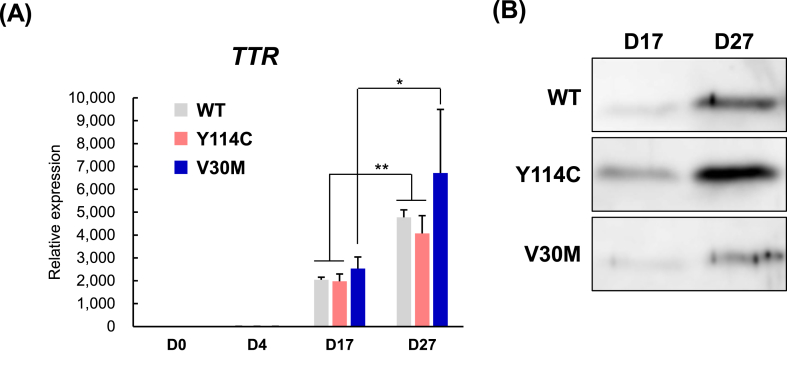
3.3. Heterozygous expression of TTR in hepatocyte like cells
Since Y114C iPS cells could be differentiated into HLCs (Fig. 2), we next sought to determine TTR expression by both real-time RT-PCR and western blotting. TTR expression at mRNA level started increasing significantly at D17 along with hepatocytes differentiation (Fig. 3A). In addition, TTR protein levels in supernatants of HLCs differentiated from WT, V30M, and Y114C iPS cells were increased from D17 to D27 (Fig. 3B). These suggest that HLCs differentiated from Y114C iPS cells is useful to investigate the mechanism underlying TTR expression. Moreover, to further evaluate the heterozygous secretions pattern of both WT TTR & ATTR Y114C in HLCs differentiated from Y114C iPS cells, we performed LC–MS/MS analysis for supernatants from HLCs differentiated from Y114C iPS cells. As expected, LC–MS/MS analysis detected only WT TTR peptides in the HLCs differentiated from WT iPS cells (Supplemental Table 1). In addition, consistent with previous study [26], both WT TTR & ATTR V30M were detected in HLCs differentiated from V30M iPS cells (Supplemental Table 2). Finally, it is first report that LC–MS/MS analysis detected both WT TTR & ATTR Y114C protein indeed expressed in the HLCs differentiated from Y114C iPS cells. Importantly, this analysis suggested that number of detected peptides derived from ATTR Y114C was lower than that of WT TTR protein in supernatants from HLCs differentiated from Y114C iPS cells (Table 1, Supplemental Fig. 3).
Fig. 3.
TTR expression in hepatocyte like cells.
(A) Human TTR mRNA expression levels were determined at D0, D4, D17 and D27 in WT, V30 M, and Y114C iPS cells by real-time RT-PCR. (B) Human TTR protein expression in the supernatant of HLCs differentiated from WT, V30M and Y114C iPS cells were determined at D17 and D27 by Western blotting (Supplemental Fig. 2). Values are expressed as the means ± S.D. of 3–4 independent experiments. *p < 0.05 and **p < 0.01 vs. D17 and D27 in WT, V30M, and Y114C iPS cells via paired t-test.
Table 1.
LC–MS/MS analysis detected WT TTR & ATTR Y114C protein.
| WT TTR peptides | number of peptides | MH + [Da] |
|---|---|---|
| RYTIAALLSPYSYSTTAVVTNPKE | 7 | 2646.89092 |
| YTIAALLSPYSYSTTAVVTNPKE | 9 | 2489.99516 |
| ATTR Y114C peptides | number of peptides | MH + [Da] |
| RYTIAALLSPCSYSTTAVVTNPKE | 2 | 2642.74875 |
| YTIAALLSPCSYSTTAVVTNPKE | 3 | 2487.89409 |
LC-MS/MS analysis for the D27 culture supernatant showed both WT TTR & ATTR Y114C at protein levels. The RYTIAALLSP Y/C SYSTTAVVTNPKE and YTIAALLSP Y/C SYSTTAVVTNPKE means 104–127 and 105–127 amino acids of TTR respectively. M + proton (H+; MH+) means the peptides operated in positive ion mode.
4. Discussion
In this study, we characterized the heterozygous Y114C iPS cells generated from patient with ATTR Tyr114Cys amyloidosis. In addition, we revealed that Y114C iPS cells could be differentiated into HLCs mainly producing TTR protein. Moreover, HLCs differentiated from Y114C iPS cells indeed expressed WT TTR & ATTR Y114C protein differently. These results suggest that Y114C iPS cells could be a potential pathological tool, which may contribute to elucidating the molecular pathogenesis of heterozygous ATTR Tyr114Cys amyloidosis.
Recent decades, attempts to establish the experimental models of ATTRv amyloidosis have not been as successful as researchers would wish [[28], [29], [30]]. Moreover, a review of previous studies about the molecular pathogenesis of ATTRv amyloidosis suggests that most of the studies investigated the pathologic effect of ATTR using non-mutant cells or an artificial high expression system [[31], [32], [33]]. Indeed, the previous experimental ATTRv amyloidosis models have simply overexpressed only ATTR Y114C and other mutations into human embryonic kidney cells, which was not disease-responsible hepatocyte cells (e.g., human embryonic kidney cells) [9,34]. As a matter of fact, ATTRv amyloidosis is an autosomal-dominant hereditary disease and those cell-based systems did not utilize heterozygous ATTR mutant cell models. Those systems highly likely not represent the physiological relevant processes in patient with ATTRv amyloidosis. Thus, we focused on the disease-specific iPS cells, exhibiting potential to differentiate into various disease-responsible cells with maintaining the disease-responsible genetic mutation. In this study, we successfully generated the heterozygous Y114C iPS cells. Our finding is the first report that heterozygous Y114C iPS cells could be differentiated into disease-responsible HLCs (Fig. 2, Fig. 3). It should be noted that our differentiation method into HLCs have confirmed the biochemical context of HLCs [26]. Differentiated HLCs showed approximately 20 μg/ml ALB protein secretion in the media, and also periodic acid-Schiff (PAS)-positive, indicating cytoplasmic glycogen storage. In addition to the comparable expression pattern of ALB and AFP expression (Fig. 2), the TTR expression was indeed confirmed at both mRNA and protein levels in the supernatant of HLCs (Fig. 3). Moreover, expression of retinol binding protein 1 (RBP1) was also confirmed by our preliminary comprehensive proteomic analysis (data not shown). Although the difference in ALB expression may be due to differentiation efficiency between among those HLCs cells [29], we thought that HLCs differentiated from Y114C iPS cells able to recreate the biochemical context of TTR synthesis. In fact, HLCs differentiated from Y114C iPS cells indeed expressed both WT TTR & ATTR Y114C protein differently (Table 1). It should be noted that a few studies have shown that disease-specific iPS cells derived from patient with ATTRv amyloidosis may have potential to be useful tools for elucidating the molecular pathogenesis. of ATTR amyloidosis. In addition to our previous study about V30M iPS cells [26], disease-specific iPS cells with other types of ATTR mutant have been generated and assessed its potential usefulness [35,36]. Unlike those reported mutations such as most common ATTR Val30Met amyloidosis, the pathogenesis of ATTR Tyr114Cys amyloidosis is still largely unknown. In this study, we revealed that HLCs differentiated from Y114C iPS cells indeed secreted low levels of ATTR Y114C, one of clinical phenotype of ATTR Tyr114Cys amyloidosis. Considering the difference from ATTR Val30Met amyloidosis, this evidence strongly suggests that different point mutations of TTR may affect the cellular responses in TTR producing cells. In fact, our preliminary comprehensive proteomic analysis showed that approximately 7000 proteins expression alterations were detected in HLCs differentiated from WT, V30M and Y114C iPS cells, respectively. Among them, it should be noted that approximately 500 proteins expression were altered only in HLCs differentiated from Y114C iPS cells (data not shown). Thus, ATTRv amyloidosis-specific iPS cells could have potential to observe and compare the changes in cellular response associated with TTR expression in different mutations of ATTRv amyloidosis. Since it is quite difficult to perform those kinds of molecular analysis in clinical specimen, Y114C iPS cells will make the various analysis of molecular pathogenesis possible, especially in TTR producing cells. Taken together, although the phenotypes in CNS have to be elucidated in the future, heterozygous Y114C iPS cells, exhibiting the disease-specific phenotypes, may have potential to contribute to elucidate the pathogenesis of patient with heterozygous ATTR Tyr114Cys amyloidosis.
As of this moment, more than 150 different point mutations of ATTR have been identified, and reported to exhibit several different phenotypes [10]. In the previous study, HLCs differentiated from V30M iPS cells, indeed expressed both WT TTR & ATTR V30M at protein level, and the ratio of TTR protein to ATTR protein is approximately 1 to 1 [26]. Alternatively, although our results showed that both WT TTR & ATTR Y114C protein were indeed expressed in the HLCs differentiated from Y114C iPS cells (Table 1). The number of detected peptides derived from ATTR Y114C protein could be lower than that of WT TTR protein (Table 1). It is well-documented that highly amyloidogenic ATTR Y114C in the blood shows low concentration, unlike ATTR Val30Met amyloidosis [15]. Our results suggests that the low serum ATTR Y114C may be occurred by secretion level, not by the clearance in the blood. Although the detailed mechanism has yet to be determined, it is noteworthy that HLCs differentiated from Y114C iPS cells indeed reflected in serum levels, one of typical clinical phenotypes of ATTR Tyr114Cys amyloidosis. This evidence may suggest the potential relationship between very low levels of ATTR Y114C in serum and mild systemic symptoms (e.g., peripheral neuropathy) in ATTR Tyr114Cys amyloidosis patients [13,37]. By contrast, ATTR Val30Met amyloidosis patients with the same level in WT TTR & ATTR V30M in serum, mainly exhibit systemic symptoms. TTR kinetic stability may be responsible for this difference because it has been reported that ATTR Y114C protein shows one of lower kinetic stability TTR. These TTR defined by the rate of TTR tetramer dissociation in vitro, such as ATTR A25T and L55P [9]. In other words, ATTR Y114C readily dissociates from tetramer to monomer and subsequently misfolds. In general, it is also known that misfolded proteins are regulated and controlled by endoplasmic reticulum-associated degradation (ERAD) [38]. It has been reported that ATF6, one of ERAD factors, suppresses destabilized TTR secretion in HEK293 overexpressed ATTR A25T [9]. And ATTR L55P amyloidosis-specific iPS cells derived HLCs upregulated ATF6 [39]. These suggested that the ERAD may also contribute to the low expression of ATTR Y114C protein in HLCs differentiated from Y114C iPS cells. In contrast to ATTR Y114C, ATTR V30M HLCs differentiated from V30M iPS cells is believed to slip through the ERAD in the same way as the WT TTR. Thus, the detailed molecular mechanisms underlying different ATTR expression pattern, utilizing heterozygous Y114C iPS cells to be further investigated.
This study has limitation. It is documented that leptomeningeal-dominant type of ATTR Tyr114Cys amyloidosis exhibits various CNS symptoms. Indeed, TTR is also known to be synthesized in the choroid plexuses of the brain. It has been reported that rat immortalized choroid plexus cells secrete destabilized ATTR L12P and D18G more than those TTR secreted from human hepatoma cells without endogenous TTR expression, respectively [40]. In fact, ATTR Y114C to WT TTR in cerebrospinal fluid from ATTR Y114C amyloidosis patients were significantly higher than those in serum [15], suggesting that secretion mechanism of choroid plexus cells is different from hepatocytes. Thus, while Y114C iPS cells indeed genetically & partially exhibited the phenotype of heterozygous ATTR Tyr114Cys amyloidosis in this study, it is also necessary to differentiate into the disease-responsible choroid plexus cells. Although the differentiation methods from iPS cells to choroid plexus has yet to established, in future, we will definitely try to differentiate Y114C iPS cells to choroid plexus cells to elucidate pathogenesis of heterozygous ATTR Tyr114Cys amyloidosis in CNS.
5. Conclusion
In the present study, we first reported the heterozygous Y114C iPS cells generated from patient with ATTR Tyr114Cys amyloidosis. Our results clearly showed that Y114C iPS cells could be differentiated into HLCs mainly producing TTR protein, and that HLCs differentiated from Y114C iPS cells indeed expressed WT TTR & ATTR Y114C protein differently, suggesting the clinical phenotype of ATTR Tyr114Cys amyloidosis. Future investigation will focus on more detailed molecular pathological dynamics of the ATTR Y114C expression to elucidate the pathogenesis of heterozygous ATTR Tyr114Cys amyloidosis.
Funding
This work was supported by Grants-in-Aid (17K19498 & 15K15006 to H.J.) from MEXT KAKENHI the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Data availability statement
Data will be made available on request.
CRediT authorship contribution statement
Kenta Ouchi: Writing – original draft, Investigation, Formal analysis, Data curation, Conceptualization. Kaori Isono: Resources, Formal analysis, Data curation. Yuki Ohya: Resources, Formal analysis, Data curation. Nobuaki Shiraki: Supervision, Methodology, Conceptualization. Masayoshi Tasaki: Supervision, Formal analysis, Data curation. Yukihiro Inomata: Supervision, Conceptualization. Mitsuharu Ueda: Supervision, Conceptualization. Takumi Era: Supervision, Conceptualization. Shoen Kume: Supervision, Conceptualization. Yukio Ando: Supervision, Conceptualization. Hirofumi Jono: Writing – review & editing, Supervision, Project administration, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e24590.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
References
- 1.Ando Y., Nakamura M., Araki S. Transthyretin-related familial amyloidotic polyneuropathy. Arch. Neurol. 2005;62:1057–1062. doi: 10.1001/ARCHNEUR.62.7.1057. [DOI] [PubMed] [Google Scholar]
- 2.Buxbaum J.N., Dispenzieri A., Eisenberg D.S., Fändrich M., Merlini G., Saraiva M.J.M., Sekijima Y., Westermark P. Amyloid nomenclature 2022: update, novel proteins, and recommendations by the international society of amyloidosis (ISA) nomenclature committee. Amyloid. 2022;29:213–219. doi: 10.1080/13506129.2022.2147636. [DOI] [PubMed] [Google Scholar]
- 3.Sekijima Y., Ueda M., Koike H., Misawa S., Ishii T., Ando Y. Diagnosis and management of transthyretin familial amyloid polyneuropathy in Japan: red-flag symptom clusters and treatment algorithm. Orphanet J. Rare Dis. 2018;13:1–17. doi: 10.1186/S13023-017-0726-X/FIGURES/5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wechalekar A.D., Gillmore J.D., Hawkins P.N. Systemic amyloidosis. Lancet. 2016;387:2641–2654. doi: 10.1016/S0140-6736(15)01274-X. [DOI] [PubMed] [Google Scholar]
- 5.Berry D.C., Noy N. 2011. Signalling by Vitamin A and Retinol-Binding Protein in Regulation of Insulin Responses and Lipid Homeostasis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gonzalez-Billault C., Plante-Bordeneuve V., Universitaires H., Mondor H., Miller F.Y., Almeida M.R., Bezerra F., Saraiva M.J. Modulation of the mechanisms driving transthyretin amyloidosis. Front. Mol. Neurosci. 2020 doi: 10.3389/fnmol.2020.592644. Www.Frontiersin.Org.13 592644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lai Z., Colón W., Kelly J.W. The acid-mediated denaturation pathway of transthyretin yields a conformational intermediate that can self-assemble into amyloid. Biochemistry. 1996;35:6470–6482. doi: 10.1021/BI952501G. [DOI] [PubMed] [Google Scholar]
- 8.Wechalekar A.D., Gillmore J.D., Hawkins P.N. Systemic amyloidosis. Lancet. 2016;387:2641–2654. doi: 10.1016/S0140-6736(15)01274-X. [DOI] [PubMed] [Google Scholar]
- 9.Sekijima Y., Wiseman R.L., Matteson J., Hammarström P., Miller S.R., Sawkar A.R., Balch W.E., Kelly J.W. The biological and chemical basis for tissue-selective amyloid disease. Cell. 2005;121:73–85. doi: 10.1016/J.CELL.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 10.Yamashita T., Ueda M., Misumi Y., Masuda T., Nomura T., Tasaki M., Takamatsu K., Sasada K., Obayashi K., Matsui H., Ando Y. Genetic and clinical characteristics of hereditary transthyretin amyloidosis in endemic and non-endemic areas: experience from a single-referral center in Japan. J. Neurol. 2018;265:134–140. doi: 10.1007/S00415-017-8640-7/FIGURES/6. [DOI] [PubMed] [Google Scholar]
- 11.Jono H., Ando Y. 2017. Towards Targets and Treatments in Transthyretin Amyloidosis; pp. 691–699. 10.1080/21678707.2017.1360181. 5. [DOI] [Google Scholar]
- 12.Ueda M. 2022. Transthyretin: its Function and Amyloid Formation. [DOI] [PubMed] [Google Scholar]
- 13.Ando Y., Almeida M.D.R., Ohlsson P.I., Ando E., Negi A., Suhr O., Terazaki H., Obayashi K., Ando M., Saraiva M.J.M. Unusual self-association properties of transthyretin Y114C related to familial amyloidotic polyneuropathy: effects on detection and quantification. Biochem. Biophys. Res. Commun. 1999;261:264–269. doi: 10.1006/BBRC.1999.1048. [DOI] [PubMed] [Google Scholar]
- 14.Nakamura M., Yamashita T., Ueda M., Obayashi K., Sato T., Ikeda T., Washimi Y., Hirai T., Kuwahara Y., Yamamoto M.T., Uchino M., Ando Y. Neuroradiologic and clinicopathologic features of oculoleptomeningeal type amyloidosis. Neurology. 2005;65:1051–1056. doi: 10.1212/01.WNL.0000178983.20975.AF. [DOI] [PubMed] [Google Scholar]
- 15.Ueda M., Misumi Y., Mizu-Guchi M., Nakamura M., Yamashita T., Sekijima Y.-S., Ota K., Shinriki S., Jono H.-R., Ikeda S.-I., Suhr O.B., Ando Y. SELDI-TOF mass spectrometry evaluation of variant transthyretins for diagnosis and pathogenesis of familial amyloidotic polyneuropathy. Clin. Chem. 2009;55:1223–1227. doi: 10.1373/clinchem.2008.118505. [DOI] [PubMed] [Google Scholar]
- 16.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/J.CELL.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 17.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/J.CELL.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 18.Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R., Slukvin I.I., Thomson J.A. Induced pluripotent stem cell lines derived from human somatic cells. Science. 1979:318. doi: 10.1126/SCIENCE.1151526/SUPPL_FILE/YU.SOM.REVISION.1.PDF. (2007) 1917–1920. [DOI] [PubMed] [Google Scholar]
- 19.Kawai S., Yoshitomi H., Sunaga J., Alev C., Nagata S., Nishio M., Hada M., Koyama Y., Uemura M., Sekiguchi K., Maekawa H., Ikeya M., Tamaki S., Jin Y., Harada Y., Fukiage K., Adachi T., Matsuda S., Toguchida J. In vitro bone-like nodules generated from patient-derived iPSCs recapitulate pathological bone phenotypes. Nat. Biomed. Eng. 2019;3(7):558–570. doi: 10.1038/s41551-019-0410-7. 3 (2019) [DOI] [PubMed] [Google Scholar]
- 20.Dimos J.T., Rodolfa K.T., Niakan K.K., Weisenthal L.M., Mitsumoto H., Chung W., Croft G.F., Saphier G., Leibel R., Goland R., Wichterle H., Henderson C.E., Eggan K. Induced pluripotent stem cells generated from patients with ALS can Be differentiated into motor neurons. Science. 1979:1218–1221. doi: 10.1126/science.1158799. https://www.science.org 321 (2008) [DOI] [PubMed] [Google Scholar]
- 21.Funakoshi S., Miki K., Takaki T., Okubo C., Hatani T., Chonabayashi K., Nishikawa M., Takei I., Oishi A., Narita M., Hoshijima M., Kimura T., Yamanaka S., Yoshida Y. Nature Publishing Group; 2015. Enhanced Engraftment, Proliferation, and Therapeutic Potential in Heart Using Optimized Human iPSC-Derived Cardiomyocytes OPEN. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.G. Aprilia Helena, T. Watanabe, Y. Kato, N. Shiraki, S. Kume, Activation of cAMP (EPAC2) signaling pathway promotes hepatocyte attachment, (123AD). 10.1038/s41598-023-39712-3. [DOI] [PMC free article] [PubMed]
- 23.Leo S., Kato Y., Wu Y., Yokota M., Koike M., Yui S., Tsuchiya K., Shiraki N., Kume S. The effect of vitamin D3 and valproic acid on the maturation of human-induced pluripotent stem cell-derived enterocyte-like cells. Stem Cell. 2023;41:775–791. doi: 10.1093/STMCLS/SXAD042. [DOI] [PubMed] [Google Scholar]
- 24.Poleganov M.A., Eminli S., Beissert T., Herz S., Il Moon J., Goldmann J., Beyer A., Heck R., Burkhart I., Barea Roldan D., Türeci Ö., Yi K., Hamilton B., Sahin U. 2015. Efficient Reprogramming of Human Fibroblasts and Blood-Derived Endothelial Progenitor Cells Using Nonmodified RNA for Reprogramming and Immune Evasion. Home.Liebertpub.Com/Hum. 26. 751–766. [DOI] [PubMed] [Google Scholar]
- 25.Imamura K., Izumi Y., Watanabe A., Tsukita K., Woltjen K., Yamamoto T., Hotta A., Kondo T., Kitaoka S., Ohta A., Tanaka A., Watanabe D., Morita M., Takuma H., Tamaoka A., Kunath T., Wray S., Furuya H., Era T., Makioka K., Okamoto K., Fujisawa T., Nishitoh H., Homma K., Ichijo H., Julien J.P., Obata N., Hosokawa M., Akiyama H., Kaneko S., Ayaki T., Ito H., Kaji R., Takahashi R., Yamanaka S., Inoue H. The Src/c-Abl pathway is a potential therapeutic target in amyotrophic lateral sclerosis. Sci. Transl. Med. 2017;9 doi: 10.1126/SCITRANSLMED.AAF3962/SUPPL_FILE/AAF3962_SM.PDF. [DOI] [PubMed] [Google Scholar]
- 26.Isono K., Jono H., Ohya Y., Shiraki N., Yamazoe T., Sugasaki A., Era T., Fusaki N., Tasaki M., Ueda M., Shinriki S., Inomata Y., Kume S., Ando Y. Generation of familial amyloidotic polyneuropathy-specific induced pluripotent stem cells. Stem Cell Res. 2014;12:574–583. doi: 10.1016/j.scr.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 27.Yoshizawa T., Karim M.F., Sato Y., Senokuchi T., Miyata K., Fukuda T., Go C., Tasaki M., Uchimura K., Kadomatsu T., Tian Z., Smolka C., Sawa T., Takeya M., Tomizawa K., Ando Y., Araki E., Akaike T., Braun T., Oike Y., Bober E., Yamagata K. SIRT7 controls hepatic lipid metabolism by regulating the ubiquitin-proteasome pathway. Cell Metab. 2014;19:712–721. doi: 10.1016/j.cmet.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 28.Berg I., Thor S., Hammarström P. Modeling familial amyloidotic polyneuropathy (transthyretin V30M) in Drosophila melanogaster. Neurodegener. Dis. 2009;6:127–138. doi: 10.1159/000213761. [DOI] [PubMed] [Google Scholar]
- 29.Pokrzywa M., Dacklin I., Hultmark D., Lundgren E. Misfolded transthyretin causes behavioral changes in a Drosophila model for transthyretin-associated amyloidosis. Eur. J. Neurosci. 2007;26:913–924. doi: 10.1111/J.1460-9568.2007.05728.X. [DOI] [PubMed] [Google Scholar]
- 30.Buxbaum J., Tagoe C., Gallo G., Reixach N., French D. 2020. The Pathogenesis of Transthyretin Tissue Deposition: Lessons from Transgenic Mice; pp. 2–6. 10.1080/13506129.2003.12088560. 10. [DOI] [PubMed] [Google Scholar]
- 31.Sousa M.M., Du Yan S., Stern D., Saraiva M.J. vol. 80. Laboratory Investigation; 2000. pp. 1101–1110. (Interaction of the Receptor for Advanced Glycation End Products (RAGE) with Transthyretin Triggers Nuclear Transcription Factor kB (NF-kB) Activation). [DOI] [PubMed] [Google Scholar]
- 32.Cardoso I., Brito M., Saraiva M.J. Extracellular matrix markers for disease progression and follow-up of therapies in familial amyloid polyneuropathy V30M TTR-related. Dis. Markers. 2008;25:37–47. doi: 10.1155/2008/549872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sato T., Susuki S., Suico M.A., Miyata M., Ando Y., Mizuguchi M., Takeuchi M., Dobashi M., Shuto T., Kai H. Endoplasmic reticulum quality control regulates the fate of transthyretin variants in the cell. EMBO J. 2007;26:2501–2512. doi: 10.1038/SJ.EMBOJ.7601685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J.J., Genereux J.C., Suh E.H., Vartabedian V.F., Rius B., Qu S., Dendle M.T.A., Kelly J.W., Wiseman R.L. Endoplasmic reticulum proteostasis influences the oligomeric state of an amyloidogenic protein secreted from mammalian cells. Cell Chem. Biol. 2016;23:1282. doi: 10.1016/J.CHEMBIOL.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giadone R.M., Rosarda J.D., Akepati P.R., Thomas A.C., Boldbaatar B., James M.F., Wilson A.A., Sanchorawala V., Connors L.H., Berk J.L., Wiseman R.L., Murphy G.J. A library of ATTR amyloidosis patient-specific induced pluripotent stem cells for disease modelling and in vitro testing of novel therapeutics. Amyloid. 2018;25:148–155. doi: 10.1080/13506129.2018.1489228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niemietz C.J., Sauer V., Stella J., Fleischhauer L., Chandhok G., Guttmann S., Avsar Y., Guo S., Ackermann E.J., Gollob J., Monia B.P., Zibert A., Schmidt H.H.J. Evaluation of therapeutic oligonucleotides for familial amyloid polyneuropathy in patient-derived hepatocyte-like cells. PLoS One. 2016;11 doi: 10.1371/journal.pone.0161455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nomura T., Ueda M., Tasaki M., Misumi Y., Masuda T., Inoue Y., Tsuda Y., Okada M., Okazaki T., Kanenawa K., Isoguchi A., Nakamura M., Obayashi K., Shinriki S., Matsui H., Yamashita T., Ando Y. New simple and quick method to analyze serum variant transthyretins: direct MALDI method for the screening of hereditary transthyretin amyloidosis. Orphanet J. Rare Dis. 2019;14:1–6. doi: 10.1186/s13023-019-1100-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakatsukasa K., Brodsky J.L. The recognition and retrotranslocation of misfolded proteins from the endoplasmic reticulum. The Authors Journal Compilation #. 2008;9:861–870. doi: 10.1111/j.1600-0854.2008.00729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giadone R.M., Liberti D.C., Matte T.M., Rosarda J.D., Torres-Arancivia C., Ghosh S., Diedrich J.K., Pankow S., Skvir N., Jean J.C., Yates J.R., Wilson A.A., Connors L.H., Kotton D.N., Wiseman R.L., Murphy G.J. Expression of amyloidogenic transthyretin drives hepatic proteostasis remodeling in an induced pluripotent stem cell model of systemic amyloid disease. Stem Cell Rep. 2020;15:515–528. doi: 10.1016/j.stemcr.2020.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Batista A.R., Sena-Esteves M., Saraiva M.J. Hepatic production of transthyretin L12P leads to intracellular lysosomal aggregates in a new somatic transgenic mouse model. Biochim. Biophys. Acta, Mol. Basis Dis. 2013;1832:1183–1193. doi: 10.1016/j.bbadis.2013.04.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.