Abstract
Background
Not only should resistance to neoadjuvant chemotherapy (NAC) be considered in patients with breast cancer but also the possibility of achieving a pathologic complete response (PCR) after NAC. Our study aims to develop 2 multimodal ultrasound deep learning (DL) models to noninvasively predict resistance and PCR to NAC before treatment.
Methods
From January 2017 to July 2022, a total of 170 patients with breast cancer were prospectively enrolled. All patients underwent multimodal ultrasound examination (grayscale 2D ultrasound and ultrasound elastography) before NAC. We combined clinicopathological information to develop 2 DL models, DL_Clinical_resistance and DL_Clinical_PCR, for predicting resistance and PCR to NAC, respectively. In addition, these 2 models were combined to stratify the prediction of response to NAC.
Results
In the test cohort, DL_Clinical_resistance had an AUC of 0.911 (95%CI, 0.814-0.979) with a sensitivity of 0.905 (95%CI, 0.765-1.000) and an NPV of 0.882 (95%CI, 0.708-1.000). Meanwhile, DL_Clinical_PCR achieved an AUC of 0.880 (95%CI, 0.751-0.973) and sensitivity and NPV of 0.875 (95%CI, 0.688-1.000) and 0.895 (95%CI, 0.739-1.000), respectively. By combining DL_Clinical_resistance and DL_Clinical_PCR, 37.1% of patients with resistance and 25.7% of patients with PCR were successfully identified by the combined model, suggesting that these patients could benefit by an early change of treatment strategy or by implementing an organ preservation strategy after NAC.
Conclusions
The proposed DL_Clinical_resistance and DL_Clinical_PCR models and combined strategy have the potential to predict resistance and PCR to NAC before treatment and allow stratified prediction of NAC response.
Keywords: multimodal ultrasound, early prediction, breast cancer, neoadjuvant chemotherapy, deep learning
Both resistance to neoadjuvant chemotherapy (NAC) and the possibility of achieving a pathologic complete response (PCR) after NAC should be considered for patients with breast cancer. This study developed 2 multimodal ultrasound deep learning models to noninvasively predict resistance and PCR to NAC before treatment.
Implications for Practice.
Multimodal ultrasound based on deep learning (DL) can predict the response of patients with breast cancer to neoadjuvant chemotherapy (NAC) before treatment. In addition, the model combined strategy can effectively predict resistance and pathologic complete response (PCR) to NAC before treatment to avoid patients with resistant from undergoing ineffective treatment and patients with PCR from undergoing surgery. The DL models can be used as a simple and effective aid for physicians to make personalized treatment plans for patients with breast cancer before treatment.
Introduction
Neoadjuvant chemotherapy (NAC) is the cornerstone of advanced breast cancer treatment because of its ability to down-stage tumors, facilitate breast-conserving surgery, and drive adjuvant treatment decisions.1,2 More importantly, patients who achieved pathologic complete response (PCR) after NAC had the best event-free survival.3,4 Unfortunately, approximately 37% of patients with breast cancer still do not benefit from NAC.5 Even 5% of patients experienced a range of negative effects, such as delayed optimal surgery, adverse effects, or increased treatment costs.6 For surgery after NAC, patients who achieve PCR can benefit from breast-conserving surgery or even skip surgery altogether. Therefore, accurate prediction of response to NAC prior to treatment holds clinical significance in improving risk stratification and the development of treatment strategies for patients with breast cancer. First, it is necessary to identify patients who are resistant to NAC (resistance to NAC) so that they can be protected from ineffective treatment and significant toxicity as early as possible and to try other treatments.7 Second, it is important to identify patients who are sensitive to NAC but unlikely to achieve PCR (sensitive and non-PCR), so that risk-adapted therapy can be used. Finally, identifying patients who can achieve PCR from NAC allows them to avoid unnecessary surgery and benefit from the traditional model of breast cancer management approach, which involves surgery after NAC.
Numerous efforts are underway to find methods that can predict response to NAC prior to or early in treatment. Imaging markers from medical images have been shown to correlate with the response of breast cancer to NAC, including magnetic resonance imaging (MRI), positron emission tomography (PET) and ultrasound (such as grayscale 2D ultrasound, ultrasound elastography, and contrast-enhanced ultrasound [CEUS]). Among these imaging modalities, ultrasound has the advantages of being more convenient, cost-effective, widely accessible, and radiation-free compared to other imaging modalities. It is also a routine clinical examination for breast cancer before NAC treatment. In addition, strain elastography (SE), a type of ultrasound elastography, is a new imaging technique to complement grayscale ultrasound by overlaying a color-coded map displaying tissue deformation distribution and reflecting the stiffness of lesions.8-12 However, the predictive performance of multimodal ultrasound (grayscale 2D ultrasound and SE) based on the expertise and knowledge of radiologists is not satisfactory(AUCs: 0.56-0.72).13-15 Recently, artificial intelligence approaches based on MRI or PET images have been used for the prediction of NAC response before treatment.16-20 Nevertheless, there is currently no research reported on the use of deep learning (DL) with multimodal ultrasound (grayscale 2D ultrasound and SE) in addressing the challenges of predicting NAC response grading.
In this study, we used multimodal ultrasound examination for breast cancer, including grayscale 2D ultrasound and SE, to build a 2-level predictive DL structure for the prediction of NAC response before treatment. In order to better meet clinical needs, the prediction task is divided into 2 tasks, as follows: (1) predict patient resistance to NAC (DL_Clinical_resistance model) and (2) predict whether patients will achieve PCR (DL_Clinical_PCR model). The ultimate goal of DL_Clinical_resistance and DL_Clinical_PCR is to assist clinicians in developing personalized breast cancer treatment strategies and even optimize treatment plans on a patient-specific basis.
Materials and Methods
Patients
This prospective study was approved by our hospital ethics committee, and informed consent was obtained from all patients.
The inclusion criteria were as follows: (a) presence of breast cancer confirmed by puncture biopsy results with no distant metastasis; (b) available clinical data (including age, volume, estrogen receptor [ER] status, progesterone receptor [PR] status, human epidermal growth factor receptor 2 [HER2], and the Ki-67 proliferation index); (c) scheduled to undergo NAC; and (d) surgery was performed after completing the entire NAC. The exclusion criteria were as follows: (a) no pathology results for NAC, (b) multifocal ipsilateral breast or multiple lesion bilateral tumors, (c) no immunohistochemical results in our hospital or in other hospitals, (d) unqualified multimodal ultrasound images, (e) did not complete NAC, (f) distant metastasis during NAC, and (g) nonstandard NAC treatment. Figure 1 shows the flowchart of patient recruitment.
Figure 1.
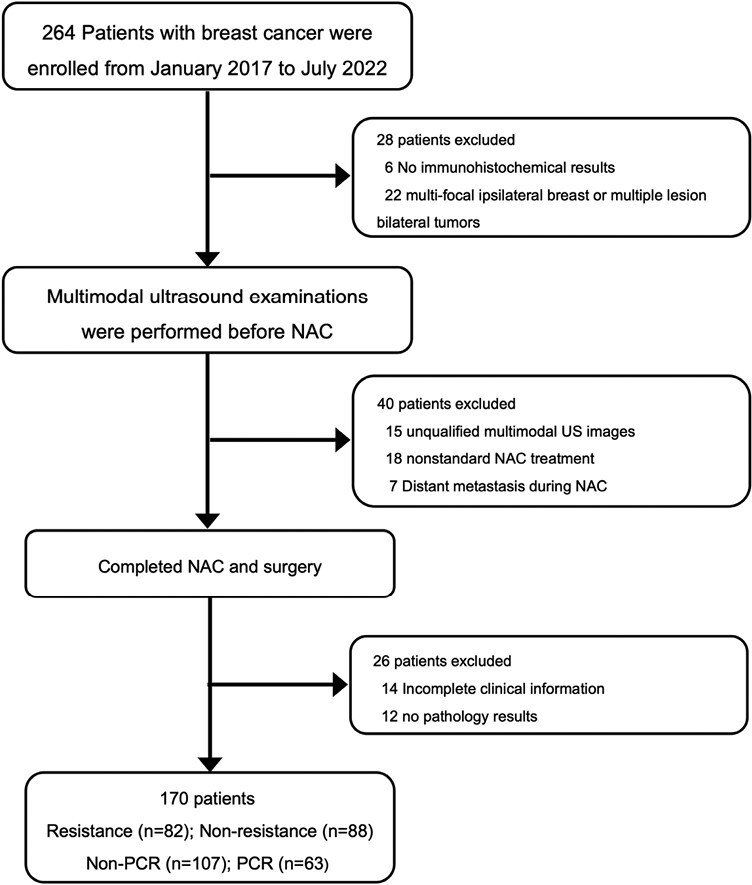
Diagram of patient recruitment process. Abbreviations: NAC: neoadjuvant chemotherapy; US: ultrasound; PCR: pathological complete response; Non-PCR: non-pathological complete response.
In this study, patients from January 2017 to March 2021 were randomly divided into a training cohort and an internal test cohort with a ratio of 3:1. And patients from April 2021 to July 2022 were used as an independent test cohort.
Multimodal Ultrasound Examinations
A radiologist with more than 20 years of ultrasound experience used a MyLab 90 ultrasound machine (Esoate, Genoa, Italy) equipped with a 4-15 MHz linear array transducer to perform a meticulous multimodal ultrasound examination of each patient within 3 days before NAC treatment. Patients took the supine position and fully exposed the breast and armpits. Two grayscale 2D ultrasound images were collected for each lesion: one was acquired at the maximum diameter of the lesion, and the other was acquired by rotating the probe 90° in the same position. The transverse and longitudinal images of the lesion were captured. The same radiologist simultaneously conducts SE examination without changing the patient’s position. The probe was lightly positioned onto the skin at the largest section of the target lesion and switched to the SE mode. Then, the radiologist applied vertically pressure to the target mass with the transducer and maintained the appropriate compression. The compression applied onto the breast was adjusted with reference to a built-in indicator on the ultrasound device that estimates the applied deformation. Radiologists captured transverse and longitudinal SE images of the lesion. (A total of 4 images were obtained for a patient, including transverse and longitudinal grayscale images of the lesion and the corresponding SE images.)
NAC and Pathological Examination
All patients received a NAC regimen based on taxanes (Docetaxel, or Docetaxel + Cyclophosphamide/Carboplatin, or Docetaxel + Cyclophosphamide + Carboplatin), anthracyclines (Epirubicin + Cyclophosphamide), or a combination of taxanes and anthracyclines (Docetaxel + Epirubicin or Docetaxel + Epirubicin + Cyclophosphamide, or Docetaxel + Epirubicin + Cyclophosphamide + Carboplatin). The duration of NAC was mainly 6 or 8 courses. In addition, patients with HER2-positive were treated with trastuzumab and/or Pertuzumab (8 mg/kg loading dose, 6 mg/kg maintenance dose). Some of the patients with HR(+)/HER2(−)received exclusive neoadjuvant endocrine therapy at the same time according to the recommendation. Treatment protocols for all patients followed national comprehensive cancer network (NCCN) guidelines.21
All patients underwent a core needle biopsy prior to NAC and immunohistochemical results, including tumor type, ER status, PR status, HER2 status, and the Ki-67 proliferation index were obtained. In addition, all patients underwent breast surgery after completing the entire NAC. According to the Miller-Payne (MP) grading system, the pathologists with 10 years of experience used surgical specimens to determine the patients’ pathological response to NAC. There are 5 levels of the MP grading system (Supplementary Table S1), where G5 was defined as PCR, and G1-G3 was defined as resistance.
The ideal NAC management for breast cancer is based on the above scenarios: (1) G5, breast preservation may be possible after NAC22; (2) G1-G3, NAC may lead to delayed surgery and increased treatment costs and early change of treatment plan7; and (3) G4, multimodal treatment or escalation of treatment is implemented.
Model Development
In this study, 2 DL models, DL_Clinical_resistance and DL_Clinical_PCR, were established for predicting the resistance and PCR to NAC before treatment. First, the rectangular box of region of interest (ROI) containing the tumor and surrounding region was annotated and cropped on grayscale ultrasound. The corresponding ROI of the elastography ultrasound image was manually cropped from the original ultrasound images. It is shown that not only single modality images can be encoded as network features, but adding other modality images can provide more and more effective information.23 Therefore, the designed DL model is a parallel model structure that contains 2 Densenet121-based networks.24 The 2 Densenet121 models shared network parameters and adopted a feature fusion strategy that can be compatible with the input images of both modality, including grayscale 2D ultrasound and stained elastography ultrasound images. Finally, the DL model outputs the classification results (Fig. 2). Three key clinicopathological information (ER, HER2, and volume) were incorporated into the last fully connected (FC) layer, instead of extracting deep features and combining them offline. The workflow of the ROI extraction, the detailed architecture of the model, and the details of the training process are described in Supplementary Material.
Figure 2.
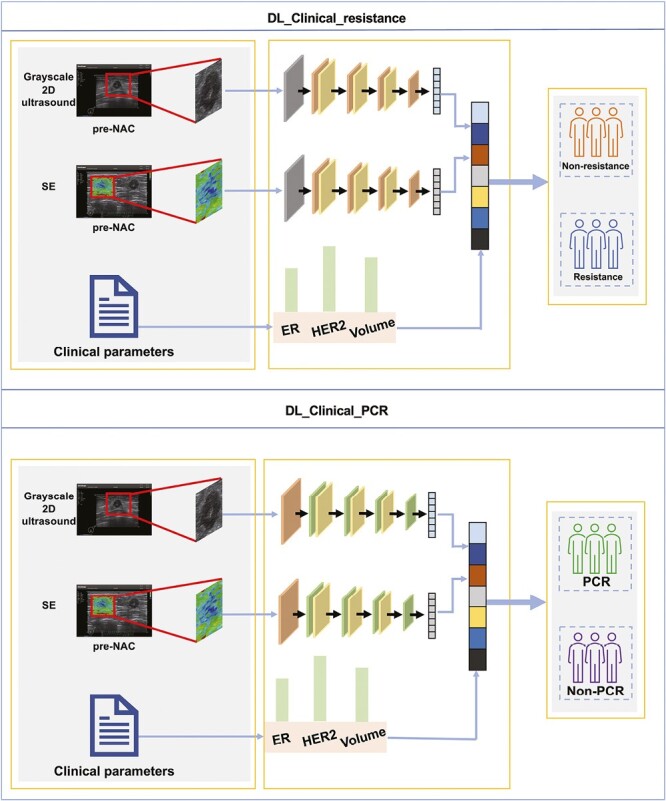
The overall pipeline of the DL_Clinical_resistance model and DL_Clinical_PCR model. The ultrasound and SE images and key clinical parameters were inputted into 2 DL models before treatment. DL_Clinical_resistance outputs the probability that a patient is resistance to NAC, and the treatment strategy is changed immediately for patients with resistance. DL_Clinical_PCR outputs the probability of patients achieving PCR after NAC, then patients with PCR can confidently proceed to NAC treatment and provide decision-making support for organ-preserving strategies. Abbreviations: DL: deep learning; NAC: neoadjuvant chemotherapy; SE: strain elastography; PCR: pathological complete response.
Statistics Analysis
All statistical analyses were performed using SPSS version 23.0 and Python version 3.7. We used the mean and SD to analyze the continuous variables and used numbers and percentages to show categorical variables. Different comparisons between groups were made using the t-test or chi-square test. The classification threshold was obtained from the receiver operating characteristic (ROC) curve based on the maximum Youden index.25 Stepwise logistic regression was used for feature selection. Comparisons between model performances were made with the Delong test. The 95% confidence interval (CI) was calculated by bootstrapping with 1000 resamples. Statistical significance was defined as P ≤ .05. The detailed statistics analysis are provided in Supplementary Material.
Results
Clinicopathological Characteristics
A total of 170 women were prospectively enrolled in this study for final analysis. The rates of PCR or resistance were not significantly different between training and test cohorts. ER and HER2 status were significantly correlated with PCR and resistance in the training cohort. However, the initial volume of the tumor was only correlated with resistance. The clinicopathological characteristics of all patients are summarized in Table 1.
Table 1.
Clinical characteristics of patients in the training and test sets.
| Characteristic | Training cohort (127) | Test cohort (43) | ||||
|---|---|---|---|---|---|---|
| Resistance (N = 61) | Non-resistance (N = 66) | P | Resistance (N = 21) | Nonresistance (N = 22) | P | |
| Age (mean, SD) | 46.80 ± 10.66 | 48.58 ± 10.22 | .341 | 48.14 ± 10.70 | 46.00 ± 10.04 | .502 |
| Volume (cm3) | 13.05 ± 12.57 | 9.15 ± 8.82 | .044 | 8.43 ± 9.96 | 7.09 ± 5.24 | .581 |
| ER | .019 | .001 | ||||
| Positive | 43 | 33 | 17 | 7 | ||
| Negative | 18 | 33 | 4 | 15 | ||
| PR | .476 | .087 | ||||
| Positive | 25 | 23 | 10 | 5 | ||
| Negative | 36 | 43 | 11 | 17 | ||
| Ki67 | .282 | .066 | ||||
| Positive | 43 | 52 | 13 | 19 | ||
| Negative | 18 | 14 | 8 | 3 | ||
| Her2 | .046 | .650 | ||||
| Positive | 19 | 32 | 11 | 10 | ||
| Negative | 42 | 34 | 10 | 12 | ||
| Molecular subtype | .023 | .502 | ||||
| HR+ and HER2− | 31 | 18 | 8 | 7 | ||
| HER2+ | 19 | 32 | 11 | 10 | ||
| Triple-negative | 11 | 16 | 2 | 5 | ||
| Non-PCR (N = 80) | PCR (N = 47) | P | Non-PCR (N = 27) | PCR (N = 16) | P | |
|---|---|---|---|---|---|---|
| Age (mean, SD) | 47.29 ± 10.26 | 48.47 ± 10.78 | .540 | 48.78 ± 11.03 | 44.13 ± 8.47 | .155 |
| Volume (cm3) | 11.67 ± 11.50 | 9.93 ± 9.87 | .388 | 7.91 ± 9.01 | 7.47 ± 5.56 | .861 |
| ER | .022 | <.001 | ||||
| Positive | 54 | 22 | 22 | 2 | ||
| Negative | 26 | 25 | 5 | 14 | ||
| PR | .295 | .295 | ||||
| Positive | 33 | 15 | 11 | 4 | ||
| Negative | 47 | 32 | 16 | 12 | ||
| Ki67 | .104 | .025 | ||||
| Positive | 56 | 39 | 17 | 15 | ||
| Negative | 24 | 8 | 10 | 1 | ||
| Her2 | .001 | .454 | ||||
| Positive | 23 | 28 | 12 | 9 | ||
| Negative | 57 | 19 | 15 | 7 | ||
| Molecular subtype | .001 | .185 | ||||
| HR+ and HER2− | 40 | 9 | 12 | 3 | ||
| HER2+ | 23 | 28 | 12 | 9 | ||
| Triple-negative | 17 | 10 | 3 | 4 |
Abbreviations: PCR: pathological complete response; Non-PCR: nonpathological complete response; ER: estrogen receptor; PR: progesterone receptor; HR: hormone receptor; HER2: human epidermal growth factor receptor 2.
Performance of DL_Clinical_resistance for Predicting the Resistance to NAC
First, we investigated the performance of clinicopathological and DL features in predicting resistance to NAC. Initial tumor volume and the status of ER and HER2 were significantly correlated with resistance and were selected as key factors used to construct the Clinical_resistance model. The AUCs for Clinical_reisitance model were 0.691 (95%CI, 0.597-0.779) and 0.697 (95%CI, 0.535-0.853) in training and test cohorts. Then, the DL_resistance model achieved higher AUCs of 0.964 (95%CI, 0.933-0.988) and 0.872 (95%CI, 0.747-0.963) in both 2 cohorts compared to the Clinical_resistance model (all P < .05).
Given that combining DL and clinical features can potentially further improve the performance of predicting resistance to NAC, we added clinical features to the last FC layer of the DL model and trained DL_Clinical_resistance model. Compared to all the models mentioned earlier, DL_Clinical_resistance achieved the best AUCs of 0.986 (95%CI, 0.971-0.997) and 0.911 (95%CI, 0.814-0.979) in 2 cohorts for resistance prediction (Delong test: all P < .05 for DL_Clinical_resistance vs. Clinical_resistance). The ROC curves and decision cures were plotted to demonstrate the comparative results of AUC in Fig. 3A and 3B. In addition, Fig. 4 shows that DL_Clinical_resistance offered high sensitivities of 0.934 (95%CI, 0.870-1.000) and 0.905 (95%CI, 0.765-1.000) with high NPV of 0.938 (95%CI, 0.878-1.000) and 0.882 (95%CI, 0.708-1.000) in the training and test cohorts, respectively. The detailed statistical results of the models are presented in Table 2.
Figure 3.
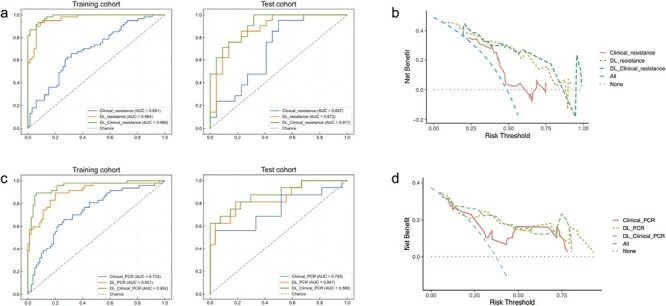
ROC curves and decision curves among clinical models, DL models, and DL_Clinical models for predicting the primary tumor PCR or resistance in the training and test cohort. (A) ROC curves of different models for predicting primary tumor resistance to NAC in the training and test cohorts. (B) Decision curves of different models for predicting primary tumor resistance to NAC in the test cohorts. (C) ROC curves of different models for predicting primary tumor PCR in the training and test cohorts. (D) Decision curves of different models for predicting primary tumor PCR in the test cohorts. Abbreviations: PCR: pathological complete response; AUC: area under the receiver operator characteristic curve; DL: deep learning.
Figure 4.
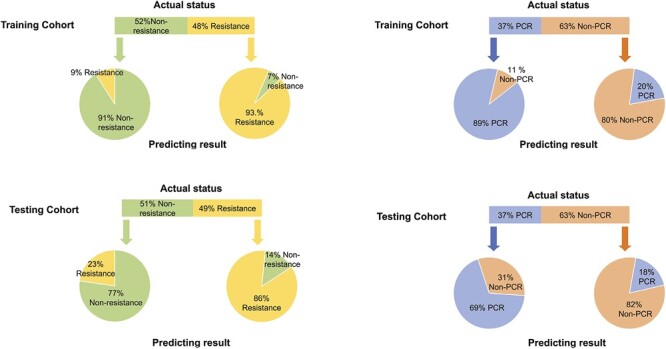
Performance of DL_Clinical_resistance and DL_Clinical_PCR models in predicting NAC response in the training cohort and test cohort. The upper panel shows the actual status of patients, and the lower panel shows the prediction results outputted by the final models in the training and the test sets.
Table 2.
Prediction performance of 3 different models for primary tumor resistance to NAC and tumor PCR in the training and test cohort.
| Models | Cohorts | AUC | ACC | SENS | SPEC | PPV | NPV |
|---|---|---|---|---|---|---|---|
| Clinical_resistance | Training | 0.691 (0.597-0.779) |
0.669 (0.583-0.748) |
0.656 (0.535-0.776) |
0.682 (0.567-0.790) |
0.656 (0.534-0.867) |
0.682 (0.571-0.791) |
| Test | 0.697 (0.535-0.853) |
0.605 (0.465-0.742) |
0.476 (0.269-0.696) |
0.727 (0.526-0.905) |
0.625 (0.385-0.866) |
0.593 (0.400-0.774) |
|
| DL_resistance | Training | 0.964 (0.933-0.988) |
0.921 (0.874-0.960) |
0.934 (0.867-0.985) |
0.909 (0.838-0.967) |
0.905 (0.825-0.968) |
0.937 (0.873-0.985) |
| Test | 0.872 (0.747-0.963) |
0.814 (0.697-0.930) |
0.857 (0.699-1.00) |
0.773 (0.583-0.950) |
0.783 (0.591-0.950) |
0.850 (0.667-1.000) |
|
| DL_Clinical_resistance | Training | 0.986 (0.971-0.997) |
0.929 (0.882-0.968) |
0.934 (0.870-1.000) |
0.924 (0.853-0.984) |
0.919 (0.841-0.983) |
0.938 (0.878-1.000) |
| Test | 0.911 (0.814-0.979) |
0.791 (0.651-0.970) |
0.905 (0.765-1.000) |
0.682 (0.474-0.869) |
0.731 (0.552-0.895) |
0.882 (0.708-1.000) |
|
| Clinical_PCR | Training | 0.733 (0.638-0.823) |
0.708 (0.630-0.787) |
0.660 (0.527-0.792) |
0.737 (0.642-0.831) |
0.596 (0.460-0.729) |
0.787 (0.693-0.871) |
| Test | 0.745 (0.563-0.902) |
0.581 (0.442-0.720) |
0.688 (0.444-0.916) |
0.519 (0.333-0.708) |
0.458 (0.261-0.655) |
0.737 (0.526-0.933) |
|
| DL_PCR | Training | 0.901 (0.836-0.953) |
0.835 (0.764-0.897) |
0.894 (0.795-0.977) |
0.800 (0.707-0.885) |
0.724 (0.604-0.836) |
0.927 (0.859-0.984) |
| Test | 0.847 (0.705-0.959) |
0.767 (0.628-0.883) |
0.688 (0.444-0.909) |
0.815 (0.654-0.958) |
0.688 (0.444-0.909) |
0.815 (0.655-0.956) |
|
| DL_Clinical_PCR | Training | 0.952 (0.909-0.984) |
0.827 (0.756-0.889) |
0.979 (0.927-1.000) |
0.738 (0.636-0.829) |
0.687 (0.577-0.796) |
0.983 (0.944-1.000) |
| Test | 0.880 (0.751-0.973) |
0.721 (0.581-0.860) |
0.875 (0.688-1.000) |
0.630 (0.444-0.815) |
0.583 (0.381-0.789) |
0.895 (0.739-1.000) |
The data in brackets are the 95% confidence intervals. Training cohort, n = 127; test cohort, n = 43.
Abbreviations: PCR: pathological complete response; DL: deep learning; AUC: area under the receiver operating characteristic curve; ACC: accuracy; SENS: sensitivity; SPEC: specificity; PPV: positive predictive value; NPV: negative predictive value.
Furthermore, the performance of DL_Clinical_resistance model was validated in different molecular subtypes of breast cancer and obtained good predictive performance in each subgroup. Supplementary Table S2 and Fig. S1A showed the detailed results.
Performance of DL_Clinical_PCR for Predicting the PCR
In addition to predicting sensitivity to NAC, we also attempted to predict whether patients could achieve PCR of the primary tumor after NAC, enabling more precision and personalized treatment. Stepwise multiple logistic regression was used to select the status of ER and HER2 as key factors in the clinical model. The initial volume of the tumor was also added to the clinical model, as it is of great interest to clinicians. However, the AUC achieved by the Clinical_PCR was unsatisfactory. In contrast, the DL_PCR model achieved much higher AUCs of 0.901 (95%CI, 0.836-0.953) and 0.847 (95%CI, 0.705-0.959) in the training and test cohorts (Delong test: P = .002 in the training cohort and P = .28 in the test cohort for DL_PCR vs. Clinical_PCR).
Similarly, we explored whether incorporating clinical features into DL models could further improve the performance of PCR prediction. Our proposed DL_Clinical_PCR model exhibits the highest AUCs of 0.952 (95%CI, 0.909-0.984) in the training cohort and 0.880 (95%CI, 0.751-0.973) in the test cohort (P = .0018 and P = .15 for DL_Clinical_PCR vs. Clinical_PCR by the Delong test on training cohort and test cohort). In addition, Fig. 3C and 3D shows the ROC curves and decision curves of the 3 different models. Meanwhile, the DL_Clinical_PCR showed a sensitivity of 0.979 (95%CI, 0.927-1.000) and NPV of 0.983 (95%CI, 0.944-1.000) in the training cohort; a sensitivity of 0.875 (95%CI, 0.688-1.000) and PPV of 0.895(95%CI, 0.739-1.000) in the test cohort as shown in Fig. 4 and Table 2.
We performed subgroup analysis based on different molecular subtypes of breast cancer to further confirm the stability of the DL_Clinical_PCR model. Detailed information is provided in Supplementary Table S3 and Supplementary Fig. S1B). DL_Clinical_PCR achieves good predictive performance in different subgroups as well.
Clinical Benefits of Combining DL_Clinical_resistance and DL_Clinical_PCR
For invasive breast cancer, the current general clinical treatment strategy is that the patients receive NAC first and undergo surgery after NAC. These would result in ineffective treatment for patients who are resistant to NAC and over-surgery for patients who achieve PCR of the primary tumor after NAC. However, by combining the predicted results of DL_Clinical_resistance and DL_Clinical_PCR, patients can be assigned to 1 of 3 groups corresponding to the 3 ideal treatment strategies: resistance, sensitive and non-PCR, and PCR. In this way, some patients who are resistance to NAC and achieve PCR can avoid ineffective treatment and overtreatment.
All 43 patients in the test cohort were used to assess the clinical benefit of combining DL_Clinical_resistance and DL_Clinical_PCR (Fig. 5). The detailed results of combining DL_Clinical_resistance and DL_Clinical_PCR were shown in Supplementary Table S4. When we implement a treatment strategy of NAC combined with surgery based on current clinical practice, this results in 86% of patients being ineffectively treated (48.8%) or overtreated (37.2%), with only 14% of patients (sensitive and non-PCR) receiving suitable treatment (NAC and surgery) (Fig. 5A). In contrast, when we combined DL_Clinical_resistance and DL_Clinical_PCR to analyze these same patients, it correctly classified 37.1% of patients to the resistance group and 25.7% to the PCR group, thus avoiding ineffective NAC and excessive surgical treatment (benefiting 62.8% of patients) (Fig. 5B). In addition, combining the 2 models ensured that 33.3% (4.6%/14%) of patients with sensitive and non-PCR received suitable NAC and surgical treatment. Ultimately, only 28% (7.1% + 6.9% + 14%) of patients were incorrectly predicted, resulting in inappropriate treatment. Therefore, combining DL_Clinical_resistance and DL_Clinical_PCR is expected to further reduce the economic cost and psychological burden on patients for the management of invasive breast cancer (28% vs. 86%).
Figure 5.
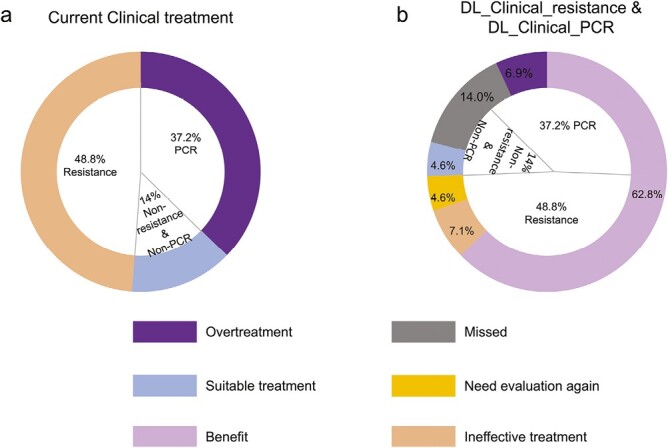
Overall benefit of combining DL_Clinical_resistance and DL_Clinical_PCR models in the test cohort. The 2 rings show the percentage of patients who received different treatment regimens in the study according to the combination of DL_Clinical_resistance and DL_Clinical_PCR or the current clinical treatment, respectively. The inner ring represents the actual percentage distribution of the status of all patients in the test cohort responding to NAC. The outer ring (A) represents the treatment based on current clinical management, resulting in 48.8% of patients experiencing ineffective treatment and 37.2% overtreatment, with only 14% of patients receiving appropriate treatment. The outer ring of (B) represents the treatment management decision made by combining DL_Clinical_resistance and DL_Clinical_PCR, where 67.4% of patients would benefit and receive suitable treatment. Overtreatment: patients with PCR were judged to be sensitive and non-PCR by the DL_Clinical_resistance and DL_Clinical_PCR models; Missed: patients with PCR were judged to be resistant and non-PCR by the DL_Clinical_resistance and DL_Clinical_PCR models or patients with sensitive and non-PCR were judged to be resistant and non-PCR by the DL_Clinical_resistance and DL_Clinical_PCR models or patients with sensitive and non-PCR were judged to be sensitive and PCR by the DL_Clinical_resistance and DL_Clinical_PCR models; suitable treatment: patients with sensitive and non-PCR were judged to be sensitive and non-PCR by the DL_Clinical_resistance and DL_Clinical_PCR models; need evaluation again: patients with resistance were judged to be resistant and PCR by the DL_Clinical_resistance and DL_Clinical_PCR models; benefits: patients with PCR were judged to be sensitive and PCR by the DL_Clinical_resistance and DL_Clinical_PCR models or patients with resistance were judged to be resistant and non-PCR by the DL_Clinical_resistance and DL_Clinical_PCR models; ineffective treatment: patients with resistance were judged to be sensitive and PCR or non-PCR by the DL_Clinical_resistance and DL_Clinical_PCR models.
Discussion
In this prospective study, we developed and validated 2 multimodal ultrasound DL models for the prediction of resistance and PCR to NAC using grayscale 2D and SE ultrasound images before NAC in patients with breast cancer. Combining DL_Clinical_resistance and DL_Clinical_PCR enables a stratified prediction of NAC response. Our results suggest that on the one hand, DL_Clinical_resistance is expected to be an effective tool to assist clinicians in adjusting the treatment plan for patients with NAC-resistance at an early stage, thus avoiding unnecessary and ineffective treatment. On the other hand, DL_Clinical_PCR can provide decision support for patients with PCR to achieve organ preservation strategies.
Patients treated with NAC tend to have the following 3 conditions: (1) patients who are resistant to NAC should stop ineffective treatment and seek alternative treatment as early as possible.7 (2) Patients sensitive to NAC but unable to achieve PCR are ideal candidates for NAC combined with surgical treatment options. And risk-adapted therapy can be used to achieve PCR as much as possible. (3) Ideally, patients achieve PCR after NAC, which could imply an extremely favorable disease-free and overall survival.3 PCR has been proposed as a surrogate early clinical endpoint for long-term survival.26 In addition, such patients are expected to avoid breast surgery. Therefore, clinicians and patients are eager to improve PCR rates. However, the vast majority of previous studies on the prediction of response to NAC have simply classified patients as non-resistance and resistance or PCR and non-PCR, which may ignore a subset of patients who respond to NAC but ultimately fail to achieve PCR.27-29 Such patients may be able to add some other treatments to improve the possibility of achieving PCR. In this study, we performed the 2 tasks of resistance and PCR prediction separately. Clinicians can refer to the prediction results of both models to develop more flexible treatment strategies. The development of the DL_Clinical_PCR model did not exclude patients who were resistant to NAC due to the limitation of the number of patients. Therefore, DL_Clinical_resistance and DL_Clinical_PCR cannot achieve stepwise prediction, but the predictive performance of combining DL_Clinical_resistance and DL_Clinical_PCR combined has demonstrated the feasibility of our graded prediction strategy.
Previous studies have shown that artificial intelligences (both DL radiomics and handcraft radiomics) based only on pre-NAC grayscale 2D ultrasound images have difficulty achieving satisfactory results in predicting response to NAC.27,28 Fernandes et al12 demonstrated that ultrasound elastography of the breast can somewhat predict the response to NAC. In addition, CEUS has also been proven to have unique value in assessing the response to NAC, as it can provide information about tumor perfusion.30,31 However, due to the limited availability of CEUS technology, we chose to utilize conventional grayscale 2D ultrasound and ultrasound elastography. The combination of these 2 modalities enables the detection of morphological information and tissue stiffness. As multimodal ultrasound DL models have been successfully applied to the prediction of breast axillary lymph node metastasis and diagnosis of lesions,23,32 it is reasonable to investigate the performance of DL models based on multimodal ultrasound images in the prediction of response to NAC in breast cancer. As expected, both DL_resistance and DL_PCR models achieved better performance. To the best of our knowledge, our study is the first attempt to use DL based on multimodal ultrasound images to predict NAC responses before treatment.
Numerous studies have confirmed that clinicopathological characteristics of patients with breast cancer including tumor size and hormone receptor status are related to NAC response.14,33-35 However, the unsatisfactory predictive performance of this method has limited its clinical application. The reasons could be, on the one hand, clinical information may only consider certain aspects of the tumor, and on the other hand, the hormonal status of patients with breast cancer before and after NAC is often altered.36 The results of the clinical model in this study (Clinical_resistance and Clinical_PCR) also reconfirmed that only using clinical predictors to predict NAC response is not reliable enough. Nevertheless, it has been found in several studies that the combination of clinicopathological information and imaging features can further improve the performance of DL models.28 Therefore, we proposed DL_Clinical_resistance and DL_Clinical_PCR models. Furthermore, unlike previous studies, the combination of clinicopathological information and DL features in this study is based entirely on the concept of using convolutional neural networks. We fused the clinical parameters in the last FC layer of the DL model, which can complement image features with more information and make the model more stable by restraining the features extracted from images.37 Thus, DL_Clinical_resistance and DL_Clinical_PCR achieved the best predictive performance, which confirmed that the deep combination of DL features and clinical information can more fully reflect tumor characteristics and heterogeneity.
Next, we analyzed the performance of DL_Clinical_resistance and DL_Clinical_PCR models. Both models achieved high sensitivities and NPVs. For DL_Clinical_ resistance, high sensitivity and NPV allow identifying as many patients as possible who are resistant to NAC without increasing the risk to avoid ineffective treatment and delay other more appropriate treatments. For DL_Clinical_PCR, high sensitivity ensures that the majority of patients with a chance of achieving PCR receive NAC and provides confidence support for implementing a step-down treatment or organ preservation strategy after NAC. In addition, high NPV also guarantees the necessity of surgical treatment for patients with non-PCR. Then, the combination of DL_Clinical_resistance and DL_Clinical_PCR better achieved a stratified prediction of NAC response. Finally, 62.8% of patients who were resistant to NAC or achieved PCR after NAC benefited from our model, and they could avoid ineffective NAC or excessive surgical treatment. It is well known that different molecular subtypes have varying PCR rates, and tumor heterogeneity in medical imaging can affect the performance of models trained on a mixture of all subtypes. Our models achieve favorable predictive performance across different subgroups, further confirming the generalizability of the model. Nevertheless, in the future, we still need to develop more accurate models based on breast cancer-specific subtypes.
The DL_Clinical_resistance and DL_Clinical_PCR models that we proposed performed well, but this study still has some limitations. First, although this study is a prospective multimodal study, the number of patients is limited, and they are all from the same hospital. Therefore, we will concentrate on multicenter studies with larger sample sizes in the future to confirm the clinical applicability and robustness of the models. Second, considering that color Doppler ultrasound or CEUS can provide further information about tumor vasculature means that the exclusion of these modalities could lead to the model performing suboptimally. Third, DL_Clinical_resistance and DL_Clinical_PCR are aimed as initial tools for patient stratification and should be used with caution. Clinicians must combine the comprehensive opinions of multiple disciplines to develop individualized treatment strategies for patients. Finally, the relationship between DL features and gene expression or biological mechanisms has not been explored. In subsequent studies, we will further explore the interpretability of DL models.
Conclusions
In conclusion, DL_Clinical_resistance and DL_Clinical_PCR models, which are based on pre-NAC multimodal ultrasound images (including grayscale 2D ultrasound and SE), were proposed to predict resistance and PCR to NAC before treatment, respectively. Furthermore, combining 2 models can stratify the response to NAC for patients with breast cancer who could benefit from optimal treatment management before NAC.
Supplementary Material
Contributor Information
Jionghui Gu, Department of Ultrasound, The First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, People’s Republic of China; The D-Lab, Department of Precision Medicine, GROW - School for Oncology and Reproduction, Maastricht University, Maastricht, The Netherlands.
Xian Zhong, The D-Lab, Department of Precision Medicine, GROW - School for Oncology and Reproduction, Maastricht University, Maastricht, The Netherlands; Department of Ultrasound, The First Affiliated Hospital of Sun Yat-Sen University, Guangzhou, People’s Republic of China.
Chengyu Fang, Department of Ultrasound, The First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, People’s Republic of China.
Wenjing Lou, Department of Ultrasound, The First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, People’s Republic of China.
Peifen Fu, Department of Breast Surgery, The First Affiliated Hospital, School of Medicine, Zhejiang University, Hangzhou, Zhejiang, People’s Republic of China.
Henry C Woodruff, The D-Lab, Department of Precision Medicine, GROW - School for Oncology and Reproduction, Maastricht University, Maastricht, The Netherlands; Department of Radiology and Nuclear Medicine, GROW - School for Oncology and Reproduction, Maastricht University Medical Centre, Maastricht, The Netherlands.
Baohua Wang, Department of Ultrasound, The First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, People’s Republic of China.
Tianan Jiang, Department of Ultrasound, The First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, People’s Republic of China.
Philippe Lambin, The D-Lab, Department of Precision Medicine, GROW - School for Oncology and Reproduction, Maastricht University, Maastricht, The Netherlands; Department of Radiology and Nuclear Medicine, GROW - School for Oncology and Reproduction, Maastricht University Medical Centre, Maastricht, The Netherlands.
Funding
This study was supported by ERC-2020-PoC: 957565-AUTO.DISTINCT, the European Union’s Horizon research and innovation programme under grant agreement: CHAIMELEON no. 952172, EuCanImage no. 952103, IMI-OPTIMA no. 101034347, AIDAVA (HORIZON-HLTH-2021-TOOL-06) no. 101057062, REALM (HORIZON-HLTH-2022-TOOL-11) no. 101095435, and EUCAIM (DIGITAL-2022-CLOUD-AI-02) no. 101100633. This study was financially supported by the program of China Scholarships Council (no. 202206320360), Scientific Research Fund of Zhejiang Provincial Education Department (no. Y202250839), the Development Project of National Major Scientific Research Instrument (no. 82027803), the National Natural Science Foundation of China (no. 81971623), and the Key Project of Natural Science Foundation of Zhejiang Province (no. LZ20H180001).
Conflict of Interest
Henry C. Woodruff declared ownership interests (minority shares) in Radiomics SA. Philippe Lambin declared the following: grants/sponsored research agreements from Radiomics SA and Convert Pharmaceuticals; minority shares in the companies Radiomics SA, Convert Pharmaceuticals, Comunicare, and LivingMed Biotech; coinventor of 2 issued patents with royalties on radiomics (PCT/NL2014/050248 and PCT/NL2014/050728), licensed to Radiomics SA; one non-issued patent on LSRT (PCT/P126537PC00, US: 17802766), licensed to Varian; 3 non-patented inventions (softwares) licensed to ptTheragnostic/DNAmito, Radiomics SA, and Health Innovation Ventures and 2 non-issued, non-licensed patents on Deep Learning-Radiomics (N2024482, N2024889); none of these entities were involved in the preparation of this article. The other authors indicated no financial relationships.
Author Contributions
Conception/design: J.G., X.Z. Provision of study material or patients: J.G., C.F., W.L., P.F., B.W. Collection and/or assembly of data: J.G., C.F., W.L., P.F., B.W. Data analysis and interpretation: J.G., X.Z., T.J., P.L. Manuscript writing: J.G., X.Z., H.C.W., B.W., T.J., P.L. Final approval of manuscript: All authors.
Data Availability
All data needed to evaluate the conclusions in the article are present in the article and/or the Supplementary Material. The code part is an open source and can be uploaded to Github for interested researchers to download. Additional data related to this article (including deidentified participant data with the data dictionary, original ultrasonographic images, study protocol, statistical analysis plan, etc.) will be available on request from the corresponding authors. A signed data use agreement and institutional review board approval will be required before the release of research data.
References
- 1. Thompson AM, Moulder-Thompson SL.. Neoadjuvant treatment of breast cancer. Ann Oncol. 2012;23(Suppl 10):x231-x236. 10.1093/annonc/mds324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Derks MGM, van de Velde CJH.. Neoadjuvant chemotherapy in breast cancer: more than just downsizing. Lancet Oncol. 2018;19(1):2-3. 10.1016/S1470-2045(17)30914-2 [DOI] [PubMed] [Google Scholar]
- 3. Cortazar P, Zhang L, Untch M, et al. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet. 2014;384(9938):164-172. 10.1016/S0140-6736(13)62422-8 [DOI] [PubMed] [Google Scholar]
- 4. Symmans WF, Wei C, Gould R, et al. Long-term prognostic risk after neoadjuvant chemotherapy associated with residual cancer burden and breast cancer subtype. J Clin Oncol. 2017;35(10):1049-1060. 10.1200/JCO.2015.63.1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Haque W, Verma V, Hatch S, et al. Response rates and pathologic complete response by breast cancer molecular subtype following neoadjuvant chemotherapy. Breast Cancer Res Treat. 2018;170(3):559-567. 10.1007/s10549-018-4801-3 [DOI] [PubMed] [Google Scholar]
- 6. Xiong Q, Zhou X, Liu Z, et al. Multiparametric MRI-based radiomics analysis for prediction of breast cancers insensitive to neoadjuvant chemotherapy. Clin Transl Oncol. 2020;22(1):50-59. 10.1007/s12094-019-02109-8 [DOI] [PubMed] [Google Scholar]
- 7. Wang Z, Lin F, Ma H, et al. Contrast-enhanced spectral mammography-based radiomics nomogram for the prediction of neoadjuvant chemotherapy-insensitive breast cancers. Front Oncol. 2021;11:605230. 10.3389/fonc.2021.605230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yuan S, Shao H, Na Z, Kong M, Cheng W.. Value of shear wave elasticity in predicting the efficacy of neoadjuvant chemotherapy in different molecular types. Clin Imaging. 2022;89:97-103. 10.1016/j.clinimag.2022.06.008 [DOI] [PubMed] [Google Scholar]
- 9. Huang JX, Lin SY, Ou Y, et al. Combining conventional ultrasound and sonoelastography to predict axillary status after neoadjuvant chemotherapy for breast cancer. Eur Radiol. 2022;32(9):5986-5996. 10.1007/s00330-022-08751-1 [DOI] [PubMed] [Google Scholar]
- 10. Liu Q, Tang L, Chen M.. Ultrasound strain elastography and contrast-enhanced ultrasound in predicting the efficacy of neoadjuvant chemotherapy for breast cancer: a nomogram integrating Ki-67 and ultrasound features. J Ultrasound Med. 2022;41(9):2191-2201. 10.1002/jum.15900 [DOI] [PubMed] [Google Scholar]
- 11. Gu J, Polley EC, Denis M, et al. Early assessment of shear wave elastography parameters foresees the response to neoadjuvant chemotherapy in patients with invasive breast cancer. Breast Cancer Res. 2021;23(1):52. 10.1186/s13058-021-01429-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fernandes J, Sannachi L, Tran WT, et al. Monitoring breast cancer response to neoadjuvant chemotherapy using ultrasound strain elastography. Transl Oncol. 2019;12(9):1177-1184. 10.1016/j.tranon.2019.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hylton NM, Blume JD, Bernreuter WK, et al. Locally advanced breast cancer: MR imaging for prediction of response to neoadjuvant chemotherapy--results from ACRIN 6657/I-SPY TRIAL. Radiology. 2012;263(3):663-672. 10.1148/radiol.12110748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vriens BE, de Vries B, Lobbes MB, et al. Ultrasound is at least as good as magnetic resonance imaging in predicting tumour size post-neoadjuvant chemotherapy in breast cancer. Eur J Cancer. 2016;52:67-76. 10.1016/j.ejca.2015.10.010 [DOI] [PubMed] [Google Scholar]
- 15. Eun NL, Son EJ, Gweon HM, Kim JA, Youk JH.. Prediction of axillary response by monitoring with ultrasound and MRI during and after neoadjuvant chemotherapy in breast cancer patients. Eur Radiol. 2020;30(3):1460-1469. 10.1007/s00330-019-06539-4 [DOI] [PubMed] [Google Scholar]
- 16. Bian T, Wu Z, Lin Q, et al. Radiomic signatures derived from multiparametric MRI for the pretreatment prediction of response to neoadjuvant chemotherapy in breast cancer. Br J Radiol. 2020;93(1115):20200287. 10.1259/bjr.20200287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu Z, Li Z, Qu J, et al. Radiomics of multiparametric MRI for pretreatment prediction of pathologic complete response to neoadjuvant chemotherapy in breast cancer: a multicenter study. Clin Cancer Res. 2019;25(12):3538-3547. 10.1158/1078-0432.CCR-18-3190 [DOI] [PubMed] [Google Scholar]
- 18. Umutlu L, Kirchner J, Bruckmann NM, et al. Multiparametric (18)F-FDG PET/MRI-based radiomics for prediction of pathological complete response to neoadjuvant chemotherapy in breast cancer. Cancers (Basel). 2022;14(7):1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Roy S, Whitehead TD, Li S, et al. Co-clinical FDG-PET radiomic signature in predicting response to neoadjuvant chemotherapy in triple-negative breast cancer. Eur J Nucl Med Mol Imaging. 2022;49(2):550-562. 10.1007/s00259-021-05489-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li P, Wang X, Xu C, et al. (18)F-FDG PET/CT radiomic predictors of pathologic complete response (pCR) to neoadjuvant chemotherapy in breast cancer patients. Eur J Nucl Med Mol Imaging. 2020;47(5):1116-1126. 10.1007/s00259-020-04684-3 [DOI] [PubMed] [Google Scholar]
- 21. Goetz MP, Gradishar WJ, Anderson BO, et al. NCCN guidelines insights: breast cancer, version 3.2018. J Natl Compr Canc Netw. 2019;17(2):118-126. 10.6004/jnccn.2019.0009 [DOI] [PubMed] [Google Scholar]
- 22. van la Parra RFD, Tadros AB, Checka CM, et al. Baseline factors predicting a response to neoadjuvant chemotherapy with implications for non-surgical management of triple-negative breast cancer. Br J Surg. 2018;105(5):535-543. 10.1002/bjs.10755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zheng X, Yao Z, Huang Y, et al. Deep learning radiomics can predict axillary lymph node status in early-stage breast cancer. Nat Commun. 2020;11(1):1236. 10.1038/s41467-020-15027-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huang G, Liu Z, Van Der Maaten L, Weinberger KQ.. Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition. 2017;2017:4700-4708. [Google Scholar]
- 25. Rücker G, Schumacher M.. Summary ROC curve based on a weighted Youden index for selecting an optimal cutpoint in meta-analysis of diagnostic accuracy. Stat Med. 2010;29(30):3069-3078. 10.1002/sim.3937 [DOI] [PubMed] [Google Scholar]
- 26. Early Breast Cancer Trialists' Collaborative Group (EBCTCG). Long-term outcomes for neoadjuvant versus adjuvant chemotherapy in early breast cancer: meta-analysis of individual patient data from ten randomised trials. Lancet Oncol. 2018;19(1):27-39. 10.1016/S1470-2045(17)30777-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Byra M, Dobruch-Sobczak K, Klimonda Z, Piotrzkowska-Wroblewska H, Litniewski J.. Early prediction of response to neoadjuvant chemotherapy in breast cancer sonography using Siamese Convolutional Neural Networks. IEEE J Biomed Health Inform. 2021;25(3):797-805. 10.1109/JBHI.2020.3008040 [DOI] [PubMed] [Google Scholar]
- 28. Jiang M, Li CL, Luo XM, et al. Ultrasound-based deep learning radiomics in the assessment of pathological complete response to neoadjuvant chemotherapy in locally advanced breast cancer. Eur J Cancer. 2021;147:95-105. 10.1016/j.ejca.2021.01.028 [DOI] [PubMed] [Google Scholar]
- 29. Wu L, Ye W, Liu Y, et al. An integrated deep learning model for the prediction of pathological complete response to neoadjuvant chemotherapy with serial ultrasonography in breast cancer patients: a multicentre, retrospective study. Breast Cancer Res. 2022;24(1):81. 10.1186/s13058-022-01580-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huang Y, Le J, Miao A, et al. Prediction of treatment responses to neoadjuvant chemotherapy in breast cancer using contrast-enhanced ultrasound. Gland Surg. 2021;10(4):1280-1290. 10.21037/gs-20-836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee YJ, Kim SH, Kang BJ, Kim YJ.. Contrast-enhanced ultrasound for early prediction of response of breast cancer to neoadjuvant chemotherapy. Ultraschall Med. 2019;40(2):194-204. 10.1055/a-0637-1601 [DOI] [PubMed] [Google Scholar]
- 32. Misra S, Jeon S, Managuli R, et al. Bi-modal transfer learning for classifying breast cancers via combined b-mode and ultrasound strain imaging. IEEE Trans Ultrason Ferroelectr Freq Control. 2022;69(1):222-232. 10.1109/TUFFC.2021.3119251 [DOI] [PubMed] [Google Scholar]
- 33. Ouldamer L, Bendifallah S, Pilloy J, et al. Risk scoring system for predicting breast conservation after neoadjuvant chemotherapy. Breast J. 2019;25(4):696-701. 10.1111/tbj.13303 [DOI] [PubMed] [Google Scholar]
- 34. Arici S, Sengiz Erhan S, Geredeli C, et al. The clinical importance of androgen receptor status in response to neoadjuvant chemotherapy in Turkish patients with local and locally advanced breast cancer. Oncol Res Treat. 2020;43(9):435-440. 10.1159/000508478 [DOI] [PubMed] [Google Scholar]
- 35. Ma Y, Zhang S, Zang L, et al. Combination of shear wave elastography and Ki-67 index as a novel predictive modality for the pathological response to neoadjuvant chemotherapy in patients with invasive breast cancer. Eur J Cancer. 2016;69:86-101. 10.1016/j.ejca.2016.09.031 [DOI] [PubMed] [Google Scholar]
- 36. Özdemir O, Zengel B, Kocatepe Çavdar D, Yılmaz C, Durusoy R.. Prognostic value of receptor change after neoadjuvant chemotherapy in breast cancer patients. Eur J Breast Health. 2022;18(2):167-171. 10.4274/ejbh.galenos.2022.2022-1-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xie Y, Zhang J, Xia Y, et al. Fusing texture, shape and deep model-learned information at decision level for automated classification of lung nodules on chest CT. Inf Fusion. 2018;42:102-110. 10.1016/j.inffus.2017.10.005 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in the article are present in the article and/or the Supplementary Material. The code part is an open source and can be uploaded to Github for interested researchers to download. Additional data related to this article (including deidentified participant data with the data dictionary, original ultrasonographic images, study protocol, statistical analysis plan, etc.) will be available on request from the corresponding authors. A signed data use agreement and institutional review board approval will be required before the release of research data.


