Abstract
Salmonella typhimurium infection of mice is an established model system for studying typhoid fever in humans. Using this model, we identified S. typhimurium genes which are absolutely required to cause fatal murine infection by testing independently derived transposon insertion mutants for loss of virulence in vivo. Of the 330 mutants tested intraperitoneally and the 197 mutants tested intragastrically, 12 mutants with 50% lethal doses greater than 1,000 times that of the parental strain were identified. These attenuated mutants were characterized by in vitro assays which correlate with known virulence functions. In addition, the corresponding transposon insertions were mapped within the S. typhimurium genome and the nucleotide sequence of the transposon-flanking DNA was obtained. Salmonella spp. and related bacteria were probed with flanking DNA for the presence of these genes. All 12 attenuated mutants had insertions in known genes, although the attenuating effects of only two of these were previously described. Furthermore, the proportion of attenuated mutants obtained in this study suggests that mutations in about 4% of the Salmonella genome lead to 1,000-fold or greater attenuation in the mouse typhoid model of infection. Most of these genes appear to be required during the early stages of a natural infection.
Bacterial pathogens require the coordinate expression of many genes in order to cause productive infection of their hosts. Some of these genes have been described and extensively characterized, often by first isolating mutants in vitro which are defective for characteristics thought to be important for virulence in vivo, e.g., the ability to invade cultured epithelial cells or to survive within phagocytic cells (12, 24). Such mutants are then tested in vivo to confirm the attenuating effect of the genetic lesion. In many cases, however, the attenuating effect of the mutation in vivo is relatively modest (13). The principal aim of this work was to identify and characterize Salmonella typhimurium genes which are among the most essential for virulence.
Salmonella is an ideal subject for this study because it is amenable to genetic manipulation and a well-characterized animal model system is available to study its pathogenicity (8, 27). Although the various Salmonella spp. are closely related, they can cause diverse human disease syndromes ranging from acute self-limiting gastroenteritis to typhoid fever, a severe and often fatal systemic illness. S. typhimurium is a major cause of food poisoning and diarrheal disease worldwide (38). In genetically susceptible mice (ItyS), however, it causes a disseminated infection similar to human typhoid. The course of oral infection in these animals is essentially linear—bacteria move from the gut via the lymphatic system to the liver and spleen, where they multiply intracellularly in macrophages until the infected animal succumbs to fatal septicemia.
Our strategy was to construct a bank of independent mutants and to screen each one directly for loss of virulence in mice. By applying a stringent definition of attenuation (a 50% lethal dose [LD50] 3 orders of magnitude greater than the parental LD50), we hoped to identify only those genes which contribute significantly to virulence. To carry out the mutagenesis, we used a transposition-defective MudJ transposon which could be temporarily complemented in cis for transposition functions (18). There are several advantages to this system: (i) the transposition of MudJ is the most random of any transposon studied (30), (ii) when the procedure is carried out as described by Hughes and Roth (18), the mutants are all independently derived, (iii) because this MudJ derivative is defective for further transposition, the insertions are stable (18), and (iv) because MudJ contains a promoterless copy of lacZ, it is possible to monitor transcription of the mutated gene, provided the insertion creates an appropriate transcriptional fusion (9).
In the course of the study, we sampled approximately 10% of the S. typhimurium genome. This allowed us to fulfill the second goal of the study, which was to estimate the number of genes required by S. typhimurium during two distinct phases of murine infection: passage from the stomach to the draining lymph nodes and penetration from these lymph nodes to the systemic circulation. Although a similar sampling approach has been used to estimate the number of genes which are essential for viability in the budding yeast, Saccharomyces cerevisiae, we believe that this is the first time such an estimate has been obtained for any bacterial pathogen and its host (7).
MATERIALS AND METHODS
Bacterial strains and cell lines.
The wild-type S. typhimurium strain ATCC 14028 was used. TT10288, an S. typhimurium strain which carries MudI and MudJ transposons, was a gift from Kelly Hughes (University of Washington, Seattle). With the exception of TT10288, which was grown at 30°C, all bacterial strains were routinely grown at 37°C on Luria-Bertani (LB) agar or in LB broth. Kanamycin and ampicillin were added to cultures where appropriate at concentrations of 60 and 100 μg/ml, respectively. Media stocks were obtained from Difco (Detroit, Mich.), and supplements were obtained from Sigma (St. Louis, Mo.).
The murine macrophage-like cell line J774.10 was kindly provided by Barry Bloom (Albert Einstein University, New York, N.Y.). These cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 4.5 g of glucose per ml, supplemented with 2 mM glutamine and 10% equine serum. The cells were passaged in the presence of penicillin (50 IU/ml) and streptomycin (50 μg/ml). They were grown in the absence of antibiotics for 24 h before assays. Chinese hamster ovary (CHO) cells were maintained in DMEM with 10% fetal calf serum and 1% nonessential amino acids. Caco-2 cells were maintained in MEM supplemented with 20% fetal calf serum, 1% nonessential amino acids, and 1 mM sodium pyruvate. All tissue culture stocks were obtained from Gibco (Gaithersburg, Md.), apart from sera which were obtained from HyClone (Logan, Utah).
Mutagenesis.
Mutagenesis was carried out according to the procedure of Hughes and Roth (18). Mutants were selected on LB agar and then screened to eliminate histidine auxotrophs (which represent homologous recombinants rather than insertion mutants) and P22 lysogens, by standard procedures (11, 21).
Identification of attenuated mutants in vivo.
Mutants were directly screened for loss of virulence in BALB/c female mice. Animals were aged 6 to 8 weeks and either were obtained from the breeding facility of the Scripps Research Institute or were purchased from Jackson Labs, Bar Harbor, Maine. For intragastric (i.g.) inoculation, 108 stationary-phase bacteria in 0.2 ml of LB broth were administered by gavage to two mice, which were then monitored for 28 days. Animals were given food and water ad libitum, and moribund animals were euthanized. If both survived, the corresponding mutation was retransduced into the wild-type background and 109 transductants were administered to two further animals. Mutants which were nonlethal at the higher dose were designated “attenuated i.g.” A dose of 103 bacteria was used for intraperitoneal (i.p.) inoculation. Bacteria were suspended in phosphate-buffered saline, pH 7.4, and administered with a 25-gauge needle in a final volume of 0.5 ml. Mutants which were nonlethal at this lower dose were retransduced and retested at a dose of 104. Those which were nonlethal at the higher dose were designated “attenuated i.p.” Mutants which did not kill animals at the lower doses but were virulent at the higher doses were described as being of intermediate virulence.
Determination of LD50s.
Four groups of four mice were inoculated with serial 10-fold dilutions of overnight bacterial cultures. Animals were monitored for 28 days, and LD50s were calculated by the method of Reed and Muench (35). Positive and negative controls were included in each experiment.
LPS profile analysis.
Alterations of the lipopolysaccharide (LPS) profile were identified by silver staining, as previously described (40).
Leakiness of the bacterial membrane.
The integrity of mutant bacterial membranes was checked as described elsewhere (3).
Measurement of resistance to sodium deoxycholate and Preimmune rabbit serum and determination of growth rate.
Approximately 105 bacteria in 100 μl of saline were added to 100 μl of either 1% sodium deoxycholate in saline, 20% normal rabbit serum, or saline. After incubation at 37°C, for 2.5 h (sodium deoxycholate) or 1 h (serum), the viability of the cultures was determined by plating. For growth rate determinations, cultures were set up in LB broth and viability was determined by plating at 1.5 and 4 h postinoculation.
Survival in murine macrophage J774.10 cells.
The ability of mutants to survive within J774.10 cells was tested as described previously (6). Bacterial viability was determined at 0 and 18 h after infection of cells.
Ability to invade epithelial cells.
Invasion assays were carried out according to the protocol of Miller and Falkow (28), with minor modifications. Bacteria were grown for 4 h, and a multiplicity of infection (MOI) of 100 to 200 was used.
Expression of β-galactosidase.
Mutants were tested for β-galactosidase activity in two ways. (i) Bacteria were plated in parallel on minimal glucose medium or LB agar supplemented with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (29), followed by overnight incubation under aerobic or anaerobic conditions. Blue colonies were scored as positive for activity. (ii) Macrophages were first infected with mutants, at an MOI of 100 to 200, unphagocytized bacteria were removed by washing and gentamicin treatment, and the cells were then overlaid with cooled fresh medium containing 0.7% Noble agar and X-Gal (0.5 mg/ml). After overnight incubation, blue colonies were scored as positive.
Determination of in vivo growth kinetics.
Bacteria were administered by gavage to mice, as described above. At each time point, four mice were sacrificed and their livers, spleens, mesenteric lymph nodes, and most-distal Peyer’s patches were aseptically removed. Organs were homogenized in sterile water in a Colworth Stomacher 80 (A. J. Seward, London, United Kingdom), and dilutions were plated to determine numbers of viable bacteria. Results are expressed as organ geometric means ± standard errors of the means.
Physical mapping of mutations.
Physical mapping of transposon insertions was carried out by contour-clamped homogeneous electric field gel analysis of XbaI and BlnI restriction digests of each mutant as previously described (23, 43). Since these restriction sites are not present in MudJ, they had to be introduced. This was achieved by first generating a nalidixic acid-resistant variant of each attenuated mutant and then using these variants as recipients in a conjugation with Escherichia coli DH5α carrying pMAP1. This suicide plasmid, which contains XbaI and BlnI sites, integrates into the MudJ element via homologous recombination with lacZ (41). The map positions of insertions were more precisely located by using flanking DNA from each of the mutant strains (see below) to probe a mapping set of Mud-P22 hybrid phage derivatives, as previously described (4).
Southern blot hybridization, mapping, and sequencing of mutated genes.
AluI, HaeIII, and EcoRI restriction digests of DNA from each mutant were probed with a 300-bp fragment from the left-hand end of phage Mu. The probe was generated by PCR from a clone of Mu using 5′ CTGTT ATTGA AATGA TTT 3′ (forward) and 5′ TGTAT TGATT CACTT GAA 3′ (reverse). Southern hybridization was carried out with the DIG DNA labelling kit (Boehringer Mannheim Biochemicals, Mannheim, Germany) (36). For additional experiments DNA flanking the insertions was first cloned by inverse PCR amplification of AluI- or HaeIII-digested chromosomal DNA using the TA cloning kit (InVitrogen, San Diego, Calif.) and then amplified with primers directly flanking the insert (33). Nucleotide sequencing was carried out by the method of Sanger et al. (37). Sequences obtained were compared to those in the GenBank and EMBL databases by using the programs BlastX and BlastN (2, 15). To determine whether closely related genes were present in other Enterobacteriaceae, the above probes were hybridized under low-stringency conditions (filters were washed at 42°C in 5× SSPE [1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA, pH 7.7]) to EcoRI restriction digests of a range of bacterial strains, including various serovars of Salmonella (among them, S. typhi), pathogenic strains of E. coli, Shigella serovars, Yersinia, and Proteus, as listed by Stojiljkovic et al. (39).
RESULTS
Identification of mutants attenuated by i.g. and i.p. infections.
We found that approximately 3% of the 330 mutants used in this study were non-histidine auxotrophs, consistent with previously published estimates of the number of genes involved in nutrient biosynthesis pathways in Salmonella (18). Histidine auxotrophs were eliminated since they represent P22 transduction of the original MudJ transposon without transposition to new chromosomal sites. The mutants were directly screened for attenuation in vivo by infection of BALB/c mice. All 330 mutants (CL1 to -647) were inoculated by the i.p. route, and the first 197 (CL1 to -429) were inoculated i.g. From a total of 12 attenuated mutants, 4 were found to be attenuated i.p. and 8 were found to be attenuated i.g. (All mutants which were of intermediate virulence or attenuated i.p. were tested i.g. Thus, mutants CL448 [attenuated i.p.], -500, -547, and -554 [intermediate virulence i.p.] were found to be attenuated orally but were not part of the systematic oral screen. They are therefore not included in the calculations of the percentage of the genome which is required for infection by the oral route.) Accurate LD50s were obtained for each attenuated mutant in Table 1. A high degree of attenuation was found for all of the mutants; the lowest LD50 within the i.p. group was more than 4 orders of magnitude higher than that of the parental strain, while, within the orally attenuated group, LD50s were also increased between 103- and 104-fold. All mutants which were attenuated i.p. were also attenuated i.g., but the converse was not true.
TABLE 1.
LD50s and in vitro characterization of avirulent mutants
| Mutant | LD50 (log10)
|
Growth in LB brotha
|
Leakiness (7.5 h)b | Resistance to:
|
Cell invasion
|
Survival in J774 cellsa | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| i.p. | i.g. | 1.5 h | 4 h | DOCc | NRSd | CHO cellsa | Caco-2 cellsa | |||
| CL13 | 6.34 | >11.81 | ++ | ++ | − | ++ | ++ | ++ | ++ | − |
| CL79 | 4.65 | >11.90 | + | + | + | +++ | + | + | + | + |
| CL230 | >7.23 | >9.52 | ++ | + | − | + | − | − | − | + |
| CL448 | 6.68 | >11.81 | + | ++ | +++ | − | − | ++ | ++ | + |
| CL98 | <4.00 | 8.90 | ++ | ++ | − | ++ | + | +++ | ++ | ++ |
| CL238 | <4.00 | >9.05 | ++ | ++ | − | ++ | ++ | ++ | ++ | ++ |
| CL287 | <4.00 | >9.13 | ++ | ++ | − | +++ | ++ | ++ | + | ++ |
| CL288 | <4.00 | >10.11 | ++ | ++ | − | +++ | +/++ | ++ | + | ++ |
| CL401 | <4.00 | >8.35 | + | ++ | + | + | +/++ | + | + | + |
| CL500 | <4.00e | >9.95 | ++ | ++ | − | ++ | ++ | ++ | ++ | ++ |
| CL547 | <4.00e | >11.87 | ++ | ++ | − | ++ | + | + | + | + |
| CL554 | <4.00e | >9.99 | ++ | ++ | − | ++ | ++ | ++ | ++ | ++ |
| 14028 | <1.00f | 5.78f | ++ | ++ | − | ++ | ++ | ++ | ++ | ++ |
++, comparable to the wild-type level; +, <50% of the wild-type level; −, <10% of the wild-type level.
Membrane leakiness was scored qualitatively by comparing the zones of clearing around colonies with that for the wild type.
In most cases there was growth of the inoculum in sodium deoxycholate (DOC), and scores were based on the level of growth observed. −, inoculum killed; +, <50% of wild-type growth; ++, growth comparable to wild-type growth; +++, growth rate exceeding wild-type growth by >50%.
Scores were based on both survival and growth of the original inoculum in normal rabbit serum (NRS), compared with those of the parental strain. −, <10% of the inoculum survived; +, <50% of the inoculum survived; +/++, the inoculum grew by <50%; ++, the inoculum grew by >50%. Comparison of the resistances of mutants to normal and heat-treated sera revealed that CL230, CL448, and CL98 are complement sensitive while CL79 and CL547 appear sensitive to a different serum component.
Between 3 and 4.
Mean for three experiments.
Correlates of attenuation.
The attenuated mutants identified in vivo were tested in a range of in vitro assays in an attempt to find a correlate(s) of attenuation. The results of these analyses are presented in Table 2. We found that almost all of the orally attenuated mutants are prototrophs with a smooth LPS profile and an intact outer membrane. Furthermore, all of the members of this group survive within macrophages. The mutants attenuated for later phases of infection (i.e., the i.p.-attenuated group) either are sensitive to surface-damaging agents or are compromised in their interactions with eukaryotic cells.
TABLE 2.
Genetic analysis of avirulent mutantsa
| Mutant | β-Galactosidase activity
|
Species specificityb | Map position (min) | Sequence homology | Reference | |||
|---|---|---|---|---|---|---|---|---|
| LBc | MinGluc,d | DMEMe | J774e | |||||
| CL13 | − | NG | + | + | 99 | deoA | Blattner et al. (5) | |
| CL79 | + | + | + | + | Salmonella | pVstm | 3′ ORF11 | Friedrich et al. (14) |
| CL230 | + | + | + | + | Salmonella | 80 | ||
| CL448 | + | + | + | + | 17 | tolB | Levengood et al. (20) | |
| CL98 | − | − | − | − | 94 | 5′ phoN | Blattner et al. (5) | |
| CL238 | − | − | − | − | 25 | o85 | Blattner et al. (5) | |
| CL287 | − | + | − | + | IC | yabJ/K | Blattner et al. (5) | |
| CL288 | + | + | + | + | 97 | mdoB | Blattner et al. (5) | |
| CL401 | − | − | − | + | 51 | fmt | Blattner et al. (5) | |
| CL500 | − | + | + | + | 34 | pdx | Blattner et al. (5) | |
| CL547 | − | + | − | + | 70 | yhdJ | Blattner et al. (5) | |
| CL554 | + | + | + | + | 54 | mdh | Blattner et al. (5) | |
| 14028 | − | − | − | − | NA | NA | NA | |
NA, not applicable; NG, no growth; IC, inconclusive.
Mutants which hybridized only with Salmonella are indicated. The remainder hybridized with at least one other enterobacterial strain.
Growth on agar containing X-Gal, scored as blue colonies. Results were similar for aerobic and anaerobic conditions.
MinGlu, minimal glucose medium.
Growth after contact with J774.10 macrophages or DMEM tissue culture medium, scored as blue wells.
Activity of MudJ-lacZ fusions.
Insertion of MudJ can generate a transcriptional fusion between an interrupted gene and lacZ, allowing the expression of the affected gene to be assessed by determination of β-galactosidase activity. We assayed this activity in our attenuated mutants under a number of different conditions (Table 2). Five mutants (CL79, -230, -448, -288, and -554) contained fusions that were expressed under every condition tested, two mutants (CL98 and -238) did not demonstrate β-galactosidase activity under any condition tested, and four mutants (CL13, -287, -500, and -547) contained fusions which were induced by growth on minimal media or in tissue culture media, suggesting perhaps that stress (in this case, nutrient limitation) is critical for regulation of these genes. One mutation appeared to be in a gene which is specifically induced by contact with J774 cells (CL401).
Characterization of selected mutants in vivo.
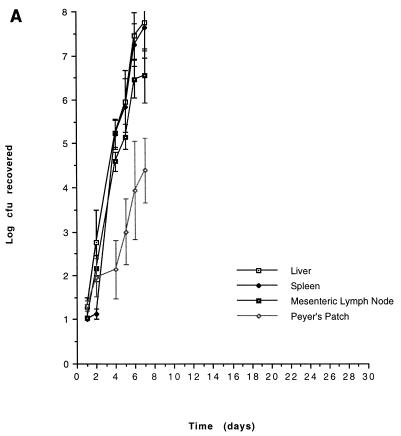
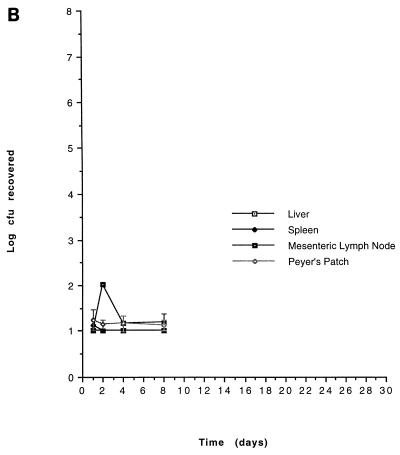
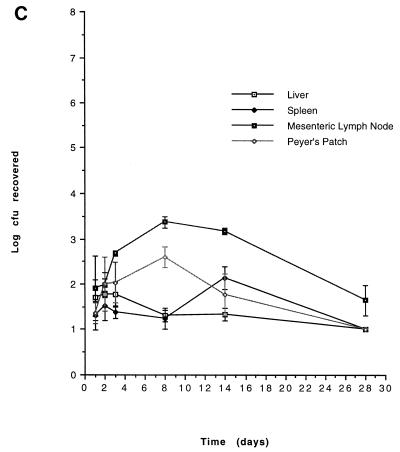
On the basis of our in vitro characterization, the in vivo growth kinetics of two mutants, CL79 and CL448 were examined. The results are shown in Fig. 1. Both CL79 and CL448 had specific blocks in infection that were consistent with phenotypes established by in vitro analyses. CL79 appears unable to penetrate beyond the gut, perhaps as a consequence of its inability to invade the lining epithelial cells of the intestine. CL448, in contrast, can successfully cross the gut, in agreement with its ability to invade epithelial cells in vitro, but is unable to cause fatal infection. Its inability to survive within macrophages in vitro or to resist the bactericidal effects of nonimmune serum also are in agreement with the observed block in infection.
FIG. 1.
In vivo growth kinetics of avirulent mutants. 14028 (A), CL79 (B), and CL448 (C) were administered at doses of log 9.17, log 10.75, and log 10.91, respectively. Results are expressed as geometric means with standard errors of the means.
Physical mapping and sequence analyses of genes identified.
In order to precisely map the mutations and identify the functions of the cognate genes, a combination of physical mapping techniques and DNA sequencing was employed. Flanking sequences from 11 of 12 mutations share DNA sequence similarity with previously identified genes (Table 2). The mutations can be divided into two groups: those with homology to genes of known function and those whose function is unknown. The first group includes the mutations in deoA, a component of the thymidine salvage pathway (5), and the mutation in CL230 which is likely to be in an LPS biosynthetic gene, although no sequence data was obtained for this mutant. The first group also includes mutations in genes with known functions whose attenuating effects were previously unknown: tolB, encoding a transport protein (20); mdoB, encoding a phosphoglycerol transferase (5); yhdJ, encoding an adenine-specific DNA methylase (5); yabJ/K, encoding a putative ABC (ATP-binding cassette) transporter and permease, respectively (5); fmt, encoding a methionyl-tRNA formyltransferase (5); pdx, encoding a pyridoxine kinase (5); and a gene encoding a putative malate oxidoreductase (5). Finally, this group also includes CL79, which contains an insertion in a gene with homology to an open reading frame on the Salmonella virulence plasmid (14). The second group is composed of mutations in genes with no known function and includes mutants CL98 and CL238.
Using DNA flanking the insertions as probes, we carried out Southern hybridization against a variety of Salmonella serovars, including S. typhi, and several other gram-negative bacteria. We found that 10 appear to be located in sequences that are conserved among the Enterobacteriaceae, while the remaining 2 either are not highly conserved or are found within sequences that are unique to Salmonella (Table 2).
DISCUSSION
Bacterial pathogenicity is thought to depend on a vast array of genes acting in concert, many of which are located in complex regulatory pathways that respond to a variety of signals within the host. Given the evidence suggesting that many genes may be involved in virulence, we attempted, for S. typhimurium, to find some of the most essential. By screening directly in vivo and setting stringent criteria for the definition of attenuation, we hoped to identify genes which play a crucial role in infection.
In a collection of 330 transposon insertion mutants, we found that 4 were highly attenuated by the i.p. route of infection. This group represents genes which are required by S. typhimurium to spread via the mediastinal lymph nodes which drain the peritoneum, to the liver and spleen. The first 197 (CL1 to -429) of the mutants were also tested i.g., and 8 (CL13, -79, -98, -230, -238, -287, -288, and -401) were found to be highly attenuated. These represent genes which would be required during a natural infection. Of these eight, we had already identified three (CL13, -79, and -230) in the i.p. screen. Thus, 5 mutants of the first 194 (197 minus 3, to avoid those already counted) were attenuated by the oral route but remained fully virulent i.p. The cognate genes in these mutants are involved in passage of the organism from the stomach to the lymphatic system.
Attenuated mutants were characterized by a range of in vitro assays in an attempt to identify one or more correlates of attenuation. We found that the clearest correlate for those which were attenuated i.p. is an inability to survive intracellularly, a result which supports the widely held belief that Salmonella is an intracellular pathogen and that the products of many genes are involved in intracellular survival. There was no obvious correlate of attenuation i.g., a finding which may reflect the wide range of host defenses the organism must overcome and perhaps underlines the complexity of Salmonella pathogenicity.
Analysis of the mutants at the DNA level revealed that of 12 mutations identified, 11 are located in sequences that have previously been reported or have homology with genes in the database. For some of these, the genetic lesion in question causes a reduction in virulence which is consistent with what is already known about Salmonella pathogenicity, e.g., the deoA mutant (CL13) is a purine auxotroph, a known attenuating mutation (32). Although CL230 has no homology to any of the genes in the database, its LPS phenotype and map location suggest that the insertion in this mutant is in an LPS biosynthesis gene, another attenuating lesion.
The insertion in CL448 (tolB) results in the leakage of cytoplasmic contents, suggesting that the mutant has a cell envelope defect which could render it susceptible to adverse environmental conditions within the host. Similarly, the insertion in CL287 lies in a region with homology to two overlapping genes of unknown function, yabJ and yabK, which are predicted to encode an ABC transporter and a permease (5), respectively.
The insertion in CL79 is found on the S. typhimurium virulence plasmid within ORF11 (14). Our sequence revealed an error in the originally reported sequence (an extra G at position 12906), the correction of which results in a reading frameshift and places the insertion in this mutant outside the ORF and downstream of ORF11, within a 200-bp inverted repeat with a long stem-loop structure. This region has homology to virulence regulatory proteins in E. coli (26) and Yersinia spp. (10). Furthermore, introduction of a clone of the inverted-repeat region surrounding this insertion into the wild-type strain resulted in a 100-fold reduction in the oral LD50, further suggesting a role for this stem-loop structure in virulence gene regulation (data not shown). The degree of attenuation exhibited by CL79 is surprising since deletion of the entire plasmid leads to <10-fold attenuation after i.p. inoculation (17).
Mutant CL547 has an insertion in an adenine-specific DNA methylase which is induced in response to several of the stress conditions we tested. Perhaps this gene could also have a regulatory role, since control of pap pilus expression by uropathogenic strains of E. coli is controlled by dam methylase (31).
The effects of other mutations on virulence, however, are less easy to understand; the disruption of mdoB (CL288), an osmoregulated transferase which adds phosphoglycerol to periplasmic oligosaccharides, also leads to attenuation. Similarly, the mutation in CL401 is in a gene which has only 30% indirect homology with the fmt gene, encoding methionyl-tRNA formyltransferase of E. coli. Unlike fmt mutants of E. coli, however, this mutant does not show a reduced growth rate in rich medium at 37°C (16). These data suggest that the interrupted gene in CL401 probably does not perform this formylation reaction as its primary function. The insertion in CL500 is in a gene with homology to an E. coli ORF, encoding a putative protein which is 51% identical to an enzyme which phosphorylates B6 vitamins. If this is the role of the Salmonella protein, it is once again difficult to suggest a possible role in virulence for the gene.
The MudJ insertion in CL554 interrupts a gene with homology to a putative malate oxidoreductase which is involved in gluconeogenesis, suggesting a role for the cognate gene in the utilization of unusual carbon sources in vivo. Alternatively, since a role in adhesion to epithelial cells for the NADP-linked malate oxidoreductase of Trichomonas vaginalis has recently been described (1), perhaps the Salmonella gene might perform a similar function.
Many of the genes described above perform what might be termed “housekeeping functions.” However, it is interesting to note that Addis and coworkers are not alone in describing cases of housekeeping genes which appear also to have a role in virulence. A further example is that of streptococcal glyceraldehyde-3-phosphate dehydrogenase, which, when located on the bacterial surface, loses its catalytic activity and functions as a plasmin receptor (25, 34). Glutamate dehydrogenase from Porphyromonas gingivalis can also be found on the surface of the organism (19), suggesting perhaps another, nonhousekeeping role for the enzyme.
For the remaining two mutants (CL98 and CL238), however, neither the sequence homologies found nor the phenotypes observed give any clues to the functions of the interrupted genes. Although the insertion in CL98 is in a region upstream of phoN, the mutant has normal acid phosphatase activity, suggesting that the insertion is unlikely to be in a region with any regulatory activity on phoN.
The size of the Salmonella genome has been estimated at 4.9 × 106 bp, and it encodes approximately 4,800 genes (22). Given that the intergenic regions are small in enteric bacteria, our sample of 197 mutants administered i.g. and 330 mutants administered i.p. is equivalent to having sampled about 4 and 7% respectively, of Salmonella genes involved in infection by these routes. We found that of the first 197 mutants screened, about 4% were avirulent i.g., and of these, 3 were also avirulent i.p. If we extrapolate to the entire genome, this percentage is equivalent to a total of approximately 200 and represents all those genes which are required for both stages of a natural infection. Comparison of the numbers of mutants found to be avirulent following i.g. and i.p. infections suggests that most virulence genes are required in the initial phase, presumably reflecting the additional obstacles an infecting organism must overcome.
Because of the stringent definition of attenuation used (LD50 increased by at least 3 orders of magnitude), our calculation above must clearly be a minimum estimate of the number of genes involved. Other factors that might limit the reliability of this figure include the facts that insertions in genes which strongly influence growth rate are less likely to be recovered, insertions in operons and regulators simultaneously inactivate multiple genes, and the insertional specificity of Mu, while certainly extremely low, may not be entirely random (42).
The large number of virulence genes suggested by this study indicates that survival in the host must involve a delicate balance of many genes acting precisely at the right time and in the appropriate environment. What is also interesting is that even if the total number of genes involved were doubled or trebled, the percentage of the genome which does not seem to be specifically required for virulence might be as high as 80%. Perhaps some of these other genes would be required for survival in different hosts or niches or if the infecting dose of organisms were lower, as it would be in nature.
ACKNOWLEDGMENTS
We thank the members of the Heffron and So Labs for advice and critical discussion.
This work was supported by NIH grant ROI AI 22933 to F.H. J. G. Kusters was supported by a fellowship from the Royal Netherlands Academy of Arts and Sciences.
REFERENCES
- 1.Addis M F, Rappelli P, Cappuccinelli P, Fiori P L. Extracellular release by Trichomonas vaginalis of a NADP+ dependent malic enzyme involved in pathogenicity. Microb Pathog. 1997;23:55–61. doi: 10.1006/mpat.1996.0128. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S, F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Amouroux C, Lazzaroni J C, Portalier R. Isolation and characterization of extragenic suppressor mutants of the tolA-876 periplasmic-leaky allele in Escherichia coli K-12. FEMS Microbiol Lett. 1991;62:305–313. doi: 10.1016/0378-1097(91)90176-b. [DOI] [PubMed] [Google Scholar]
- 4.Benson N R, Goldman B S. Rapid mapping in Salmonella typhimurium with Mud-P22 prophages. J Bacteriol. 1992;174:1673–1681. doi: 10.1128/jb.174.5.1673-1681.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blattner F R, Plunkett G R, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 6.Buchmeier N A, Heffron F. Intracellular survival of wild-type Salmonella typhimurium and macrophage-sensitive mutants in diverse populations of macrophages. Infect Immun. 1989;57:1–7. doi: 10.1128/iai.57.1.1-7.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burns N, Grimwade B, Ross-Macdonald P B, Choi E Y, Finberg K, Roeder G S, Snyder M. Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae. Genes Dev. 1994;8:1087–1105. doi: 10.1101/gad.8.9.1087. [DOI] [PubMed] [Google Scholar]
- 8.Carter P B, Collins F M. The route of enteric infection in normal mice. J Exp Med. 1974;139:1189–1203. doi: 10.1084/jem.139.5.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casadaban M J, Cohen S. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc Natl Acad Sci USA. 1979;76:4530–4533. doi: 10.1073/pnas.76.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cornelis G, Sluiters C, Lambert de Rouvroit C, Michiels T. Homology between VirF, the transcriptional activator of the Yersinia virulence regulon, and AraC, the Escherichia coli arabinose operon regulator. J Bacteriol. 1989;171:254–262. doi: 10.1128/jb.171.1.254-262.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis R W, Botstein D, Roth J R. Advanced bacterial genetics: a manual for genetic engineering. Cold Spring Harbor, New York, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 12.Fields P I, Swanson R V, Haidaris C G, Heffron F. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc Natl Acad Sci USA. 1986;83:5189–5193. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finlay B B, Starnbach M N, Francis C L, Stocker B A D, Chatfield S, Dougan G, Falkow S. Identification of TnphoA mutants of Salmonella that are unable to pass through a polarized MDCK epithelial cell monolayer. Mol Microbiol. 1988;2:757–766. doi: 10.1111/j.1365-2958.1988.tb00087.x. [DOI] [PubMed] [Google Scholar]
- 14.Friedrich M J, Kinsey N E, Vila J, Kadner R J. Nucleotide sequence of a 13.9kb segment of the 90kb virulence plasmid of Salmonella typhimurium: the presence of fimbrial biosynthetic genes. Mol Microbiol. 1993;8:543–558. doi: 10.1111/j.1365-2958.1993.tb01599.x. [DOI] [PubMed] [Google Scholar]
- 15.Gish W, States D J. Identification of protein coding regions by database similarity search. Nat Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- 16.Guillon J-M, Mechulam Y, Schmitter J-M, Blanquet S, Fayat G. Disruption of the gene for Met-tRNAfMet formyltransferase severely impairs growth of Escherichia coli. J Bacteriol. 1992;174:4294–4301. doi: 10.1128/jb.174.13.4294-4301.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gulig P A, Curtiss R., III Plasmid-associated virulence of Salmonella typhimurium. Infect Immun. 1987;55:2891–2901. doi: 10.1128/iai.55.12.2891-2901.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hughes K, Roth J. Transitory cis complementation: a method for providing transposition functions to defective transposons. Genetics. 1988;119:9–12. doi: 10.1093/genetics/119.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joe A, Murray C S, McBride B C. Nucleotide sequence of a Porphyromonas gingivalis gene encoding a surface-associated glutamate dehydrogenase and construction of a glutamate dehydrogenase-deficient isogenic mutant. Infect Immun. 1994;62:1358–1368. doi: 10.1128/iai.62.4.1358-1368.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levengood S K, Webster R E. Nucleotide sequences of the tolA and tolB genes and localization of their products, components of a multistep translocation system in Escherichia coli. J Bacteriol. 1989;171:6600–6609. doi: 10.1128/jb.171.12.6600-6609.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levine M. Mutations in the temperate phage P22 and lysogeny in Salmonella. Virology. 1957;3:22–41. doi: 10.1016/0042-6822(57)90021-1. [DOI] [PubMed] [Google Scholar]
- 22.Liu S-L, Hessel A, Sanderson K E. The XbaI-BlnI-CeuI genomic cleavage map of Salmonella typhimurium LT2 determined by double digestion, end labelling, and pulsed-field gel electrophoresis. J Bacteriol. 1993;175:4104–4120. doi: 10.1128/jb.175.13.4104-4120.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu S-L, Sanderson K E. A physical map of the Salmonella typhimurium LT2 genome made by using XbaI analysis. J Bacteriol. 1992;174:1662–1672. doi: 10.1128/jb.174.5.1662-1672.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lodge J, Douce G R, Amin I I, Bolton A J, Martin G D, Chatfield S, Dougan G, Brown N L, Stephen J. Biological and genetic characterization of TnphoA mutants of Salmonella typhimurium TML in the context of gastroenteritis. Infect Immun. 1995;63:762–769. doi: 10.1128/iai.63.3.762-769.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lottenberg R, Broder C C, Boyle M D P. Cloning, sequence analysis, and expression in Escherichia coli of a streptococcal plasmin receptor. J Bacteriol. 1992;174:5204–5210. doi: 10.1128/jb.174.16.5204-5210.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lundrigan M D, Friedrich M J, Kadner R J. Nucleotide sequence of the Escherichia coli porin thermoregulatory gene envY. Nucleic Acids Res. 1989;17:800. doi: 10.1093/nar/17.2.800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mackaness G B, Blanden R V, Collins F M. Host-parasite relations in mouse typhoid. J Exp Med. 1966;124:573–583. doi: 10.1084/jem.124.4.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller V L, Falkow S. Evidence for two genetic loci in Yersinia enterocolitica that can promote invasion of epithelial cells. Infect Immun. 1988;56:1242–1248. doi: 10.1128/iai.56.5.1242-1248.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller F D, Hershberger C L. A quantitative beta-galactosidase alpha-complementation assay for fusion proteins containing human insulin B-chain peptides. Gene. 1984;29:247–250. doi: 10.1016/0378-1119(84)90185-9. [DOI] [PubMed] [Google Scholar]
- 30.Mizuuchi K, Craigie R. Mechanism of bacteriophage Mu transposition. Annu Rev Genet. 1986;20:385–429. doi: 10.1146/annurev.ge.20.120186.002125. [DOI] [PubMed] [Google Scholar]
- 31.Nou X, Skinner B, Braaten B, Blyn D, Hirsch D, Low D. Regulation of pyelonephritis-associated pili phase-variation in Escherichia coli: binding of the PapI and the Lrp regulatory proteins is controlled by DNA methylation. Mol Microbiol. 1993;7:545–553. doi: 10.1111/j.1365-2958.1993.tb01145.x. [DOI] [PubMed] [Google Scholar]
- 32.O’Callaghan D, Maskell D, Liew F W, Easmon C F, Dougan G. Characterization of aromatic- and purine-dependent Salmonella typhimurium: attenuation, persistence, and ability to induce protective immunity in BALB/c mice. Infect Immun. 1988;5:419–423. doi: 10.1128/iai.56.2.419-423.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ochman H, Medhora M M, Garza D, Hartl D L. Amplification of flanking DNA by inverse PCR. In: Innis M A, Gelfand D H, Sninsku J J, White T J, editors. PCR protocols. San Diego, Calif: Academic Press; 1990. pp. 219–227. [Google Scholar]
- 34.Pancholi V, Fischetti V A. Glyceraldehyde-3-phosphate dehydrogenase on the surface of group A streptococci is also an ADP-ribosylating enzyme. Proc Natl Acad Sci USA. 1993;90:8154–8158. doi: 10.1073/pnas.90.17.8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reed L J, Muench H. A simple method for estimating fifty percent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, New York, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stainer R Y, Ingraham J L, Wheelis M L, Painter P R. General microbiology. 5th ed. London, United Kingdom: MacMillan; 1987. [Google Scholar]
- 39.Stojiljkovic I, Schonherr R, Kusters J G. Identification of the hopG gene, a component of Escherichia coli K-12 type II export system, and its conservation among different pathogenic Escherichia coli and Shigella isolates. J Bacteriol. 1995;177:1892–1895. doi: 10.1128/jb.177.7.1892-1895.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsai C M, Frasch C E. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem. 1982;119:115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]
- 41.Tsolis R M, Bäumler A J, Stojiljkovic I, Heffron F. Fur regulon of Salmonella typhimurium: identification of new iron-regulated genes. J Bacteriol. 1995;177:4628–4637. doi: 10.1128/jb.177.16.4628-4637.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang X, Higgins N P. “Muprints” of the lac operon demonstrate physiological control over the randomness of in vivo transposition. Mol Microbiol. 1994;12:665–677. doi: 10.1111/j.1365-2958.1994.tb01054.x. [DOI] [PubMed] [Google Scholar]
- 43.Wong K K, McClelland M. A BlnI restriction map of the Salmonella typhimurium LT2 genome. J Bacteriol. 1992;174:1656–1661. doi: 10.1128/jb.174.5.1656-1661.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]