Abstract
Microbiomes are generally characterized by high diversity of coexisting microbial species and strains that remains stable within a broad range of conditions. However, under fixed conditions, microbial ecology conforms with the exclusion principle under which two populations competing for the same resource within the same niche cannot coexist because the less fit population inevitably goes extinct. To explore the conditions for stabilization of microbial diversity, we developed a simple mathematical model consisting of two competing populations that could exchange a single gene allele via horizontal gene transfer (HGT). We found that, although in a fixed environment, with unbiased HGT, the system obeyed the exclusion principle, in an oscillating environment, within large regions of the phase space bounded by the rates of reproduction and HGT, the two populations coexist. Moreover, depending on the parameter combination, all three major types of symbiosis obtained, namely, pure competition, host-parasite relationship and mutualism. In each of these regimes, certain parameter combinations provided for synergy, that is, a greater total abundance of both populations compared to the abundance of the winning population in the fixed environments. These findings show that basic phenomena that are universal in microbial communities, environmental variation and HGT, provide for stabilization of microbial diversity and ecological complexity.
BACKGROUND
The extensive recent efforts in metagenomics have led to the realization that microbiomes, for example, the well-studied gut microbial communities, are surprisingly stable on the species level over long periods of time [1–3]. Changes in microbiome content caused by such factors as antibiotics intervention or diet change can lead to obesity, inflammatory bowel disease and other pathological conditions, so that microbiome stability is essential for human health [4]. Bacterial and archaeal species within the microbiome have metabolic networks that complement each other, and are thought to cooperate [5–7]. However, there are also indications of strong interspecies competition in complex microbial communities such as soil [8].
With the development of single cell metagenomics, finer structure of microbiomes was discovered, demonstrating that most of the constituent bacterial species are represented by multiple, closely related strains [9–12]. The coexistence of multiple strains has been shown to be common, and moreover, strain composition tends to be stable over extended periods of time [10, 13, 14]. It is generally assumed that the observed stability of the strain composition can be accounted for by models with no competition between strains [15]. Whereas prevalence of cooperation has been demonstrated for different microbial species, metabolic networks of strains of the same species are closely similar, so that competition for resources is expected to occur. Indeed, attempts on administration of probiotics have shown that bacteria, occupying the same niche as the species that is already dominant in the microbiome, are eliminated [16, 17].
A key process in the evolution of prokaryotes is the extensive horizontal gene transfer (HGT) that occurs via transformation, transduction or conjugation (reviewed in [18]). The size of the transferred DNA fragments varies from several hundred nucleotides to segments of many kilobases encompassing multiple genes. Horizontal gene transfer is thought to be essential for the survival of microbial populations [19] and appears to be largely responsible for rapid adaptation to new environments and even for the emergence of major clades of bacteria and archaea with distinct lifestyles, such as acetoclastic methanogens or extreme halophiles [20, 21]. The rate of HGT is highly non-uniform across the tree of life and across environments [22]. Indeed, there is growing evidence that HGT rate can be influenced by the environmental changes. For example, it has been shown that natural competence of the bacterium Staphylococcus aureus is induced by reactive oxygen species, antibiotics and host defenses [23, 24]. Importantly, the HGT rate is much higher between closely related organisms compared to distantly related ones thanks, primarily, to the high rate of homologous recombination [25–27].
In recent years, microbiome stability and variation have become a major problem in microbiology, with important implications for human health [28, 29]. Horizontal gene transfer plays a crucial role in the preservation of genome diversity [30–36] but the effects of environmental variations on the microbiome composition remain poorly understood. Evolutionary processes in populations in time-varying environments can drastically differ from those in fixed environments [37–48]. The complexity of microbial communities including multiple strains that persist for extended periods of time and widespread HGT among them call for developing theoretical models describing the relationships between strains within complex microbial communities, that are expected to compete with each other, but also constantly exchange genes via HGT.
Here we describe the simplest possible model that includes competition and HGT between two microbial populations differing by a single gene allele in a fixed or oscillating environment. The time-varying environment drastically changes the evolutionary scenarios observed in a fixed environment. We show that in this setting environmental variation plays a crucial role in the preservation of genome diversity. The coarse-graining modeling presented here allows us to uncover different types of symbiotic relationships that arise due to environmental variation, spanning all possible scenarios, from pure competition to mutualism [49–52].
MODEL OF MICROBIAL POPULATIONS DYNAMICS
We explore a dynamic evolutionary scenario where two populations engage in both within- and between -population competition for shared resources. The two populations differ by a single gene allele, resulting in differential fitness. HGT plays a key role in the model, mediating the exchange of genetic material between the two populations by copying the gene from the donor and replacing the corresponding gene in the recipient. For instance, consider the case when population harbors allele 1, and population carries allele −1. If is the donor and is the recipient, then the allele from replaces that in , and as a result, the recipient changes its type , and the population sizes of and increase and decrease by one, respectively. The donor and recipient entities are selected randomly within their respective populations. The rates of gene transfer vary depending on whether or is the donor. Inactivation of genes by mutations is disregarded.
To capture the dynamics of this HGT-mediated interaction mathematically, we derive a system of differential equations that describes the continuous variation of population sizes over time:
| (1) |
| (2) |
where and are population sizes of and at time , respectively. and are reproduction rates of and , respectively. and are within- and between- population competition rates. is the balance of gene transfer between and types, i.e corresponds to the positive gene flow from to , when the HGT rate from to is greater than the HGT rate from to . The rates describing the competition, both within and between two populations, are assumed to be positive , , and the fitness of each population is a decreasing function of the presence of competing population, corresponding to a purely competitive ecosystem in the absence of HGT. The detailed derivation of these equations from elementary processes, based on previous work [53, 54], is provided in the Additional File1. For the purpose of the present manuscript, we will describe the system of (1, 2) by the equivalent representation through the total abundance of both populations and fractions of each population , , such that . From (1,2), we obtain the following dynamical system representing the time variations of total abundance of both populations and the fraction of each population:
| (3) |
| (4) |
Equation (3) represents well-known replicator dynamics in evolutionary game theory [55–59]. However, the crucial difference between (3,4) and standard replicator dynamics is the dependence of fitness functions on the total population size . The fitness of each population can be represented as the expected payoff obtained from a symmetric game with the payoff matrix shown in Fig.1. The payoffs of each strategy/population and depend on the total abundance of both populations. The time variation of the total abundance of both populations is defined by the mean fitness of both populations. In the equilibrium states, when the right-hand sides of both equations (3,4) nullify, the mean fitness and the fitness of each population represented in the equilibrium state nullify as well. Solving the right hand-side of (3,4) with respect to the fractions of each population (taking into an account the normalization ) and total population abundance, we find 4 possible non-trivial outcomes: , , – if , along , then depending on the stability of equilibrium the outcome is either coexistence of both populations or bistability, where extinction of a population depends on the initial condition, and the interior unstable equilibrium separates basins of attraction of two stable equilibrium states representing extinction of either or populations. The trivial equilibrium state, corresponding to the extinction of both populations, is always unstable for positive reproduction rates , .
FIG. 1: Evolutionary dynamics of two competing populations with HGT in a fixed and in a fast-oscillating environment.
Left, fixed/averaged environment; right, oscillating environment. The coarse-grained time variations of the fractions of each population and the total abundance of both populations are described by the replicator dynamics with density dependent payoff matrices. The fitness of each population is comprised of two terms obtained from two different evolutionary games. These evolutionary games represent the interaction between the two populations in the fixed/averaged and varying environments (left and right matrices, respectively). In the left matrix, and are reproduction rates of the populations and in the averaged/fixed environment, respectively. The competitions within and between the two populations are given by , rates. is the gene transfer balance in the averaged/fixed environment. and represent environmental variations in the model, these are defined by the product of oscillatory parts of reproduction and gene transfer balance rates, defined in (11). is the total abundance of both populations.
The stability of equilibrium states with the trivial composition of the total population is defined by the following conditions:
| (5) |
for and , respectively. For , the conditions (5) link between-populations and within-populations competition with the balance of the reproduction rates. Indeed for , a given population outcompetes the opponent if the competition within the population of the winner is less severe than the competition between-populations.
If both conditions in (5) are satisfied simultaneously, along with , then one obtains a bistable dynamical system, where the outcome of competition depends on both the initial abundance and fractions of both populations. The last condition is linked to the sign of the determinant of the community matrix, represented via the competition and gene transfer balance rates
| (6) |
Coexistence is obtained, once both conditions in (5) are violated simultaneously along with
| (7) |
that is the .
So far, we analyzed the model for general competition and reproduction rates, without imposing any other condition on the rates except for being positive. Therefore, our initial assumption is that populations and differ only by a single gene allele, and we further assume that the alleles of this gene affect only the reproduction rates and , whereas the rates describing the competition within and between populations are independent of the gene allele variation, and are equal, . Thus, for equal reproduction rates and balanced gene transfer between two populations , the total abundance of both populations is always equal to . Coexistence of both populations could not be achieved in the case of balanced gene transfer and unequal reproduction rates , inasmuch as it is impossible to violate both conditions in (5) simultaneously. This impossibility of coexistence between populations corresponds to the exclusion principle which states that, if both populations compete for the same resources, then, one of them will be eliminated, under equal competition rates ; in our setting, the exclusion principle applies in the case of balanced gene transfer [60–62].
In the case of unbalanced gene transfer coexistence of the two competing populations is possible if
| (8) |
that is, both conditions in (5) are violated and (7) is satisfied simultaneously, under equal competition rates .
The above conclusions were obtained for a fixed environment. We now consider the same evolutionary process in a fluctuating environment. We assume that the environment oscillates faster than the characteristic timescale of the population dynamics, that is, both populations experience many changes of the environment before reaching the dynamic equilibrium. The environmental variations are incorporated in the model by assuming that the reproduction rates and the gene transfer balance rate are periodical function , and with period , where , describes fast variations of the environment. Here, is the slow-time, that represents coarse-grained variations in competing populations. The coarse-graining procedure is based on the elimination of the fast-oscillating terms by identifying the dynamical impact of these terms on the slow-time behavior. That is, the fast-varying terms introduce an ”effective potential” in the slow-time behavior, and these fast-oscillating terms are themselves affected by the quantities varying in the slow-time. The time dependent reproduction and gene transfer balance rates are represented as the sum of fixed and oscillatory parts and , where the time averages of oscillatory parts nullify over a period of environmental variations, that is and . That is, and represent the reproduction and gene transfer balance rates, respectively, in the averaged/fixed environment. Considering the evolutionary processes with averaged rates yields the system (3, 4) and the conclusions obtained for the fixed environment.
The derivation of the slow-time behavior is given in the Additional File1; here, we briefly outline the assumptions and implementation steps of this derivation. We seek the solution for the composition and total abundance of both populations as a sum of slow varying and oscillating parts, that is and , respectively. The fast oscillating parts yield zero average over the period of environmental variations . Note that the fast oscillating quantities and depend on the slow varying counterparts and . Putting the expressions of time-dependent rates , together with and back into (3,4), expanding the right hand side over , , seeking the fast oscillating parts as a power series with as the order parameter, that is and , separating the quantities varying in different time scales, averaging over a period of environmental variations and keeping the terms up to , we find the slow time behavior of and
| (9) |
| (10) |
Thus, in the oscillating environment, the coarse-grained variations of the fraction of each population and total abundance of both populations, again, are given by the replicator dynamics, but the fitness of each population obtains an additional term due to the oscillating environment (Fig.1). Note, that , is given by the average/fixed rates, as in (3,4). The new terms ϕi are expressed through the oscillating rates of reproduction and gene transfer balance as follows
| (11) |
In (11), and denote the primitives of and , respectively, that is . The existence and stability of possible equilibria of (9,10) are described by the conditions (5,7), where the between-population competition rates are substituted as follows and . The initial conditions for (9,10) are found by solving the following system of algebraic equations and , where , are initial conditions for (9,10), , are the initial conditions for (3,4) with time-dependent payoffs and , are the leading order oscillating terms proportional to . The latter terms are represented through , and time-dependent rates and (see Supplementary Information for further details).
RESULTS
Environmental oscillations enable coexistence of competing populations
In the present model, oscillations in the environment induce a new evolutionary game between the competing populations (Fig.1). The emergence of the new game results from the coarse-graining of the effects of environmental variations on the competing populations. Specifically, the fast-oscillating terms induce an ”effective potential”, a new game that alters the slow-time dynamics of the fractions of the two populations and total population abundance of both populations. The coarse-grained variations of the total population structure and abundance are defined by additive fitnesses obtained from two distinct games, one of which represents the evolutionary process in the fixed/averaged environment, and the other, emerging game represents the coarse-grained impact of the oscillating environment (11). The fitness of each population in the new game is expressed through the overlap in the oscillations of gene transfer balance rate and the reproduction rate of the competitor over the period of environmental variations. The impact of the environmental variations is trivial if the oscillating part of the reproduction rates and gene transfer rate is the same, that is if , , then , . To substantially influence population growth and composition compared to the fixed environment, environmental variations must differentially affect the reproduction and gene transfer balance rates. The new payoffs , nullify when for both the bounded oscillations of reproduction and gene transfer rates, as it follows from (11). That is, if environmental oscillations are too fast, then only the averages of reproduction and gene transfer balance determine the evolutionary outcome.
The payoff structure of the emerged game is simple for any positive population abundance . These payoffs are fixed once the time dependence of reproduction and gene transfer balance rate is known. Then, either there is a dominant strategy in the game (that is, either or always win in the new game)–if and have the same sign (for example, dominates if , ), or there is a non-trivial equilibrium for any if and have different signs [56, 58, 63].
Environmental oscillation can result in coexistence of the two populations in a regime for which it is impossible in a fixed environment, that is, when gene transfer is balanced between competing populations and the allele difference affects only the reproduction rates . In a fixed/averaged environment, the exclusion principle applies so that only the population with the higher reproduction rate survives, giving the total abundance of both populations as .
Assuming that and gene transfer is balanced on average , we substitute , in (5,7) and find the following conditions for the existence and stability of the coexistence of the two populations in the coarse-grained dynamics (9,10)
| (12) |
| (13) |
Let us assume that the average reproduction rates of both populations are almost equal, 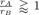 . Then, from (12), it follows that, in a varying environment coexistence is possible for and , whereas, from (5), it follows that in a fixed environment coexistence is impossible if and . From (12) it follows that environmental variations dampen the between-population competition such that the competition within a population becomes more severe than the competition between the populations. This is the case because environmental variation shifts the between-population competition rates and , as follows from emerged fitness terms (11), thus reducing between-population competition compared to the within-population competition.
. Then, from (12), it follows that, in a varying environment coexistence is possible for and , whereas, from (5), it follows that in a fixed environment coexistence is impossible if and . From (12) it follows that environmental variations dampen the between-population competition such that the competition within a population becomes more severe than the competition between the populations. This is the case because environmental variation shifts the between-population competition rates and , as follows from emerged fitness terms (11), thus reducing between-population competition compared to the within-population competition.
The total abundance of both populations in the coexisting equilibrium state, obtained from the coarse-grained representation of the oscillating environment, is
| (14) |
The conditions (12) and (13) provide , ensuring that the numerator and denominator are both positive, respectively.
The phase space of interactions between populations
Environmental oscillations can result not only in coexistence but also in a synergistic interaction between the two competing populations for given reproduction rates and . By synergy, we refer to the situations when the presence of the two populations in the equilibrium state increases the total abundance of both populations compared to the total abundance in the equilibrium state in a fixed environment . The synergy emerges, because the presence of both populations increases the utilization of available resources in the environment. Comparing, the total abundance of both populations in the varying environment-induced coexistence regime (14) and that in the fixed/averaged environment , we find that
| (15) |
Assuming , we now consider different regions of the parameter space bounded by and , where (12) is satisfied, corresponding to the different scenarios. The separation of these regions must be complemented with (13) for the coexistence equilibrium to be stable and the total abundance of the two populations to remain positive.
Coexistence without synergy: pure competition and parasitism:
and , region I in Fig.2a.
FIG. 2: Phase space for possible outcomes of the coarse-grained description of two competing populations in an oscillating environment (9,10), for different values of environment-induced fitness terms.
(a), partitioning of the phase space into distinct regions (see color code). Regions I, II, and III correspond to the coexistence of the two populations. In the regions II and III, there is synergy between the competing populations, that is, in the unique equilibrium state, the total abundance of both populations is greater than that in the fixed environment (for the considered values of the model parameters, outcompetes in the fixed environment). In regions IV, outcompetes , like in a fixed environment, V is the region of bistability, where either or wins depending on the initial state, and in region VI, outcompetes , opposite to the outcome in the fixed environment. Regions IV, V and VI are defined by violations of one or more of the conditions (12,13). N/A represents the region where the coarse-grained approach fails, that is (12) holds, but (13) is violated.
(b), detailed structure of the regions I, II, III and N/A. The hatched areas represent the regions where the fitnesses of the two populations are differently affected by the presence of the competitor. The solid black curves show the total abundance of both population defined by (14), increasing from I to III. The model parameters are , , and .
In this region, the fitness of , the winner in the fixed environment, is lower than it would be in the latter case. Indeed, the total fitness of is , where (Fig.1). Conversely, the population obtains a positive fitness surplus due to the environmental oscillations . In the emerged game, always has an advantage over . Thus, dominates over in a fixed environment, but has the advantage in the new game induced by the oscillating environment. In this region, there is coexistence but no synergy between two populations, that is, the total abundance of the two populations is lower than that in the fixed environment with same parameters, .
The coarse-grained approach allows us to identify distinct sub-regions of different symbiotic relations between the two populations, that is, define the impact of the opponent on the fitness of a population. Region I includes two sub-regions that correspond, respectively, to competition and parasitism. In the competitive sub-region and , the fitness of each population is a decreasing function on the fraction of the competitor, that is and . The latter quantity changes its sign for , so in the corresponding sub-region, is a parasite of , dashed sub-region of I in Fig.2b. Due to environmental oscillations, the above derivatives can change their signs in the case of time-dependent rates, hence, the type of the symbiotic relation between the two populations cannot be inferred without coarse-graining.
Synergy between populations: pure competition:
and , region II in Fig.2a. The payoffs of the emerged game are positive , , in contrast to region I. Thus, the two populations coexist due to the balance between the two games. The fitness of each population is a decreasing function of the competitor’s fraction , , hence, the interaction is purely competitive. Nevertheless, the total abundance of both populations is greater than that in the fixed environment , as follows from (15), corresponding to increased utilization of the available resources.
Synergy: parasitism and mutualism:
, , and , corresponding to parasitism and mutualism, respectively, in two sub-regions of region III (Fig.2a,2b).
In the sub-region , the fitness of is positively affected by the presence of the competitor , that is , whereas the fitness of is negatively impacted by , that is , so that is a parasite of . Conversely, in the sub-region, , fitness of is negatively impacted by , whereas positively affects the fitness of . Accordingly, the host-parasite relationship is reversed.
The symbiosis between the two populations is mutualistic when . In this sub-region of region III, the fitness of each population is an increasing function of the fraction of the other population , (2b). The total abundance of both populations is greater than that in the fixed environment in both parasitic and mutualistic symbiosis, that is, both types of symbiosis are synergistic.
The total abundance of both populations (14), , increases from region I to region III. The total abundance of both populations in the fixed environment corresponds to line, thus, by substituting in (14), one recovers . In the fixed environment, only population survives, so that the total abundance equals to the abundance of , whereas in the oscillating environment, both and are present in equilibrium with fractions and , respectively.
The coarse-grained approach fails as one gets closer to the curve , where , whereas the time-dependent solution is bounded (N/A regions in Fig.2). Unbounded growth corresponds to violation of (13) that holds as equality. Below the curve defined by , . The fraction of each population satisfy , . Thus, there is a non-trivial composition of both populations, but, there is no physical level of total abundance. This is one of the differences from the well-known replicator dynamics, which deals with the composition of the total population only. The reason behind the failure is the unbounded growth of both populations (see Supplementary Information for details on the limitations of the coarse-grained approach).
Figure 3 illustrates the solutions for the fractions and total abundance of two populations, for both time-dependent rates and the coarse-grained behavior (9,10).
FIG. 3: Fractions and total abundance of the two populations.
(a), fraction of the population for different values of and .
(b), total abundance of both populations for different values of and .
Oscillating curves show the solutions of (3,4) with time-dependent reproduction and gene transfer balance rates. (region I in Fig.2; blue curve), (region II; orange curve) and (region III; green curve). The time dependent rates are as follows , and , where , and (−2.2, 2.2) for the blue, orange and green curves, respectively. The remaining model parameters are , , , and . The dashed line in (b) represents the total abundance of both populations in the fixed environment .
The ordering of fractions and total abundance does not follow a uniform pattern across the different scenarios of interaction between the populations. In the case of pure competition with synergy, the equilibrium fraction of population surpasses that in the mutualistic scenario. However, the total abundance of both populations is greater in the mutualistic scenario than in the pure competitive case. The scenario with no synergy (region I in Fig.2) yields the smallest total abundance for both populations as well as, for the fraction of population . These observations imply complex relationships between competition, synergy, and mutualistic symbiosis, which affect both population fractions and overall abundance.
So far, we addressed the emergence of coexistence between two competing populations due to fast environmental variations. In other parts of the phase space, however, the environmental variations can preserve the outcome of the competition in the fixed environment, that is, outcompetes (region IV in Fig.2), or reverse it (region VI) resulting in the domination of and extinction of . The former scenario occurs if and , that is, the inequality (12) is reversed for . Conversely, outcompetes if the inequality is reversed for , whereas that for holds. Finally, the bistable dynamics (region V) is observed if (12) and (13) are simultaneously violated.
Unbalanced gene transfer in a fixed environment can result in stable coexistence of the two populations (8). In this case, environmental variations can alter the composition and the total abundance of the two populations and even destroy the coexistence of the two competing populations. Indeed, substituting , and in (5, 7), we obtain the conditions for the stable coexistence of both populations with unbalanced gene transfer in the oscillating environment:
| (16) |
| (17) |
The conditions (16) and (17) are the counterparts of (12) and (13), respectively, for the case of unbalanced gene transfer in a fixed environment. From (16,17) and (8), together, it follows that the environmental variations can change the composition and the total abundance of the two populations in the coexisting equilibrium. Environmental oscillations destroy the coexistence of the two populations observed in the fixed environment if any of the conditions (16,17) is violated, but (8) holds.
The limitations of the model are discussed in the Supplementary Information. There, we compare the solutions obtained by numerically integrating the system (3,4) with time-dependent rates and the coarse grained counterparts obtained via (9,10). The comparison is based on the Euclidean distance of the time-averaged fractions of both populations obtained via (3,4) and those obtained from (9,10). The difference is negligible for the regions IV, V, VI, but not for the coexistence regimes. Nevertheless, even in the worst case scenario, both methods show the coexistence of both populations although the fractions of the two populations at equilibrium differ. We also compare the relative error of total population abundances. The relative error increases as one approaches the boundary where (N/A), thus increasing the discrepancy between the results of the coarse-grained approach and the time average of the solution obtained via time-dependent rates, which remains bounded in general.
DISCUSSION
In this work, we developed a mathematical model to explore the role of HGT in the interaction between two cohabiting populations in an oscillating vs a fixed environment. We explored what seems to be the simplest possible model in which two populations differed by a single gene allele, such that each entity in the pool carried only one allele of the given gene that is subject to HGT through copying the gene in the donor cell, and then, substituting the existing gene of the recipient cell, which corresponds to homologous recombination [14, 18]. Notwithstanding the ultimate simplicity of this scheme, it is biologically realistic, for example, in the context of acquisition of multi-drug-resistance [64]. We start with the stochastic description of all possible interactions between two populations, namely, reproduction and death of each population, competition within and between populations, and gene transfer between the populations. Then, we take the continuous limit and obtain the deterministic description of the time variations of the size of each population (see Additional File1, [53, 54]). The dynamical system obtained by this procedure is a variation of the well-known replicator dynamics [55–59], with varying total abundance of both populations. The composition and the total abundance of both populations in the equilibrium states are found from the rest points of this system.
By examining this mathematical model, we show that stable coexistence of the two populations in a fixed environment is unattainable within the assumption that the gene allele replacement via HGT only affects the reproduction rate, the competing abilities of both populations are the same within and between populations, and HGT between the populations is balanced, that is, there is no preferred direction in the gene flow between the populations. These results reflect the classic competitive exclusion (Gauze) principle according to which, in a spatially homogeneous environment, a population with an inferior reproduction rate will inevitably go extinct when competing with a fitter population for the same resource, within the same niche [60–62]. Notable, however, if the HGT is unbalanced, that is, the rates of gene transfer in the two directions are unequal, the exclusion principle does not apply anymore, and the two populations can coexist.
Evolutionary outcomes in a time-varying environment can drastically differ from those in the fixed environment, and despite the simplicity of our model, the emerging dynamics is complex, with the outcomes critically depending on the parameter combination. Environmental oscillations are incorporated into the model by assuming that the reproduction and gene transfer rates are time-periodic functions, with the implicit assumption that these rates are affected by the changes in the environment. The oscillations were set up such that the averages across environmental variations coincided with the constant rates in the fixed environment, providing for a fair comparison of the dynamics in the two types of environment.
The environmental oscillations occur on a fast time scale, whereby the populations are exposed to numerous changes in the environment before approaching any potential equilibrium. We adopted a coarse-graining approach to account for the environmental variations whereby the solution for the composition and total abundance of both populations was obtained as a combination of two components, one delineating the slow-time (coarse grained) behavior and the other capturing the oscillating component [44, 45]. By averaging over the period of environmental variations and retaining the first two leading-order terms of the varying quantities, we obtained the replicator dynamics model with varying total abundance that features fitness contributions derived from two distinct games. The first game represents the fixed/averaged environment whereas the second one arises due to environmental fluctuations, with the payoffs in this game determined by the intersection of the oscillating components of the reproduction and gene transfer equilibrium rates. These new payoffs are such that the fitness of a particular population is defined by the average of the product of the competitor’s reproduction rate and the gene transfer equilibrium rate between the populations throughout environmental oscillations. For the emergence of the new game, reproduction and gene transfer balance rates must be differentially affected by the environment. Notably, variations either in the reproduction rates or in the gene transfer rate alone did not result in new evolutionary scenarios compared to the fixed environment case.
The emerged game maintains a straightforward payoff structure: once the environmental variations of reproduction and gene transfer rates are given, then either one of the strategies consistently dominates, or there exists a unique non-trivial equilibrium for any given total abundance of both populations. With these adjusted fitness terms, stable coexistence between the two populations becomes possible within a broad range of model parameters. Moreover, depending on the combination of the time-dependent reproduction and gene transfer balance rates, the co-existence of the two populations manifests as all major types of symbiotic relationships. Two populations can coexist in a purely competitive symbiosis, where the fitness of each is negatively impacted by the presence of the other population; a host-parasite relationship, whereby the impact of the second population is positive for one and negative for the other population; and in mutualistic symbiosis, where the presence of the other population is reciprocally beneficial.
We further analyzed the behavior of the total abundance of both populations and defined the regions of synergistic interactions between the competing populations. In this case, synergy is observed, that is, the total abundance of the two populations at the stable coexistence in the oscillating environment is greater than the total abundance in the fixed environment, that is, the population size of the winner at equilibrium, under the same model parameter values. As could be expected, mutualism necessarily entails synergy and yields the greatest total abundance of both populations among all the regimes by increasing the resource utilization in the environment. Notably, however, under certain combinations of the reproduction and HGT rates, the synergistic effect was observed also in the cases of parasitism and even pure competitive symbiosis.
Outside of the coexistence regimes, the outcomes of the competition between two populations in a stable environment can persist in the oscillating environment such that the winner in the fixed environment wins in the oscillating environment as well. However, in another region of the phase space, the outcome can be reversed due to environmental oscillations. Moreover, there is also a bistability regime where one or the other population goes extinct depending on the initial conditions, a scenario precluded in the fixed environment assuming equal competing abilities and balanced gene transfer. Despite our prior discussion on coexistence and potential outcomes in an oscillating environment with balanced gene transfer in the fixed environment, the introduction of oscillations can notably alter the composition at the coexistence equilibrium, even if both populations coexist in the fixed environment due to non-zero gene transfer balance. Environmental variations, in this case, can even destroy the stable coexistence of the two populations, attained in the fixed environment with unbalanced HGT.
From the methods point of view, it is worth pointing out that coarse graining was an essential ingredient of the present analysis because without it, the symbiotic relationships between two populations would be impossible to elucidate because the fitness gradients can change their signs throughout environmental oscillation due to the oscillating rates.
From the biological standpoint, environmental oscillations provide for the synergy between the two populations by lowering the intensity of the inter-population competition below the level of the intra-population competition. This work shows that stabilization of strain diversity and increase of ecological complexity via HGT in an oscillating environment is an intrinsic feature of even the simplest microbiomes that emerges under minimal assumptions on the basic processes occurring within a microbial community. Given that both environmental variation and HGT are ubiquitous phenomena that affect any microbiome [65, 66], these conclusions appear to be broadly applicable. Notably, all emerging coexistence regimes provide for synergy between the populations indicating that HGT in an oscillating environment is favorable for a microbial community perceived as an integral whole.
Evidently, the model used in this work is grossly (and deliberately) over-simplified. Real microbiomes encompass interactions among thousands of microbial strains and species, HGT of multiple genes via different routes and many other processes [14, 50, 66]. Nevertheless, the general principle established here should apply, amplified by the microbiome complexity. Characterization of the conditions for stabilization of microbiome diversity and the factors that can perturb it is crucial for understanding the role of the microbiome in health and diseases as well as the ecology of microbial communities.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are thankful to Armen E. Allahverdyan and Koonin group members for valuable discussions.
Funding Statement
The authors are supported through the Intramural Research Program of the National Library of Medicine, National Institutes of Health.
Footnotes
SUPLLEMENTARY INFORMATION
The manuscript contains supplementary material. Additional File1 contains derivations and analysis of (1,2) and (9,10), and limitations of the proposed model.
ETHICS APPROVAL AND CONSENT OF PARTICIPATE
Not applicable.
COMPETING INTERESTS
The authors declare they have no competing interests.
AVAILABILITY OF DATA AND MATERIALS
This is a theoretical work that did not involve data analysis.
References
- [1].Chen Lianmin, Wang Daoming, Garmaeva Sanzhima, Kurilshikov Alexander, Vila Arnau Vich, Gacesa Ranko, Sinha Trishla, Segal Eran, Weersma Rinse K., Wijmenga Cisca, Zhernakova Alexandra, and Fu Jingyuan. The long-term genetic stability and individual specificity of the human gut microbiome. Cell, 184(9):2302–2315.e12, April 2021. ISSN 0092-8674. doi: 10.1016/j.cell.2021.03.024. [DOI] [PubMed] [Google Scholar]
- [2].Mehta Raaj S., Abu-Ali Galeb S., Drew David A., Lloyd-Price Jason, Subramanian Ayshwarya, Lochhead Paul, Joshi Amit D., Ivey Kerry L., Khalili Hamed, Brown Gordon T., DuLong Casey, Song Mingyang, Nguyen Long H., Mallick Himel, Rimm Eric B., Izard Jacques, Huttenhower Curtis, and Chan Andrew T. Stability of the human faecal microbiome in a cohort of adult men. Nat Microbiol, 3(3):347–355, March 2018. ISSN 2058-5276. doi: 10.1038/s41564-017-0096-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Huttenhower Curtis, Gevers Dirk, Knight Rob, Abubucker Sahar, Badger Jonathan H., Chinwalla Asif T., Creasy Heather H., Earl Ashlee M., FitzGerald Michael G., Fulton Robert S., Giglio Michelle G., Hallsworth-Pepin Kymberlie, Lobos Elizabeth A., Madupu Ramana, Magrini Vincent, Martin John C., Mitreva Makedonka, Muzny Donna M., Sodergren Erica J., Versalovic James, Wollam Aye M., Worley Kim C., Wortman Jennifer R., Young Sarah K., Zeng Qiandong, Aagaard Kjersti M., Abolude Olukemi O., Allen-Vercoe Emma, Alm Eric J., Alvarado Lucia, Andersen Gary L., Anderson Scott, Appelbaum Elizabeth, Arachchi Harindra M., Armitage Gary, Arze Cesar A., Ayvaz Tulin, Baker Carl C., Begg Lisa, Belachew Tsegahiwot, Bhonagiri Veena, Bihan Monika, Blaser Martin J., Bloom Toby, Bonazzi Vivien, Paul Brooks J., Buck Gregory A., Buhay Christian J., Busam Dana A., Campbell Joseph L., Canon Shane R., Cantarel Brandi L., Chain Patrick S. G., Chen I-Min A., Chen Lei, Chhibba Shaila, Chu Ken, Ciulla Dawn M., Clemente Jose C., Clifton Sandra W., Conlan Sean, Crabtree Jonathan, Cutting Mary A., Davidovics Noam J., Davis Catherine C., DeSantis Todd Z., Deal Carolyn, Delehaunty Kimberley D., Dewhirst Floyd E., Deych Elena, Ding Yan, Dooling David J., Dugan Shannon P., Michael Dunne Wm, Scott Durkin A., Edgar Robert C., Erlich Rachel L., Farmer Candace N., Farrell Ruth M., Faust Karoline, Feldgarden Michael, Felix Victor M., Fisher Sheila, Fodor Anthony A., Forney Larry J., Foster Leslie, Di Francesco Valentina, Friedman Jonathan, Friedrich Dennis C., Fronick Catrina C., Fulton Lucinda L., Gao Hongyu, Garcia Nathalia, Giannoukos Georgia, Giblin Christina, Giovanni Maria Y., Goldberg Jonathan M., Goll Johannes, Gonzalez Antonio, Griggs Allison, Gujja Sharvari, Haake Susan Kinder, Haas Brian J., Hamilton Holli A., Harris Emily L., Hepburn Theresa A., Herter Brandi, Hoffmann Diane E., Holder Michael E., Howarth Clinton, Huang Katherine H., Huse Susan M., Izard Jacques, Jansson Janet K., Jiang Huaiyang, Jordan Catherine, Joshi Vandita, Katancik James A., Keitel Wendy A., Kelley Scott T., Kells Cristyn, King Nicholas B., Knights Dan, Kong Heidi H., Koren Omry, Koren Sergey, Kota Karthik C., Kovar Christie L., Kyrpides Nikos C., La Rosa Patricio S., Lee Sandra L., Lemon Katherine P., Lennon Niall, Lewis Cecil M., Lewis Lora, Ley Ruth E., Li Kelvin, Liolios Konstantinos, Liu Bo, Liu Yue, Lo Chien-Chi, Lozupone Catherine A., Dwayne Lunsford R., Madden Tessa, Mahurkar Anup A., Mannon Peter J., Mardis Elaine R., Markowitz Victor M., Mavromatis Konstantinos, McCorrison Jamison M., McDonald Daniel, McEwen Jean, McGuire Amy L.,McInnes Pamela, Mehta Teena, Mihindukulasuriya Kathie A., Miller Jason R., Minx Patrick J., Newsham Irene, Nusbaum Chad, O’Laughlin Michelle, Orvis Joshua, Pagani Ioanna, Palaniappan Krishna, Patel Shital M., Pearson Matthew, Peterson Jane, Podar Mircea, Pohl Craig, Pollard Katherine S., Pop Mihai, Priest Margaret E., Proctor Lita M., Qin Xiang, Raes Jeroen, Ravel Jacques, Reid Jeffrey G., Rho Mina, Rhodes Rosamond, Riehle Kevin P., Rivera Maria C., Rodriguez-Mueller Beltran, Rogers Yu-Hui, Ross Matthew C., Russ Carsten, Sanka Ravi K., Sankar Pamela, Fah Sathirapongsasuti J.,Schloss Jeffery A., Schloss Patrick D., Schmidt Thomas M., Scholz Matthew, Schriml Lynn, Schubert Alyxandria M., Segata Nicola, Segre Julia A., Shannon William D., Sharp Richard R., Sharpton Thomas J., Shenoy Narmada, Sheth Nihar U., Simone Gina A., Singh Indresh, Smillie Christopher S., Sobel Jack D., Sommer Daniel D., Spicer Paul, Sutton Granger G., Sykes Sean M., Tabbaa Diana G., Thiagarajan Mathangi, Tomlinson Chad M., Torralba Manolito, Treangen Todd J., Truty Rebecca M., Vishnivetskaya Tatiana A., Walker Jason, Wang Lu, Wang Zhengyuan, Ward Doyle V., Warren Wesley, Watson Mark A., Wellington Christopher, Wetterstrand Kris A., White James R., Wilczek-Boney Katarzyna, Wu YuanQing, Wylie Kristine M., Wylie Todd, Yandava Chandri, Ye Liang, Ye Yuzhen, Yooseph Shibu, Youmans Bonnie P., Zhang Lan, Zhou Yanjiao, Zhu Yiming, Zoloth Laurie, Zucker Jeremy D., Birren Bruce W., Gibbs Richard A., Highlander Sarah K., Methé Barbara A., Nelson Karen E., Petrosino Joseph F., Weinstock George M., Wilson Richard K., Owen White, and The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature, 486 (7402):207–214, June 2012. ISSN 1476-4687. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Gilbert Jack A., Blaser Martin J., Gregory Caporaso J., Jansson Janet K., Lynch Susan V., and Knight Rob. Current understanding of the human microbiome. Nat Med, 24(4):392–400, April 2018. ISSN 1546-170X. doi: 10.1038/nm.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Freilich Shiri, Zarecki Raphy, Eilam Omer, Segal Ella Shtifman, Henry Christopher S., Kupiec Martin, Gophna Uri, Sharan Roded, and Ruppin Eytan. Competitive and cooperative metabolic interactions in bacterial communities. Nat Commun, 2(1):589, December 2011. ISSN 2041-1723. doi: 10.1038/ncomms1597. [DOI] [PubMed] [Google Scholar]
- [6].Levy Roie and Borenstein Elhanan. Metabolic modeling of species interaction in the human microbiome elucidates community-level assembly rules. Proceedings of the National Academy of Sciences, 110(31):12804–12809, July 2013. doi: 10.1073/pnas.1300926110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zelezniak Aleksej, Andrejev Sergej, Ponomarova Olga, Mende Daniel R., Bork Peer, and Patil Kiran Raosaheb. Metabolic dependencies drive species co-occurrence in diverse microbial communities. Proceedings of the National Academy of Sciences, 112(20):6449–6454, May 2015. doi: 10.1073/pnas.1421834112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Machado Daniel, Maistrenko Oleksandr M., Andrejev Sergej, Kim Yongkyu, Bork Peer, Patil Kaustubh R., and Patil Kiran R. Polarization of microbial communities between competitive and cooperative metabolism. Nat Ecol Evol, 5(2):195–203, February 2021. ISSN 2397-334X. doi: 10.1038/s41559-020-01353-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lo Ian, Denef Vincent J., VerBerkmoes Nathan C., Shah Manesh B., Goltsman Daniela, DiBartolo Genevieve, Tyson Gene W., Allen Eric E., Ram Rachna J., Chris Detter J., Richardson Paul, Thelen Michael P., Hettich Robert L., and Banfield Jillian F. Strain-resolved community proteomics reveals recombining genomes of acidophilic bacteria. Nature, 446(7135):537–541, March 2007. ISSN 1476-4687. doi: 10.1038/nature05624. [DOI] [PubMed] [Google Scholar]
- [10].Zhao Shijie, Lieberman Tami D., Poyet Mathilde, Kauffman Kathryn M., Gibbons Sean M., Groussin Mathieu, Xavier Ramnik J., and Alm Eric J. Adaptive Evolution within Gut Microbiomes of Healthy People. Cell Host & Microbe, 25(5):656–667.e8, May 2019. ISSN 1931-3128. doi: 10.1016/j.chom.2019.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Bickhart Derek M., Kolmogorov Mikhail, Tseng Elizabeth, Portik Daniel M., Korobeynikov Anton, Tolstoganov Ivan, Uritskiy Gherman, Liachko Ivan, Sullivan Shawn T., Shin Sung Bong, Zorea Alvah, Andreu Victòria Pascal, Panke-Buisse Kevin, Medema Marnix H., Mizrahi Itzhak, Pevzner Pavel A., and Smith Timothy P. L. Generating lineage-resolved, complete metagenome-assembled genomes from complex microbial communities. Nat Biotechnol, 40(5): 711–719, May 2022. ISSN 1546-1696. doi: 10.1038/s41587-021-01130-z. [DOI] [PubMed] [Google Scholar]
- [12].Olm Matthew R., Crits-Christoph Alexander, Bouma-Gregson Keith, Firek Brian A., Morowitz Michael J., and Banfield Jillian F. inStrain profiles population microdiversity from metagenomic data and sensitively detects shared microbial strains. Nat Biotechnol, 39(6):727–736, June 2021. ISSN 1546-1696. doi: 10.1038/s41587-020-00797-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lloyd-Price Jason, Mahurkar Anup, Rahnavard Gholamali, Crabtree Jonathan, Orvis Joshua, Brantley Hall A., Brady Arthur, Creasy Heather H., McCracken Carrie, Giglio Michelle G., McDonald Daniel, Franzosa Eric A., Knight Rob, White Owen, and Huttenhower Curtis. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature, 550(7674):61–66, October 2017. ISSN 1476-4687. doi: 10.1038/nature23889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Van Rossum Thea, Ferretti Pamela, Maistrenko Oleksandr M., and Bork Peer. Diversity within species: Interpreting strains in microbiomes. Nat Rev Microbiol, 18(9):491–506, September 2020. ISSN 1740-1534. doi: 10.1038/s41579-020-0368-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wolff Richard, Shoemaker William, and Garud Nandita. Ecological stability emerges at the level of strains in the human gut microbiome. Mbio, 14(2):e02502–22, 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Martínez Inés, Maldonado-Gomez Maria X, Gomes-Neto Joao Carlos, Kittana Hatem, Ding Hua, Schmaltz Robert, Joglekar Payal, Cardona Roberto Jiménez, Marsteller Nathan L, Kembel Steven W, Benson Andrew K, Peterson Daniel A, Ramer-Tait Amanda E, and Walter Jens. Experimental evaluation of the importance of colonization history in early-life gut microbiota assembly. eLife, 7:e36521, September 2018. ISSN 2050-084X. doi: 10.7554/eLife.36521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Segura Munoz Rafael R., Mantz Sara, Martínez Ines, Li Fuyong, Schmaltz Robert J., Pudlo Nicholas A., Urs Karthik, Martens Eric C., Walter Jens, and Ramer-Tait Amanda E. Experimental evaluation of ecological principles to understand and modulate the outcome of bacterial strain competition in gut microbiomes. ISME J, 16(6):1594–1604, June 2022. ISSN 1751-7370. doi: 10.1038/s41396-022-01208-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Arnold Brian J., Huang I.-Ting, and Hanage William P. Horizontal gene transfer and adaptive evolution in bacteria. Nat Rev Microbiol, 20(4):206–218, April 2022. ISSN 1740-1534. doi: 10.1038/s41579-021-00650-4. [DOI] [PubMed] [Google Scholar]
- [19].Koonin Eugene V. Horizontal gene transfer: Essentiality and evolvability in prokaryotes, and roles in evolutionary transitions. F1000Res, 5:F1000 Faculty Rev–1805, July 2016. ISSN 2046–1402. doi: 10.12688/f1000research.8737.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Fournier Gregory P. and Peter Gogarten J. Evolution of Acetoclastic Methanogenesis in Methanosarcina via Horizontal Gene Transfer from Cellulolytic Clostridia. Journal of Bacteriology, 190(3):1124–1127, February 2008. doi: 10.1128/jb.01382-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].López-García Purificación, Zivanovic Yvan, Deschamps Philippe, and Moreira David. Bacterial gene import and mesophilic adaptation in archaea. Nat Rev Microbiol, 13(7):447–456, July 2015. ISSN 1740-1534. doi: 10.1038/nrmicro3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Aminov Rustam I. Horizontal Gene Exchange in Environmental Microbiota. Front Microbiol, 2:158, July 2011. ISSN 1664-302X. doi: 10.3389/fmicb.2011.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Cordero Mar, García-Fernández Julia, Acosta Ivan C., Yepes Ana, Avendano-Ortiz Jose, Lisowski Clivia, Oesterreicht Babett, Ohlsen Knut, Lopez-Collazo Eduardo, Förstner Konrad U., Eulalio Ana, and Lopez Daniel. The induction of natural competence adapts staphylococcal metabolism to infection. Nat Commun, 13(1):1525, March 2022. ISSN 2041-1723. doi: 10.1038/s41467-022-29206-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Thi Le Thuy Nguyen, Romero Veronica Medrano, and Morikawa Kazuya. Cell wall-affecting antibiotics modulate natural transformation in SigH-expressing Staphylococcus aureus. J Antibiot, 69(6):464–466, June 2016. ISSN 1881-1469. doi: 10.1038/ja.2015.132. [DOI] [PubMed] [Google Scholar]
- [25].Fraser Christophe, Hanage William P, and Spratt Brian G. Recombination and the nature of bacterial speciation. Science, 315(5811):476–480, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Dixit Purushottam D, Pang Tin Yau, and Maslov Sergei. Recombination-driven genome evolution and stability of bacterial species. Genetics, 207(1):281–295, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Bobay Louis-Marie and Ochman Howard. Biological species are universal across life’s domains. Genome Biology and Evolution, 9(3):491–501, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Bucci Vanni, Bradde Serena, Biroli Giulio, and Xavier Joao B. Social interaction, noise and antibiotic-mediated switches in the intestinal microbiota. PLoS computational biology, 8(4):e1002497, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Coyte Katharine Z, Schluter Jonas, and Foster Kevin R. The ecology of the microbiome: networks, competition, and stability. Science, 350(6261):663–666, 2015. [DOI] [PubMed] [Google Scholar]
- [30].Novozhilov Artem S, Karev Georgy P, and Koonin Eugene V. Mathematical modeling of evolution of horizontally transferred genes. Molecular biology and evolution, 22(8):1721–1732, 2005. [DOI] [PubMed] [Google Scholar]
- [31].Dohi Marina and Mougi Akihiko. A coexistence theory in microbial communities. Royal Society Open Science, 5 (9):180476, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Saakian David B, Koonin Eugene V, and Cheong Kang Hao. Key role of recombination in evolutionary processes with migration between two habitats. Physical Review E, 100(3):030401, 2019. [DOI] [PubMed] [Google Scholar]
- [33].Takeuchi Nobuto, Cordero Otto X, Koonin Eugene V, and Kaneko Kunihiko. Gene-specific selective sweeps in bacteria and archaea caused by negative frequency-dependent selection. BMC biology, 13:1–11, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wang Teng, Weiss Andrea, Aqeel Ammara, Wu Feilun, Lopatkin Allison J, David Lawrence A, and You Lingchong. Horizontal gene transfer enables programmable gene stability in synthetic microbiota. Nature chemical biology, 18 (11):1245–1252, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Pompei Simone, Bella Edoardo, Weitz Joshua S, Grilli Jacopo, and Lagomarsino Marco Cosentino. Metacommunity structure preserves genome diversity in the presence of gene-specific selective sweeps under moderate rates of horizontal gene transfer. PLOS Computational Biology, 19(10):e1011532, 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Coyte Katharine Z, Stevenson Cagla, Knight Christopher G, Harrison Ellie, Hall James PJ, and Brockhurst Michael A. Horizontal gene transfer and ecological interactions jointly control microbiome stability. PLoS biology, 20 (11):e3001847, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Levins Richard. Coexistence in a variable environment. The American Naturalist, 114(6):765–783, 1979. [Google Scholar]
- [38].Chesson Peter Land Warner Robert R. Environmental variability promotes coexistence in lottery competitive systems. The American Naturalist, 117(6):923–943, 1981. [Google Scholar]
- [39].Levins Richard. Evolution in changing environments: some theoretical explorations. Princeton University Press, 1968. [Google Scholar]
- [40].Burkart Tom, Willeke Jan, and Frey Erwin. Periodic temporal environmental variations induce coexistence in resource competition models. Physical Review E, 108(3):034404, 2023. [DOI] [PubMed] [Google Scholar]
- [41].Steinmetz Bnaya, Meyer Immanuel, and Shnerb Nadav M. Evolution in fluctuating environments: A generic modular approach. Evolution, 76(11):2739–2757, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Babajanyan SG, Koonin Eugene V, and Cheong Kang Hao. Can environmental manipulation help suppress cancer? non-linear competition among tumor cells in periodically changing conditions. Advanced Science, 7(16):2000340, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Babajanyan SG, Lin Wayne, and Cheong Kang Hao. Cooperate or not cooperate in predictable but periodically varying situations? cooperation in fast oscillating environment. Advanced Science, 7(21):2001995, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Allahverdyan Armen E and Hu Chin-Kun. Replicators in a fine-grained environment: adaptation and polymorphism. Physical review letters, 102(5):058102, 2009. [DOI] [PubMed] [Google Scholar]
- [45].Allahverdyan Armen E, Babajanyan Sanasar G, and Hu Chin-Kun. Polymorphism in rapidly changing cyclic environment. Physical Review E, 100(3):032401, 2019. [DOI] [PubMed] [Google Scholar]
- [46].Babajanyan SG, Koonin EV, and Allahverdyan AE. Thermodynamic selection: mechanisms and scenarios. New journal of physics, 24(5):053006, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Benincà Elisa, Ballantine Bill, Ellner Stephen P, and Huisman Jef. Species fluctuations sustained by a cyclic succession at the edge of chaos. Proceedings of the National Academy of Sciences, 112(20):6389–6394, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Roca Carlos P, Cuesta José A, and Sánchez Angel. Time scales in evolutionary dynamics. Physical review letters, 97(15):158701, 2006. [DOI] [PubMed] [Google Scholar]
- [49].Lee Isaiah Paolo A, Eldakar Omar Tonsi, Peter Gogarten J, and Andam Cheryl P. Bacterial cooperation through horizontal gene transfer. Trends in Ecology & Evolution, 37(3):223–232, 2022. [DOI] [PubMed] [Google Scholar]
- [50].Palmer Jacob Dand Foster Kevin R. Bacterial species rarely work together. Science, 376(6593):581–582, 2022. [DOI] [PubMed] [Google Scholar]
- [51].Kehe Jared, Ortiz Anthony, Kulesa Anthony, Gore Jeff, Blainey Paul C, and Friedman Jonathan. Positive interactions are common among culturable bacteria. Science advances, 7(45):eabi7159, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Foster Kevin R and Bell Thomas. Competition, not cooperation, dominates interactions among culturable microbial species. Current biology, 22(19):1845–1850, 2012. [DOI] [PubMed] [Google Scholar]
- [53].McKane Alan J and Newman Timothy J. Predator-prey cycles from resonant amplification of demographic stochasticity. Physical review letters, 94(21):218102, 2005. [DOI] [PubMed] [Google Scholar]
- [54].McKane Alan J and Newman Timothy J. Stochastic models in population biology and their deterministic analogs. Physical Review E, 70(4):041902, 2004. [DOI] [PubMed] [Google Scholar]
- [55].Taylor Peter D and Jonker Leo B. Evolutionary stable strategies and game dynamics. Mathematical biosciences, 40 (1–2):145–156, 1978. [Google Scholar]
- [56].Hofbauer Josef and Sigmund Karl. Evolutionary games and population dynamics. Cambridge university press, 1998. [Google Scholar]
- [57].Sandholm William H. Population games and evolutionary dynamics. MIT press, 2010. [Google Scholar]
- [58].Weibull Jörgen W. Evolutionary game theory. 1997. [Google Scholar]
- [59].Logofet DO. Stability of Biological Communities. Mir Publishers, 1983. [Google Scholar]
- [60].Gause G.F. The Struggle for Existence. Dover Books on Biology Series. Dover Publications, Incorporated, 2003. ISBN 9780486495200. URL https://books.google.com/books?id=v01OToAhJboC. [Google Scholar]
- [61].Murray James D. Mathematical biology i: an introduction, 2002. [Google Scholar]
- [62].Hardin Garrett. The competitive exclusion principle: an idea that took a century to be born has implications in ecology, economics, and genetics. science, 131(3409):1292–1297, 1960. [DOI] [PubMed] [Google Scholar]
- [63].Robinson David and Goforth David. The topology of the 2×2 games: a new periodic table, volume 3. Psychology Press, 2005. [Google Scholar]
- [64].Perron Gabriel G., Lee Alexander E. G., Wang Yun, Huang Wei E., and Barraclough Timothy G. Bacterial recombination promotes the evolution of multi-drug-resistance in functionally diverse populations. Proceedings of the Royal Society B: Biological Sciences, 279(1733):1477–1484, April 2012. ISSN 0962-8452. doi: 10.1098/rspb.2011.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Ferreiro Aura, Crook Nathan, Gasparrini Andrew J, and Dantas Gautam. Multiscale evolutionary dynamics of host-associated microbiomes. Cell, 172(6):1216–1227, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Sarkar Amar, McInroy Cameron JA, Harty Siobhán, Raulo Aura, Ibata Neil GO, Valles-Colomer Mireia, Johnson Katerina V-A, Brito Ilana L, Henrich Joseph, Archie Elizabeth A, et al. Microbial transmission in the social microbiome and host health and disease. Cell, 187(1):17–43, 2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This is a theoretical work that did not involve data analysis.





