Abstract
Carcinoembryonic antigen (CEA)-related cell adhesion molecule 6 (CEACAM6) is a cell adhesion protein of the CEA family of glycosyl phosphatidyl inositol anchored cell surface glycoproteins. A wealth of research has demonstrated that CEACAM6 is generally upregulated in pancreatic adenocarcinoma, breast cancer, non-small cell lung cancer, gastric cancer, colon cancer and other cancers and promotes tumor progression, invasion and metastasis. The transcriptional expression of CEACAM6 is regulated by various factors, including the CD151/TGF-β1/Smad3 axis, microRNA (miR)-146, miR-26a, miR-29a/b/c, miR-128, miR-1256 and DNA methylation. In addition, the N-glycosylation of CEACAM6 protein at Asn256 is mediated by α-1,6-mannosylglycoptotein 6-β-N-acetylglucosaminyltransferase. In terms of downstream signaling pathways, CEACAM6 promotes tumor proliferation by increasing levels of cyclin D1 and cyclin-dependent kinase 4 proteins. CEACAM6 can activate the ERK1/2/MAPK or SRC/focal adhesion kinase/PI3K/AKT pathways directly or through EGFR, leading to stimulation of tumor proliferation, invasion, migration, resistance to anoikis and chemotherapy, as well as angiogenesis. This article provides a review of the expression pattern, biological function and relationship with prognosis of CEACAM6 in cancer. In summary, CEACAM6 may be a valuable diagnostic biomarker and potential therapeutic target for human cancers exhibiting overexpression of CEACAM6.
Keywords: CEACAM6, biomarker, cancer
1. Introduction
Carcinoembryonic antigen (CEA)-related cell adhesion molecules (CEACAMs) have 12 members with diverse functions and biological processes, such as cell adhesion, intracellular and intercellular signaling and shaping of tissue architecture (1,2). All members are highly glycosylated proteins and typically consist of one variable (V)-like Ig domain, known as the N domain, except for CEACAM16, which has two N domains. However, they differ in the number of constant C2-like Ig domains and the method of membrane anchorage. CEACAM5, CEACAM6, CEACAM7 and CEACAM8 are linked to the membrane through a glycosyl phosphatidyl inositol (GPI) linkage (3,4), while CEACAM1, CEACAM3, CEACAM4, CEACAM19, CEACAM20 and CEACAM21 are anchored to the cellular membrane through transmembrane domains. CEACAM16, on the other hand, is a secreted version without any membrane anchorage. The cytoplasmic domain of CEACAM1 contains immunoreceptor tyrosine-based inhibition motifs, while CEACAM3, CEACAM4, CEACAM19 and CEACAM20 carry immunoreceptor tyrosine-based activation motifs. The extracellular domains, comprising the N domain and C2-like Ig domains, have a crucial role in the functionality of CEACAMs. These domains act as both homophilic and heterophilic intercellular adhesion molecules (5), and also act as receptors for human and rodent pathogens (6,7). CEACAM1, CEACAM3, CEACAM6 and CEACAM8 are expressed on human neutrophils and the presence of related antibodies has been shown to increase leukocyte adhesion to human endothelial cells (3). These CEACAM proteins exist as dimers and oligomers, allowing them to form multiple associations with other partners on the cell membrane. This ability to interact with various partners helps regulate important cellular functions. CEACAM6, also termed CD66c (formerly nonspecific cross-reacting antigen), is expressed in a variety of normal tissues such as neutrophils (8) and columnar epithelial and goblet cells of the colon (9). It is upregulated in numerous oncological processes, such as those of pancreatic, lung and colon cancers. CEACAM6-associated carcinogenesis mainly includes promotion of abnormal proliferation, invasion, migration, angiogenesis, anti-apoptosis and resistance to chemotherapeutic drugs. Tumor model studies showed that targeting CEACAM6 had a positive effect on the treatment and diagnosis of tumors with high expression of CEACAM6 (10). The present article reviews the expression, regulatory function and mechanism of CEACAM6 in various cancers. Based on this, the potential clinical value of CEACAM6 in the diagnosis, treatment and prognosis of cancer was also discussed.
2. Overview of CEACAM6
The CEACAM6 gene is located at the 19q13.2 site of human genome and the coding region is composed of 6 exons with only one transcript encoding 344 amino acids in total. CEACAM6 consists of an N-terminus Ig-like V-type domain, 2 N-terminus IgC-like domains and a membrane-linked glycoprotein. The extracellular N-terminus Ig-like V-type domain is necessary for homophilic and heterophilic intercellular adhesion (10-12). CEACAM6 is anchored in the cell surface by the transmembrane domain of the membrane-linked glycoproteins.
In response to changes in the environment, numerous cell surface molecules, including adhesion molecules, can transmit information from the environment to the cell interior through signal transduction. Glycosylation may regulate membrane protein folding, which alters receptor activation and changes epitope exposure for antibody recognition (13). This provides a clue that CEACAM6 does not have transmembrane or intracellular structural domains and affects intracellular signaling. For instance, neutrophil CEACAM6 signaling regulates CD11/CD18 adhesion activity, leading to increased neutrophil adhesion to human umbilical vein endothelial cells (14,15). N-domain sequences determine the specificity of the CEACAM6 for interaction with itself or other CEACAMs, including CEACAM8, CEACAM1 and CEA (12,16), and the bacterial proteins (12). CEACAM6 can form a homodimeric complex or a heterodimeric complex with CEACAM8 through homodimerization via its Ig-like V-type domain (17). CEACAM8 is highly expressed in developing neutrophil progenitors, while CEACAM6 is highly expressed in type II pneumocytes. The interaction between developing neutrophils and type II pneumocytes in Coronavirus disease 2019 involves crosstalk mediated by CEACAM8-CEACAM6 (18). CEACAM6 blockade by efficient inhibition of the CEACAM6 and CEACAM1 interaction reactivates the antitumor response of T cells (19). In addition, N-glycosylated CEACAM6 interacts with the EGFR in oral squamous cell carcinoma (OSCC) cells and regulates intracellular signaling for tumor invasion, migration and metastasis (20).
GPI-anchored proteins, known as 'lipid rafts' (21), are abundant in ligands and effectors. The presence of CEACAM6 in the lipid raft region and its involvement in signaling align with these findings, resulting in enhanced cell adhesion, cell differentiation and anoikis (22). CEACAM6 in pancreatic adenocarcinoma cells interacts with αvβ3 integrins, which in turn increases the adhesion of extracellular matrix components, leading to cell invasion and metastasis (23). Besides, integrin αvβ3, as a cell surface receptor, participates in signaling transduction pathways in cancer cell proliferation and metastasis (24).
3. Role of CEACAM6 in cancer
A substantial body of evidence has now confirmed that CEACAM6 has a momentous role in the carcinogenesis and clinical features of various types of cancer. It has been reported that CEACAM6 is upregulated in a variety of cancers, including non-small-cell lung carcinoma (NSCLC) (25), pancreatic adenocarcinoma (26-28) and colorectal carcinoma (CRC) (29). In this section, the effect of CEACAM6 in 12 types of cancer was summarized. Overall, high expression of CEACAM6 is positively correlated with the development of these cancers. Furthermore, the specific mechanisms involved were summarized in this section (Tables I-III; Fig. 1).
Table I.
Roles of CEACAM6 in various cancers.
| Author, year | Cancer type | Cell/tissue | Expression change | Function in cancer carcinogenesis | (Refs.) |
|---|---|---|---|---|---|
| Liu, 2022 | Cholangiocarcinoma | Cholangiocarcinoma tissues, RBE | + | Oncogene/diagnostic marker | (89) |
| Tian, 2020 | GBC | Peritumoral tissue, gallbladder carcinoma tissue, SGC-996, GBC-SD | + | Oncogene | (92) |
| Rose, 2016 | Extrahepatic cholangiocarcinoma | Bile | + | Oncogene/diagnostic marker | (91) |
| Ieta, 2006 | Intrahepatic cholangiocarcinoma | Cancer tissue, noncancerous tissue, TFK-1, HuCC-T1, MEC | + | Oncogene, prognostic marker, marker of chemoresistance to gemcitabine | (90) |
| Liu, 2016 | Colorectal cancer | Normal colon tissue to adenoma and cancer tissue | + | Oncogene | (53) |
| Jantscheff, 2003 | Colorectal cancer | Colorectal cancer | + | Prognosis marker | (50) |
| Ilantzis, 2012 | Colon carcinogenesis | SW-1222, Caco-2 | / | Oncogene | (29) |
| Schölzel, 2000 | Colorectal tumors | Hyperplastic polyps and adenoma normal mucosa | + | Oncogene | (49) |
| An, 2021 | Gastric cancer | Gastric mucosa, cancer/AGS, MKN-45 | + | Oncogene, diagnostic marker | (64) |
| Ru, 2017 | Gastric cancer | Gastric mucosa, cancer | + | Oncogene, prognostic marker, therapeutic target (improvement of radiosensitivity) | (63) |
| Roy, 2016 | Gastric cancer | Gastric mucosa, cancer | + | Oncogene, diagnostic marker | (61) |
| Zang, 2015 | Gastric cancer | SCG-7901, MKN-45 | + | Oncogene | (72) |
| Zang, 2014 | Gastric cancer | Tumor tissues/SCG-7901, MKN-45, MKN-28, SNU-16 | + | Oncogene, diagnostic marker, therapeutic target | (65) |
| Zang, 2014 | Gastric cancer | Gastric tissues/SCG-7901, MKN-45, MKN-28 | + | Oncogene, diagnostic marker | (66) |
| Deng, 2017 | Gastric cancer | Gastric tissues | + | Oncogene, diagnostic marker, prognostic marker | (62) |
| Zhao, 2017 | Gastric cancer | Peripheral blood | + | Oncogene, diagnostic marker, prognostic marker | (79) |
| Ferlizza, 2021 | Lung adenocarcinoma | Cerebrospinal fluid/A549 | + | Oncogene, diagnostic marker | (60) |
| Son, 2019 | Lung adenocarcinoma | A549 | + | Oncogene/therapeutic target | (33) |
| Du, 2020 | Lung adenocarcinoma | A549/DDP, A549 | + | Oncogene, therapeutic target (improvement of chemosensitivity) | (31) |
| Tsang, 2013 | Breast cancer | Tissues | + | Oncogene, diagnostic marker | (80) |
| Lewis-Wambi, 2008 | Breast cancer | MCF-7:5C/MCF-7/MCF-7:2A | + | Oncogene | (85) |
| Koh, 2020 | Breast cancer | MCF-7 | / | Oncogene | (88) |
| Balk-Møller, 2014 | Breast cancer | SKBR3, MCF7 | / | Tumor suppressor | (81) |
| Duxbury, 2005 | Pancreatic cancer | Normal pancreas, pancreatic adenocarcinoma | + | Oncogene, prognostic marker | (39) |
| Yan, 2016 | Pancreatic cancer | Tissues/BxPC-3, SW1990, MiA PaCa-2 | + | Oncogene, diagnostic marker, prognostic marker | (26) |
| Wang, 2009 | Pancreatic cancer | PaTu8988t, Suit-2 | / | Oncogene, therapeutic target | (47) |
| Pandey, 2019 | Pancreatic cancer | HPAF-II PC | + | Oncogene, therapeutic target | (28) |
| Kurlinkus, 2021 | Pancreatic cancer | Pancreatic tissue samples, blood serum | + | Oncogene, prognostic marker (prediction of chemoresistance properties) | (27) |
| Duxbury, 2004 | Pancreatic cancer | MiAAR, MiaPaCa2 | / | Oncogene, therapeutic target | (41) |
| Duxbury, 2004 | Pancreatic cancer | Capan2 | / | Oncogene, therapeutic target | (48) |
| Duxbury, 2004 | Pancreatic cancer | Capan2, BxPC3 | / | Oncogene, therapeutic target | (46) |
| Duxbury, 2004 | Pancreatic cancer | BxPC3 | / | Oncogene | (23) |
| Duxbury, 2004 | Pancreatic cancer | BxPC3 | / | Oncogene | (45) |
| Chen, 2013 | Pancreatic cancer | Pancreatic cancer tissue/PANC-1, CFPAC-1 | + | Oncogene | (43) |
| Chiang, 2018 | OSCC | Tissue/OEC-M1, OC-2,HSC-3 | + | Oncogene, therapeutic target | (20) |
| Cameron, 2012 | HNSCC | Tissue/Detroit 562 | + | Oncogene, therapeutic target | (97) |
| Bednarek, 2018 | LSCC | LSCC cell lines | / | Tumor-suppressive | (96) |
| Wang, 2018 | Osteosarcoma | Osteosarcoma tissues/Mg-63, U-2 Os | + | Oncogene, diagnostic marker, therapeutic target | (98) |
| Steiner, 2019 | Plasma cell disorders | Peripheral blood | + | Oncogene, diagnostic marker, therapeutic target | (101) |
| Lasa, 2008 | ALL | Leukemic samples | + | / | (99) |
| Kanderová, 2010 | ALL | NALM-24, BCP-ALL | / | / | (101) |
| Zhu, 2019 | ccRCC | 786-O, A498 | + | Oncogene | (102) |
Expression change means a change in CEACAM6 expression from normal tissue to tumor. +, increase in expression; /, information not available; CEACAM6, carcinoembryonic antigen-related cell adhesion molecule 6; HNSCC, head and neck squamous cell carcinoma; LSCC, laryngeal squamous cell carcinoma; OSCC, oral squamous cell carcinoma; ALL, acute lymphoblastic leukemia; BCP-ALL, B-cell precursor acute lymphoblastic leukemia; ccRCC, clear cell renal cell carcinoma; GBC, gallbladder cancer.
Table II.
Upstream genes or molecules of carcinoembryonic antigen-related cell adhesion molecule 6 in cancers.
| Author, year | Cancer type | Molecule/complex | Direction of regulatory effect | (Refs.) |
|---|---|---|---|---|
| Tsang, 2013 | Breast cancers | pSMAD3 | + | (80) |
| Du, 2020 | Lung adenocarcinoma | miR-146a, miR-26a | - | (31) |
| Chen, 2013 | Pancreatic cancer | miR-29a/b/c | - | (43) |
| Han, 2008 | Gastric cancer | Smad3 | + | (73) |
| Yang, 2021 | Colorectal cancer | TGFβ1, CD151 | + | (51) |
| Chuang, 2020 | Cervical cancer | miR-128 | - | (120) |
| Chu, 2022 | Gastric cancer | miR-1256 | - | (74) |
+, positive regulation; -, negative regulation; pSMAD3, phosphorylated SMAD3; miR, microRNA.
Table III.
Downstream molecules or pathways of carcinoembryonic antigen-related cell adhesion molecule 6 in cancers.
| Author, year | Cancer type | Target or associated molecule/complex | (Refs.) |
|---|---|---|---|
| Liu, 2022 | Cholangiocarcinoma | SRC/PI3K/AKT signaling pathway | (89) |
| Tian, 2020 | GBC | MMP2, vimentin, BCL-2, BAX | (92) |
| Chiang, 2018 | Oral squamous cell carcinoma | EGFR | (20) |
| Yan, 2016 | Pancreatic carcinoma | Cyclin D1 and CDK4 | (26) |
| Zang, 2015 | Gastric cancer | FAK signaling | (72) |
| Zang, 2014 | Gastric cancer | C-SRC phosphorylation | (65) |
| Zang, 2014 | Gastric cancer | PI3K/AKT signaling pathway | (66) |
| Cameron, 2012 | Head and neck cancer | PI3K/AKT | (97) |
| Kanderová, 2010 | ALL | Erk1/2, Akt, and p38 MAPK phosphorylation | (100) |
| Duxbury, 2004 | Pancreatic adenocarcinoma | AKT phosphorylation (Ser-473) | (41) |
| Duxbury, 2004 | Pancreatic adenocarcinoma | AKT, C-SRC | (48) |
| Duxbury, 2004 | Pancreatic adenocarcinoma | C-SRC | (46) |
| Duxbury, 2004 | Pancreatic adenocarcinoma | FAK phosphorylation (p125) | (45) |
FAK, focal adhesion kinase; CDK, cyclin-dependent kinase; ALL, acute lymphoblastic leukemia; GBC, gallbladder cancer; SRC, steroid receptor coactivator.
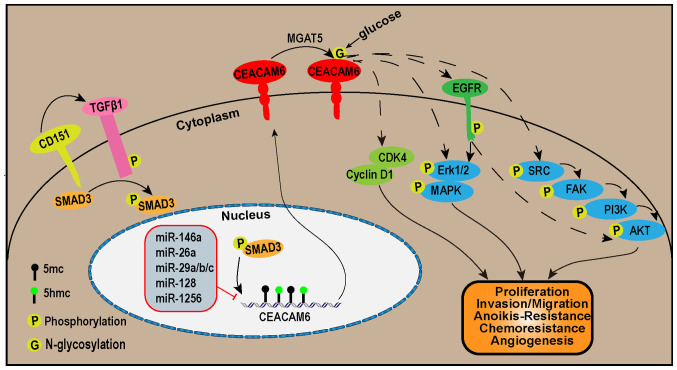
Figure 1.
The CEACAM6 signaling pathways. In the upstream of the CEACAM6 signaling pathways, CD151 promotes SMAD3 phosphorylation via TGF-β1. This leads to the phosphorylation of SMAD3, which in turn promotes the transcription of CEACAM6. On the other hand, miR-146, miR-26a, miR-29a/b/c, miR-128 and miR-1256 inhibit the transcription of CEACAM6. The transcriptional expression of CEACAM6 is also regulated by promoter DNA methylation. In addition, MGAT5 facilitates N-glycosylation at Asn256 of the CEACAM6 protein. Downstream of the CEACAM6 signaling pathways, CEACAM6 promotes tumor proliferation by increasing cyclin D1 and CDK4 protein levels. CEACAM6 activates the ERK1/2/MAPK or SRC/FAK/PI3K/AKT pathways either directly or via EGFR to stimulate tumor proliferation, invasion, migration, resistance to anoikis and chemotherapy, as well as angiogenesis. FAK, focal adhesion kinase; MGAT5, α-1,6-mannosylglycoptotein 6-β-N-acetylglucosaminyltransferase; CDK, cyclin-dependent kinase; CEACAM6, carcinoembryonic antigen-related cell adhesion molecule 6.
NSCLC
CEACAM6 expression was detected by immunohistochemical assessments to be higher in lung adenocarcinomas (LUAD) than squamous tumors (25). Furthermore, exosome testing of patients' serum revealed that LUAD had higher levels of CEACAM6 compared to squamous cell carcinoma (30). This is probably the reason why the study of CEACAM6 in lung cancer is mainly in NSCLC. CEACAM6 expression is increased in NSCLC compared to normal tissues, and CEACAM6 levels are associated with tumor progression and metastasis (25).
CEACAM6 is closely associated with chemoresistance in NSCLC. It is one of the most prominently upregulated genes in the cisplatin-resistant A549/cisplatin cell line when compared to the parental A549 cell line. Overexpression of CEACAM6 leads to increased IC50 values of cisplatin, as well as enhanced cell proliferation, invasion and migration (31). CEACAM6 induces upregulation of N-cadherin, vimentin, SOX2, POU class 5 homeobox 1 and active ras homolog family member A, while causing the downregulation of E-cadherin. Furthermore, microRNA (miR)-146a and miR-26a have been identified as potential regulators of CEACAM6, negatively affecting its expression and enhancing the sensitivity of NSCLC to cisplatin (31). Furthermore, CEACAM6 triggers the activation of Src-focal adhesion kinase (FAK) signaling and hinders anoikis through homologous interactions in LUAD (32). Treatment with small interfering RNA targeting CEACAM6 (siCEACAM6) alone resulted in tumor growth inhibition rates of up to 35.5%, which were significantly enhanced by up to 47% when combined with cisplatin in murine xenograft models (33). In a mouse model of a LUAD A549 cell-derived xenograft, treatment with anti-CEACAM6 antibody alone resulted in a 40% inhibition of tumor growth. However, when the antibody was combined with paclitaxel, the inhibition of tumor growth was significantly enhanced, reaching up to 80% (34).
CEACAM6 was demonstrated to promote NSCLC progression and metastasis (25,35). CEACAM6 has a role in leptomeningeal metastases of NSCLC and cell-free RNA testing by lumbar puncture cerebrospinal fluid helps detect brain metastases from patients with NSCLC (sensitivity, 88.24%; specificity, 100%) (36). By enzyme-linked immunosorbent assay (ELISA) of serum, CAECAM6 levels were found to be higher in patients with LUAD than in healthy controls, with the highest levels in patients with leptomeningeal metastasis (37).
Pancreatic adenocarcinoma
The incidence of pancreatic adenocarcinoma is increasing and ~50% of patients have advanced disease at diagnosis; however, available cytotoxic therapies for advanced disease are modestly effective (38). CEACAM6 has been identified as a valuable biomarker for pancreatic adenocarcinomas. Multiple studies have demonstrated its presence in tumor specimens and serum of patients, as opposed to healthy individuals; this discovery emphasizes the potential usefulness of CEACAM6 in the diagnosis and monitoring of pancreatic adenocarcinomas (26-28). Tumoral CEACAM6 appears to be present early in the development of pancreatic adenocarcinoma. CEACAM6 expression was more prevalent in pancreatic intraepithelial neoplasia lesions and negative tumoral CEACAM6 expression was associated with the absence of lymph node metastases, lower disease stage and longer postoperative survival (39).
In the aspects of diagnosis and prognosis of pancreatic adenocarcinoma, CEACAM6 has manifested its potential value. CEACAM6 overexpression is universally a poor prognostic marker in KRAS-mutant and wild-type pancreatic adenocarcinoma and is overexpressed in primary and metastatic basal and classical pancreatic adenocarcinoma subtypes (28). Of note, serum concentrations of CEACAM6 demonstrated their potential value for assessing disease-free survival (DFS) and overall survival (OS) of patients with pancreatic adenocarcinoma after radical and adjuvant chemotherapy (27). This provides an important tool for making individualized treatment plans and for monitoring the disease process in patients with pancreatic adenocarcinoma.
CEACAM6 is a potential therapeutic target in pancreatic adenocarcinoma, and it affects the fibrotic reaction, immune regulation and energy metabolism (28). Anti-CEACAM6 antibody markedly suppressed pancreatic adenocarcinoma tumor growth and enhanced tumor apoptosis in a nude mouse xenograft model (40). Anoikis resistance was associated with increased CEACAM6 expression. CEACAM6 knockdown in pancreatic adenocarcinoma cell lines increased susceptibility to caspase-mediated anoikis, and this effect was abrogated by caspase inhibitor Z-Val-Ala-Asp-fluoromethyl ketone (41). Recombinant plasmids knocking down CEACAM6 also led to a favorable anti-tumor phenotype in SW1990 pancreatic adenocarcinoma cells (42). As the positive regulator of epithelial-to-mesenchymal transition (EMT), miR-29a/b/c-specific target CEACAM6 regulates its expression at the post-transcriptional level (43). In a mouse xenograft model, ASPER-29 treatment significantly blocked the metastasis of BxPC-3 pancreatic adenocarcinoma cells to lung and liver tissues (44).
Mechanically, CEACAM6 promoted proliferation of pancreatic adenocarcinoma by increased cyclin D1 and cyclin-dependent kinase (CDK)4 protein levels, as confirmed by knockdown of endogenous CEACAM6 in BxPC-3 and SW1990 cells and overexpression of CEACAM6 in MIA PaCa-2 cells (26). CEACAM6 signaling by cross-linking with its antibody reduced the phosphorylation of caveolin-1, leading to reduced recruitment of C-terminal Src kinase (Csk) to the membrane. Csk negatively regulates phosphorylation of c-Src 527 tyrosine residues, leading to reduced tyrosine phosphorylation at a variety of downstream targets (45). The role of c-Src in mediating increases in αvβ3 integrin binding activity was observed. This increase in activity enhances cellular adhesion to its classical ligand, vitronectin, as well as to fibronectin. As a result, c-Src affects cell adhesion, migration, proliferation and survival (23). Treatment with anti-avβ3 integrin almost completely abolished the increase in vitronectin adhesion following CEACAM6 crosslinking. Both 4-amino-5-(4-chlorophenyl)-7-(t-butyl) pyrazolo(3,4-d) pyrimidine, an inhibitor of c-Src family kinases, and c-Src knockdown markedly attenuated the enhanced cellular adhesion to vitronectin following CEACAM6 crosslinking (23). In addition, CEACAM6 can modulate pancreatic adenocarcinoma cellular invasiveness by c-Src-dependent modulation of matrix metalloproteinase (MMP)-9 activity (46). CEACAM6 can antagonize the Src signaling pathway, downregulate cancer cell cytoskeleton proteins and block adenovirus trafficking to the nucleus of human pancreatic adenocarcinoma cells, which decreases the antitumor effect of an oncolytic adenovirus (47). CEACAM6 in pancreatic adenocarcinoma cell lines promoted Akt phosphorylation (Ser-473) under anchorage-independent conditions (41), and increased Akt kinase activities (48). Akt kinase is both necessary and sufficient to induce insulin-like growth factor I receptor (IGF-IR) upregulation (48). IGF-IR enhances the expression of MMP-2 by interacting with its ligand IGF-I.
Poorer OS after radical treatment and adjuvant chemotherapy in patients with pancreatic adenocarcinoma was significantly dependent on elevated CEACAM6 blood levels (17.0 vs. 12.6 months, P=0.017) (27). This suggests that CEACAM6 is a promising new biomarker for patients with pancreatic adenocarcinoma, with important prognostic value and predictive properties of chemotherapy resistance. Real-time monitoring of CEACAM6 serum concentrations may improve individualized treatment approaches for patients with pancreatic adenocarcinoma (27).
CRC
CEACAM6 expression is upregulated in proliferative polyps and early adenomas, indicating early molecular events in colorectal tumor progression (49). CEACAM6 expression was increased in CRC and its level on the CRC cell surface was inversely correlated with the degree of cellular differentiation (29). After analyzing the tissue microarrays from a randomized controlled clinical trial of adjuvant fluorouracil-based chemotherapy, which consisted of 243 paraffin-embedded biopsies, it was observed that CEACAM6 expression in CRC served as an independent prognostic factor. In addition, it was found that CEACAM6 overexpression was associated with poorer OS and DFS (50). Mechanistically, it was discovered that CD151, through TGFβ1, upregulates CEACAM6 expression, thereby promoting CRC progression (51). TGF-β1 and TGF-β receptor 1 are highly expressed in a variety of tumors, such as breast cancer, colon cancer, gastric cancer and hepatocellular carcinoma (52). The expression of CEACAM6 is enhanced by CD151 or TGFβ1. This presents a potential avenue for developing innovative targeted therapies aimed at CD151 or TGFβ1 to address tumors with upregulated CEACAM6.
Immunohistochemical analysis and reverse transcription-quantitative PCR (RT-qPCR) were used to analyze 78 colon cancer specimens, which included 40 cases of stage I-II and 38 cases of stage III-IV. In addition, 30 cases of colonic adenoma and 12 healthy controls were included in the study (53). The findings revealed a gradual increase in the expression of CEACAM6 from normal colonic mucosa to colonic adenoma to colonic cancer. Of note, forkhead box (FOX)P3 exhibited a similar expression pattern to CEACAM6 (53). In a 1,2-dimethylhydantoin-induced colorectal cancer model in rats, immunization of rats with recombinant attenuated Salmonella containing the CEACAM6 and 4-1BB ligand genes reduced the number of FOXP3 cells and inhibited colorectal carcinogenesis (54). FOXP3 as a transcription factor is a crucial regulator in the development and function of regulatory T cells (Treg). Numerous studies have indicated that FOXP3 promotes carcinogenesis (55,56). The relationship between CEACAM6 and FOXP3 in the development of colon cancer needs to be further explored.
For the early screening of patients with CRC and healthy individuals, the potential markers CEACAM6, tetraspanin 8, galectin 4 and collagen type I α2 chain (the four genes in combination are known as CELTiC) were examined. The validation of these candidate markers was conducted through RT-qPCR using blood samples from 67 patients and 67 healthy controls. The sensitivity and specificity of the two groups were determined to be 92 and 67%, respectively (57). The CELTiC panel was subsequently analyzed using a nonparametric test and multinomial logistic model in a population of fecal immunochemical test (FIT)-positive subjects, who were compared with patients with CRC and healthy individuals, confirming its ability to identify patients with high-risk lesions, and appeared able to discriminate false-positive FIT and low-risk patients (non-advanced adenoma and polyps) (58). Furthermore, CELTiC was able to distinguish between patients with CRC and false-positive FIT participants (59). The biggest advantage is that RT-qPCR is highly sensitive and specific and more efficient and low-cost for disease screening (60).
Gastric cancer (GC)
The expression of CEACAM6 was found to be higher in early GC compared to normal tissue, suggesting that upregulation of CEACAM6 may be an early event in the development of human GC (61). CEACAM6 mRNA and protein levels were detected by RT-qPCR and immunohistochemistry in 75 cases of primary gastric adenocarcinoma, 20 cases of paraneoplastic tissues and normal gastric mucosal tissues. Analysis revealed the level of CEACAM6 was not associated with tumor size, histological differentiation or tumor subtype (62). However, patients with high CEACAM6 expression had a significantly shorter median survival time after surgery than patients with low CEACAM6 expression (17 vs. 43 months; log-rank test P=0.046), suggesting that CEACAM6 is associated with poor clinical prognosis (63,64). Furthermore, elevated CEACAM6 in GC is significantly associated with lymph node metastasis, distant metastasis (63) and advanced stage (62).
In terms of basic research, forced CEACAM6 expression in GC cells (MKN-45, SGC-7901) promoted migration and invasion in vitro and athymic mice, whereas migration and invasion of GC cells (MKN-28, SNU-16) were suppressed by knockdown of CEACAM6 (65). Regarding the mechanism, CEACAM6 induced EMT in GC, by causing increases in the EMT markers N-cadherin, vimentin and Slug, while E-cadherin expression was decreased (66). The Snail family of zinc finger transcription factors, including Snail and Slug, is involved in EMT during development. Slug was first described as a transcription factor expressed in cells undergoing EMT during gastrulation and neural crest emergence in chickens (67-71). Furthermore, E-cadherin expression was negatively associated with the depth of tumor invasion, lymph node metastasis and TNM stage in GC tissues (66). CEACAM6 also increased the levels of phosphorylated (p-) AKT, which is involved in the progression of a variety of human tumors. LY294002, a PI3K inhibitor, could reverse CEACAM6-induced EMT via mesenchymal-epithelial transition (66). CEACAM6 elevated MMP-9 activity in GC cells, and anti-MMP-9 antibody could reverse the increasing invasion and migration induced by CEACAM6 (66). CEACAM6 increased the levels of p-Src (65), p-PI3K, p-Akt and MMP9 proteins, and when Src signaling was inhibited, CEACAM6 failed to restore the upregulation of these proteins, indicating that p-Src is at a more upstream position in this signaling pathway (64). CEACAM6 also enhances tubule formation in human umbilical vein endothelial cells and promotes the formation of angiogenic mimetics in GC cells. Mechanistically, CEACAM6 induces the phosphorylation of FAK and paxillin in GC cells (SCG-7901, MKN-45), and the tubule and angiogenic mimetic formation induced by CEACAM6 is reduced by the FAK inhibitor Y15 (72). This provides a potential target for anti-vascular treatment of CEACAM6-overexpressing tumors. In addition, SMAD3-mediated TGFβ signaling increased the activity of CEACAM6 promoter and induced the expression of CEACAM6 in GC cell lines (SNU638, SNU484) (73). Circ_0008035 regulated the expression of CEACAM6 by targeting miR-1256, thereby promoting the development of GC (74). There are reports that tamoxifen could be a therapeutic alternative for patients with GC with CEACAM6 overexpression (63).
CEACAM6 was also found to play a role in gastric infection with Helicobacter pylori and its carcinogenesis (61). Infection with H. pylori is the strongest risk factor for the development of GC, and cytotoxin-associated gene A (CagA) protein is the most important bacterial oncoprotein (75). Helicobacter pylori conserved outer membrane adhesin (HopQ) may interact with CEACAM6 of the human gastric cells to induce the development of gastric ulcers and cancers by transferring CagA oncoprotein or inducing activation of the type-IV secretion system to initiate and maintain inflammatory reactions (76). Infection of wild-type MKN28 cells by a CagA and live CagA wild-type H. pylori strain in vitro resulted in upregulation of CEACAM6 (61). Further studies revealed that neutrophils interacted with H. pylori in a HopQ-dependent manner, markedly promoting CagA translocation and phosphorylation and prolonging survival within neutrophils (77). It suggests that H. pylori infects the stomach through CEACAM6, which in turn promotes gastric expression of CEACAM6 to exacerbate the infection, and promotes inflammation through neutrophils, which explains why elevated expression of CEACAM6 is present in the early stages of GC.
For early detection of GC, anti-CEACAM6 probe-assisted fluorescence endoscopy may be a potential option for the diagnosis of precancerous lesions (61,64). Furthermore, CEACAM6 may be a predictor for patients with advanced GC, and higher expression of CEACAM6 was found to be associated with short postoperative survival time of patients with GC (62). In addition, combination diagnosis of GC with multiple molecules, including CEACAM6, is increasingly reported. CEACAM6, CEAMCAM5, EpCAM and CA72-4 form a versatile set of markers for robust discrimination of GC from adjacent normal tissue (78). CEACAM6 combined with ITGB1 and CYR61 in peripheral blood of patients with GC is more sensitive than CEA, IGF-IR, cytokeratin 20 and S100A4 for early diagnosis of metastasis and recurrence (79).
Breast cancer
In a multicenter study, CEACAM6 expression was detected in 37.1% (312/840) of primary breast cancers (80). The human EGFR2 (HER2)-positive subtype showed the highest CEACAM6 expression (62.7%) and other subtypes showed relatively low expression (21.8-37.5%) (80). This was also confirmed in another study; HER2-positive subtype breast cancers showed a high proportion of CEACAM6 (83%), and this phenomenon was observed in 13 established breast cancer cell lines (81). Of note, significantly worse OS was found in high nodal stage HER2-positive cancers with CEACAM6 positivity (80). HER2-overexpressing breast cancer cells SK-BR-3 were treated with TGFβ or EGF, resulting in SMAD3 phosphorylation and CEACAM6 expression (80). A previous study showed that the CD151/TGF-β1/Smad3 axis has a role in promoting breast cancer progression, leading to a more advanced tumor stage, lymph node metastasis, distant metastases and poorer prognosis (82). It suggested that CEACAM6 is a target gene for SMAD3-mediated TGFβ signaling (73). HER2 signaling synergistically regulates transcription of SMAD target genes and signaling pathways with TGFβ (83). This suggests that CEACAM6 expression in breast cancer is associated with HER2 and TGFβ signaling.
It was also reported that CEACAM6 was associated with tamoxifen resistance in human breast cancer (1,84). In the breast cancer cell lines MCF-7:5C and MCF-7:2A, CEACAM6 was overexpressed compared to wild-type MCF-7 cells, and acquired tamoxifen resistance lead to invasion and migration by upregulation of p-Akt and p-c-Src (85). CEACAM6 was found to have a significantly lower expression in trastuzumab-responsive than trastuzumab-resistant breast cancers (86). Thus, CEACAM6 may contribute to trastuzumab resistance. CEACAM6 may function differently in a context-dependent manner in breast cancers. CEACAM6 expression was higher in breast cancer with bone metastases than other metastatic sites (87). After MCF-7 cells were induced to become breast cancer stem cells (BCSCs), the expression of CEACAM6 was increased, while knockdown of CEACAM6 in BCSCs reduced its proliferation (88). However, CEACAM6 expression was significantly lower in basal-like breast cancers, the other aggressive breast cancer subtype with poor outcome (80). This indicates the heterogeneity of CEACAM6 function in different cancer subtypes.
Cholangiocarcinoma
CEACAM6 was overexpressed in highly differentiated and less differentiated cholangiocarcinoma tissues compared with para-cancerous tissues, and the expression level of CEACAM6 was negatively associated with the degree of differentiation of cholangiocarcinoma (89). Patients with high CEACAM6 expression showed a significantly poorer DFS rate than those with low CEACAM6 expression (90). Silencing CEACAM6 inhibited cell viability, invasion and migration but promoted cell apoptosis in the human cholangiocarcinoma cell line RBE. Mechanistically, CEACAM6 knockdown decreased the expression of Bcl-2, N-cadherin, MMP-2 and MMP-9, Twist1, vimentin, VEGFA and intercellular adhesion molecule-1 (ICAM-1), but increased the expression of BAX and cleaved caspase-3, caspase-8 and caspase-9 and E-cadherin. Besides, CEACAM6 knockdown reduced the expression of the SRC/PI3K/AKT signaling pathway (89). Besides, overexpression of CEACAM6 caused resistance to gemcitabine in cholangiocarcinoma. Among cholangiocarcinoma cell lines, TFK-1 with high CEACAM6 expression was more resistant to gemcitabine than HuCC-T1 and MEC with low CEACAM6 expression. This was confirmed by CEACAM6 overexpression in HuCC-T1 and knockdown reduction in TFK-1 (90).
CEACAM6 was overexpressed in cholangiocarcinoma (89), and high CEACAM6 levels showed a significantly poorer DFS rate (90). A study confirmed the diagnostic value of the CEACAM6 concentration in bile for cholangiocarcinoma (91). Bile from 83 patients (42 with benign disease, 41 with cholangiocarcinoma) was collected at the time of index operation. The concentration of CEACAM6 was quantified by sandwich ELISA and correlated to the pathological diagnosis. Patients in the benign group had lower median biliary CEACAM6 levels than patients in the malignant group (7.5 vs. 40 ng/ml; P≤0.001). Receiver operating characteristic curve analysis determined CEACAM6 to be a positive predictor of cholangiocarcinoma with a CEACAM6 level >14 ng/ml associated with 87.5% sensitivity and 69.1% specificity.
Gallbladder cancer (GBC)
In human GBC, CEACAM6 gene expression was significantly greater in GBC tissues than in peritumoral tissues and its positive expression was associated with poor prognosis. CEACAM6 knockdown in GBC-SD and SGC-996 cell lines inhibited GBC cell proliferation, migration and invasion but promoted cell apoptosis. Mechanistically, CEACAM6 knockdown inhibited migration and invasion by decreasing MMP2 and vimentin, promoted cell apoptosis by increasing BAX and decreasing BCL-2, and inhibited proliferation by increasing the cell population in the G0/G1 phase and reducing it in G2/M and S phases (92).
Ovarian neoplasms
CEACAM6 overexpression was demonstrated in 13/16 (81%) borderline, low-grade and high-grade invasive mucinous ovarian neoplasms (MON), compared to 5/50 (10%) serous and 1/5 (20%) benign mucinous samples (93). Another study found that mucinous ovarian adenocarcinomas had almost 3-fold more CEACAM6 than serous ovarian adenocarcinomas by analysis of immunohistochemical results from tissue microarrays (25). This result is consistent with the cell lines. CEACAM6 was expressed in 2 of 3 mucinous cancer cell lines and was not expressed in any of the 2 normal human ovarian surface epithelium, 7 serous cancer and 2 clear cell cancer cell lines (93). Of note, there was no difference in CEACAM6 expression amongst various grades of MON, including borderline, low-grade and high-grade MON, at the mRNA or protein level (93). This raises the possibility that aberrant CEACAM6 expression may occur early in mucinous ovarian cancer tumorigenesis. In addition, high expression of CEACAM6 may lead to carboplatin resistance in ovarian cancer (94). Interestingly, CEACAM6 expression was noted in mucinous secretions by immunohistochemistry, and this raises the possibility that CEACAM6 may be useful as a serum marker for MON (93).
In patient-derived xenograft models, the recurrence and acquisition of drug resistance in carboplatin-sensitive high-grade ovarian cancer was attended by elevated expression of CEACAM6, crystallin αB and SOX2 genes at the protein and RNA levels (94). This suggests that CEACAM6 has a role in drug resistance and recurrence in high-grade ovarian cancer.
Head and neck squamous cell carcinoma (HNSCC)
Bioinformatics analysis revealed that CEACAM6 gene expression was downregulated in most squamous carcinoma tissues, such as laryngeal, hypopharyngeal and oropharyngeal carcinomas, compared to adjacent normal tissues (95). With consistent results, laryngeal squamous cell carcinoma (LSCC) showed downregulated CEACAM6 expression (96). Expression microarrays of 16 LSCC samples (11 cell lines and 5 primary tumors) indicated downregulation of CEACAM6 compared to non-tumor controls. It was further confirmed by RT-qPCR of 25 LSCC cell lines that the CEACAM6 gene was downregulated in recurrent, advanced and high-grade tumors compared to controls. Mechanistically, bisulfite pyrophosphate sequencing identified 9/25 (36%) LSCC cell lines with DNA hypermethylation in the CEACAM6 promoter region (mean DNA methylation, >78%). In addition, 5-aza-2-deoxycytidineinduced DNA methylation inhibition restored CEACAM6 expression at the mRNA level in both LSCC cell lines. However, another study showed the opposite result: CEACAM6 expression was significantly increased in highly tumorigenic HNSCC cell lines relative to less tumorigenic HNSCC cell lines. Mechanistically, CEACAM6 enhances tumor-initiating activity and tumor growth by activating AKT and inhibiting caspase-3-mediated cell death (97). In addition, glycosylated CEACAM6 was found to be a tumor marker associated with recurrence in patients with early-stage OSCC (20). Mechanistically, N-glycosylation at Asn256 of CEACAM6 mediated by α-1,6-mannosylglycoptotein 6-β-N-acetylglucosaminyltransferse was found to promote OSCC cell invasion and migration, cytoskeletal rearrangement and metastasis. This effect was attributed to the interaction between glycosylated CEACAM6 and EGFR, which enhanced EGFR activation, clustering and intracellular signaling cascades. In addition, treatment with anti-CEACAM6 antibody inhibited the oncological effects and EGF-induced signaling in cells overexpressing CEACAM6.
Osteosarcoma
CEACAM6 expression was found to be significantly upregulated in metastatic osteosarcoma tissues compared to nonmetastatic tissues. Furthermore, the upregulation of CEACAM6 was strongly associated with the presence of lung metastasis. Survival analysis revealed that patients with osteosarcoma with high CEACAM6 expression had significantly shorter OS and lung metastasis-free survival compared to those with low CEACAM6 expression. In addition, knockdown of CEACAM6 was observed to inhibit osteosarcoma cell migration, invasion and EMT (98).
Acute leukemias
Acute leukemias showed overexpression of CEACAM6 compared with normal granulocytes (99). In acute lymphoblastic leukemia cells, cross-linking with anti-CEACAM6 antibody can imitate CEACAM6 signaling. Mechanistically, CEACAM6 signaling promotes ERK1/2, Akt and P38 MAPK phosphorylation and integrin upregulation, as well as enhanced integrin ligand binding, including vascular cell adhesion molecule-1 and ICAM-1 (100).
CEACAM6 is a potential diagnostic marker for plasma cell diseases (101). CEACAM6 levels in peripheral blood are higher in plasma cell disorders than in healthy controls. A marked difference of CEACAM6 in peripheral blood was observed between healthy controls (median, 15.2 pg/ml; interquartile range, 12.1-17.1 pg/ml), monoclonal gammopathy of unknown significance patients (median, 19.0; interquartile range, 16.4-22.5 pg/ml), newly diagnosed patients with multiple myeloma (median, 18.0; interquartile range, 13.4-21.2 pg/ml) and patients with relapsed/refractory multiple myeloma (median, 18.9; interquartile range, 16.5-24.1 pg/ml) (P<0.001).
Urinary tract cancers
CEACAM6 expression was found to be upregulated in clear cell renal cell carcinoma (ccRCC) through RT-qPCR, immunohistochemical staining and western blot analysis of ccRCC tumor tissues and cell lines. Furthermore, patients with relatively low CEACAM6 levels exhibited longer OS. Overexpression of CEACAM6 was found to enhance proliferation and migration in ccRCC cells. Conversely, depletion of CEACAM6 through short hairpin RNA was observed to modulate these changes. Further investigation revealed that the activation of the ERK/AKT signaling pathway has a crucial role in these processes. In addition, blocking the PI3K/AKT pathway was able to negate the effects of CEACAM6 overexpression (102).
Furthermore, urine examination for urothelial carcinoma of the bladder (UCB) revealed elevated expression of CEACAM1, CEACAM5 and CEACAM6, which may be used as a potential biomarker for the screening of UCB of the bladder (103).
4. Molecular therapy of targeting CEACAM6
The molecular heterogeneity of tumor cells has likely contributed to the lack of effective targeted therapies (104). Optimal proteins to target using theragnostic agents must exhibit high membrane expression on cancerous tissue with low expression on healthy tissue to afford improved disease outcomes with minimal off-target effects and toxicities (105). CEACAM6 has emerged as a highly specific marker for invasive cancers (106) and shows great potential as a target for effective cancer therapy (107). High expression of CEACAM6 is associated with unfavorable OS and DFS in different malignancies (108). Targeting CEACAM6 with antibodies has been reported in numerous tumors and has a significant role in tumor treatment, including directly blocking the intracellular signaling it causes, attenuating drug resistance and mediating the accumulation of drugs in tumor cells.
CEACAM6 has been identified as a target candidate for chimeric antigen receptor T (CAR-T) cell therapy for immunotherapy of pancreatic adenocarcinoma (109). Binding and cross-linking of CEACAM6 to cytotoxic T cells inhibits its activation, resulting in T-cell nonresponse. Blockade of CEACAM6 on the surface of myeloma cells by specific monoclonal antibodies or knockdown of the CEACAM6 gene restores T-cell responsiveness to malignant plasma cells (110). In vitro experiments have shown that the humanized CEACAM6 blocking antibody BAY1834942 is equally or more effective than blocking programmed cell death 1 (PD-1) ligand 1 in activating T-cell anti-tumor responses, and that this effect is enhanced when combined with anti-PD-1 antibodies (19). CEACAM6 ligand vaccine demonstrated good immune antitumor effects in a rat colorectal cancer model via mechanisms including promoting antitumor immune response, increasing the number of CD45RO+ memory T cells, decreasing the number of FOXP3+ cells and promoting type 1 T-helper cell polarization (111). CEACAM6 as a subtype-specific tumor surface antigen demonstrates the potential for CAR-T cell and antibody-drug coupled therapy in breast cancer (112).
Humanized monoclonal antibody NEO-201, with a potential target of CEACAM6, exhibits specificity for a variety of cancers, including colon, pancreatic and mucinous ovarian cancer tissues, as well as cell lines, and showed no cross-reactivity with surrounding healthy tissues (113). It has also shown good anti-tumor effects in in vitro cellular assays and in vivo animal models. Functional analysis revealed that NEO-201 mediates antibody-dependent cellular cytotoxicity killing against human ovarian and CRC cell lines in vitro. Anti-CEACAM6 single domain antibody (sdAb) showed good anti-tumor effects in pancreatic adenocarcinoma cell models, including reduction of cell proliferation, invasion, and metastasis, and inhibition of angiogenesis (114). Antibodies designed against human CEACAM6 in a mouse xenograft model of pancreatic adenocarcinoma inhibited tumor growth (40,115) and enhanced tumor cell apoptosis (40), and were more effective when used in combination with gemcitabine (115). Optimization of antibodies allows for better results in in vivo experiments. Heavy chain antibody 2A3-mFc is superior to the single domain antibody and the full-length antibody regarding tumor detection and pharmacokinetics (116). Multivalent antibodies show higher affinity and therapeutic efficacy. The developed bivalent sdAb and quadrivalent sdAb anti-CEACAM6 antibodies inhibited tumor invasion and migration in the NSCLC cell line A549. In addition, these antibodies also inhibited tumor growth in A549-derived xenograft models (104).
Antibodies coupled to special carriers help enhance their function and improve the therapeutic effect. CEACAM6-targeting albumin-based nanoparticles allowed the delivery of drugs to metastatic anoikis-resistant tumor cells in vitro and in vivo. Based on the expression of CEACAM6 in a variety of tumors, this may be used to target various types of metastatic tumor cells (117). It was demonstrated that polyethylene glycol-modified iron oxide nanoparticles triple single chain antibodies (scAbs) (scAb mucin 4, scAbCEACAM6 and sc variable fragments CD44v6) had an excellent performance in the diagnosis and treatment of pancreatic adenocarcinoma (118). It significantly shortened the MRI T2-weighted signal intensity both in vitro and in vivo and also showed a favorable anti-pancreatic adenocarcinoma effect. In addition, the delivery of siCEACAM6 to LUAD using the pH low insertion peptide has therapeutic potential as a unique cancer treatment approach (33). At least it has shown good effects in vitro cellular models and in vivo nude mice xenograft tumor models.
Anoikis is the apoptotic response induced in normal cells by inadequate or inappropriate adhesion to substrate. CEACAM6 is highly expressed in metastatic anoikis-resistant tumor cells (119). Treatment with anti-CEACAM6 monoclonal antibody clone 8F5 to A549 cells decreased cellular CEACAM6 expression and reversed anoikis resistance (34). Furthermore, the utilization of human serum albumin nanomedicine targeted CEACAM6 by delivering an encapsulated chemotherapeutic drug, adriamycin, which effectively targeted circulating metastatic anoikis-resistant tumor cells (117).
CEACAM6, as a molecule specifically expressed in a variety of epithelial tumors, has a role in tumor development. Targeting CEACAM6 has also demonstrated favorable therapeutic effects in cellular and animal models. Studies related to cellular and animal models of CEACAM6 molecular targeting therapy are listed in Table IV, including tumor type, treatment method and treatment effect. It is expected that in the future, clinical research and applications of targeted CEACAM6 will provide better results.
Table IV.
CEACAM6 targeted therapy in cancers.
| Author, year | Cancer type | Method of treatment | Models | Effect of treatment | (Refs.) |
|---|---|---|---|---|---|
| Wu, 2021 | NSCLC | Bivalent sdAb (2Ab) tetravalent sdAb (4Ab) | A549 cells, murine xenograft models | Inhibition of migration and invasion | (104) |
| Zou, 2021 | Pancreatic cancer | Triple scAbs | Murine xenograft models | Shortening of the MRI T2-weighted signal intensity, favorable anti-pancreatic cancer effect | (118) |
| Zeligs, 2020 | Ovarian, colorectal carcinoma | NEO-201 | OV90 and LST174T cells, primary and metastatic ovarian cancer models | Decreases cell viability | (113) |
| Son, 2019 | Lung adenocarcinoma | pHLIP-siCEACAM6 | Murine xenograft models | Tumour growth inhibition up to 35.5%. Tumour growth inhibition up to 47% combined with cisplatin | (33) |
| Lee, 2018 | Metastatic anoikis-resistant tumor cells | tHSA-NPs | Murine xenograft models | Effective inhibition of lung metastasis of malignant circulating anoikis-resistant tumor cells. Detection of anti-anoikis tumor cells by photoacoustic imaging | (117) |
| Hong, 2015 | Lung adenocarcinoma | 8F5 | Murine xenograft models | Tumor growth inhibition of 40%, tumor growth inhibition of up to 80% in combination with paclitaxel for 8F5 | (34) |
| Cheng, 2014 | Pancreatic cancer | 2A3, 2A3-Fc | BxPC3 cells | Inhibited MMP-9 activity by 33%, BxPC3 invasion by 73%, and capillary length by 21 and 49% | (114) |
| Niu, 2012 | Pancreatic cancer | 2A3, 2A3-mFc, 9A6 | BxPC3 cells | 2A3-mFc is superior to 2A3 and 9A6 in terms of tumor detection and pharmacokinetics | (116) |
| Riley, 2009 | Pancreatic cancer | scFvs | Murine xenograft models | Tumor growth inhibition, enhanced in combination with gemcitabine. Promotes apoptosis and inhibits angiogenesis and proliferation | (115) |
| Duxbury, 2004 | Pancreatic cancer | By114 | BxPC3 cells, murine xenograft models | Inhibits tumor growth and enhances tumor apoptosis | (40) |
| Blumenthal, 2005 | Pancreatic, breast, and colonic cancer | MN-15, MN-3, MN-14 | Human pancreatic, breast and colonic cancer cells. GW-39 cells, human colonic micrometastasis model | Decreased adhesion of tumor cells to endothelial cells by 49-58%. Treatment of colonic micrometastasis model with MN-3 Fab' or MN-15 Fab' increased their survival | (121) |
NSCLC, non-small cell lung cancer; Triple scAbs, scAbMUC4, scAbCEACAM6, scFvCD44v6; pHLIP-siCEACAM6, small inhibitory RNA targeting CEACAM6 to a peptide with a low pH-induced transmembrane structure; tHSA-NPs, human serum albumin-based nanoparticles (with photoacoustic imaging capability, which target CEACAM6); scFvs, humanized anti-CEACAM6 single chain variable fragments; Ab, antibody; MUC, mucin; CEACAM6, carcinoembryonic antigen-related cell adhesion molecule 6.
5. Conclusions
Accumulating evidence has indicated the important roles of CEACAM6 in the pathogenesis, clinical manifestations and management of cancer, including proliferation, invasion, metastasis, angiogenesis, anoikis resistance, drug resistance, diagnosis and prognosis. Upstream of CEACAM6 signaling, CD151 promotes SMAD3 phosphorylation via TGFβ1. This leads to the phosphorylation of SMAD3, which in turn promotes the transcription of CEACAM6. In order to investigate the functions of CD151 and the TGFβ/SMAD3 axis in different types of tumors, further experiments using various cellular and animal tumor models are needed. On the other hand, miR-146, miR-26a, miR-29a/b/c, miR-128 and miR-1256 inhibit the transcription of CEACAM6. The transcriptional expression of CEACAM6 is also regulated by promoter DNA methylation. In addition, α-1,6-mannosylglycoptotein 6-β-N -acetylglucosaminyltransferse facilitates N-glycosylation at Asn256 of the CEACAM6 protein. Downstream of CEACAM6 signaling, CEACAM6 promotes tumor proliferation by increasing cyclin D1 and CDK4 protein levels. CEACAM6 activates the ERK1/2/MAPK or SRC/FAK/PI3K/AKT pathways either directly or via EGFR to stimulate tumor proliferation, invasion, migration, resistance to anoikis and chemotherapy, as well as angiogenesis. CEACAM6 has shown favorable results in basic tumor research in diagnosis and targeted therapy, with potential translational application value. Clinical experiments based on large sample size and further in-depth mechanistic research are the future research directions of CEACAM6. CEACAM6 may provide new ideas for the treatment of tumors.
Acknowledgments
Not applicable.
Abbreviations
- CEACAM6
carcinoembryonic antigen-related cell adhesion molecule 6
- OSCC
oral squamous cell carcinoma
- HNSCC
head and neck squamous cell carcinoma
- LSCC
laryngeal squamous cell carcinoma
- ALL
acute lymphoblastic leukemia
- ccRCC
clear cell renal cell carcinoma
- BCP-ALL
B-cell precursor ALL
- ELISA
enzyme-linked immunosorbent assay
- CEA
carcinoembryonic antigen
- GPI
glycosyl phosphatidyl inositol
- DFS
disease-free survival
- OS
overall survival
- EMT
epithelial-to-mesenchymal transition
- IGF-IR
IGF-I receptor
- IGF-I
insulin-like growth factor I
- CRC
colorectal carcinoma
- GBC
gallbladder cancer
- MON
mucinous ovarian neoplasm
- EGFR
epidermal growth factor receptor
- ICAM-1
intercellular adhesion molecule-1
- RT-qPCR
reverse transcription-quantitative polymerase chain reaction
- LUAD
lung adenocarcinoma
- Csk
C-terminal Src kinase
- FIT
fecal immunochemical test
- CELTiC
the four genes CEACAM6, tetraspanin 8, galectin 4 and collagen type I α2 chain in combination
- CagA
cytotoxin-associated gene A
- HopQ
Helicobacter pylori conserved outer membrane adhesin HopQ
- UCB
urothelial carcinoma of the bladder
- CAR-T
chimeric antigen receptor T
- pHLIP
pH low insertion peptide
Funding Statement
This work was supported by the National Natural Science Foundation of China (grant nos. 81974438 and 82173069).
Availability of data and materials
Not applicable.
Authors' contributions
GW and YC conceived the study and wrote the manuscript. DW, FX, QW, WL and JC conducted literature search/selection and data extraction. YC (corresponding author) revised and edited the manuscript. All authors have read and approved the final manuscript. Data authentication is not applicable.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Beauchemin N, Arabzadeh A. Carcinoembryonic antigen-related cell adhesion molecules (CEACAMs) in cancer progression and metastasis. Cancer Metastasis Rev. 2013;32:643–671. doi: 10.1007/s10555-013-9444-6. [DOI] [PubMed] [Google Scholar]
- 2.Zid M, Drouin G. Gene conversions are under purifying selection in the carcinoembryonic antigen immunoglobulin gene families of primates. Genomics. 2013;102:301–309. doi: 10.1016/j.ygeno.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 3.Naghibalhossaini F, Yoder AD, Tobi M, Stanners CP. Evolution of a tumorigenic property conferred by glycophosphatidyl-inositol membrane anchors of carcinoembryonic antigen gene family members during the primate radiation. Mol Biol Cell. 2007;18:1366–1374. doi: 10.1091/mbc.E06-10-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wautier JL, Wautier MP. Old and new blood markers in human colorectal cancer. Int J Mol Sci. 2022;23:12968. doi: 10.3390/ijms232112968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Obrink B. CEA adhesion molecules: Multifunctional proteins with signal-regulatory properties. Curr Opin Cell Biol. 1997;9:616–626. doi: 10.1016/S0955-0674(97)80114-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuespert K, Pils S, Hauck CR. CEACAMs: Their role in physiology and pathophysiology. Curr Opin Cell Biol. 2006;18:565–571. doi: 10.1016/j.ceb.2006.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Voges M, Bachmann V, Naujoks J, Kopp K, Hauck CR. Extracellular IgC2 constant domains of CEACAMs mediate PI3K sensitivity during uptake of pathogens. PLoS One. 2012;7:e39908. doi: 10.1371/journal.pone.0039908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuijpers TW, Hoogerwerf M, van der Laan LJ, Nagel G, van der Schoot CE, Grunert F, Roos D. CD66 nonspecific cross-reacting antigens are involved in neutrophil adherence to cytokine-activated endothelial cells. J Cell Biol. 1992;118:457–466. doi: 10.1083/jcb.118.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frängsmyr L, Baranov V, Hammarström S. Four carcinoembryonic antigen subfamily members, CEA, NCA, BGP and CGM2, selectively expressed in the normal human colonic epithelium, are integral components of the fuzzy coat. Tumour Biol. 1999;20:277–292. doi: 10.1159/000030075. [DOI] [PubMed] [Google Scholar]
- 10.Oikawa S, Inuzuka C, Kuroki M, Arakawa F, Matsuoka Y, Kosaki G, Nakazato H. A specific heterotypic cell adhesion activity between members of carcinoembryonic antigen family, W272 and NCA, is mediated by N-domains. J Biol Chem. 1991;266:7995–8001. [PubMed] [Google Scholar]
- 11.Yamanaka T, Kuroki M, Kinugasa T, Matsuo Y, Matsuoka Y. Preparation and characterization of two human carcinoembryonic antigen family proteins of neutrophils, CD66b and c, in silkworm larvae. Protein Expr Purif. 1996;7:438–446. doi: 10.1006/prep.1996.0065. [DOI] [PubMed] [Google Scholar]
- 12.Kuroki M, Abe H, Imakiirei T, Liao S, Uchida H, Yamauchi Y, Oikawa S, Kuroki M. Identification and comparison of residues critical for cell-adhesion activities of two neutrophil CD66 antigens, CEACAM6 and CEACAM8. J Leukoc Biol. 2001;70:543–550. [PubMed] [Google Scholar]
- 13.Xu C, Ng DTW. Glycosylation-directed quality control of protein folding. Nat Rev Mol Cell Biol. 2015;16:742–752. doi: 10.1038/nrm4073. [DOI] [PubMed] [Google Scholar]
- 14.Skubitz KM, Campbell KD, Skubitz AP. CD66a, CD66b, CD66c, and CD66d each independently stimulate neutrophils. J Leukoc Biol. 1996;60:106–117. doi: 10.1002/jlb.60.1.106. [DOI] [PubMed] [Google Scholar]
- 15.Skubitz KM, Campbell KD, Skubitz AP. Synthetic peptides from the N-domains of CEACAMs activate neutrophils. J Pept Res. 2001;58:515–526. doi: 10.1034/j.1399-3011.2001.00931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oikawa S, Inuzuka C, Kuroki M, Matsuoka Y, Kosaki G, Nakazato H. Cell adhesion activity of non-specific cross-reacting antigen (NCA) and carcinoembryonic antigen (CEA) expressed on CHO cell surface: Homophilic and heterophilic adhesion. Biochem Biophys Res Commun. 1989;164:39–45. doi: 10.1016/0006-291x(89)91679-3. [DOI] [PubMed] [Google Scholar]
- 17.Bonsor DA, Günther S, Beadenkopf R, Beckett D, Sundberg EJ. Diverse oligomeric states of CEACAM IgV domains. Proc Natl Acad Sci USA. 2015;112:13561–13566. doi: 10.1073/pnas.1509511112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang R, Meng T, Zha Q, Cheng K, Zhou X, Zheng J, Zhang D, Liu R. The predicting roles of carcinoembryonic antigen and its underlying mechanism in the progression of coronavirus disease 2019. Crit Care. 2021;25:234. doi: 10.1186/s13054-021-03661-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pinkert J, Boehm HH, Trautwein M, Doecke WD, Wessel F, Ge Y, Gutierrez EM, Carretero R, Freiberg C, Gritzan U, et al. T cell-mediated elimination of cancer cells by blocking CEACAM6-CEACAM1 interaction. Oncoimmunology. 2022;11:2008110. doi: 10.1080/2162402X.2021.2008110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiang WF, Cheng TM, Chang CC, Pan SH, Changou CA, Chang TH, Lee KH, Wu SY, Chen YF, Chuang KH, et al. Carcinoembryonic antigen-related cell adhesion molecule 6 (CEACAM6) promotes EGF receptor signaling of oral squamous cell carcinoma metastasis via the complex N-glycosylation. Oncogene. 2018;37:116–127. doi: 10.1038/onc.2017.303. [DOI] [PubMed] [Google Scholar]
- 21.Friedrichson T, Kurzchalia TV. Microdomains of GPI-anchored proteins in living cells revealed by crosslinking. Nature. 1998;394:802–805. doi: 10.1038/29570. [DOI] [PubMed] [Google Scholar]
- 22.Ordonez C, Zhai AB, Camacho-Leal P, Demarte L, Fan MM, Stanners CP. GPI-anchored CEA family glycoproteins CEA and CEACAM6 mediate their biological effects through enhanced integrin alpha5beta1-fibronectin interaction. J Cell Physiol. 2007;210:757–765. doi: 10.1002/jcp.20887. [DOI] [PubMed] [Google Scholar]
- 23.Duxbury MS, Ito H, Ashley SW, Whang EE. c-Src-dependent cross-talk between CEACAM6 and alphavbeta3 integrin enhances pancreatic adenocarcinoma cell adhesion to extracellular matrix components. Biochem Biophys Res Commun. 2004;317:133–141. doi: 10.1016/j.bbrc.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 24.Cheng TM, Chang WJ, Chu HY, De Luca R, Pedersen JZ, Incerpi S, Li ZL, Shih YJ, Lin HY, Wang K, et al. Nano-strategies targeting the integrin αvβ3 network for cancer therapy. Cells. 2021;10:1684. doi: 10.3390/cells10071684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blumenthal RD, Leon E, Hansen HJ, Goldenberg DM. Expression patterns of CEACAM5 and CEACAM6 in primary and metastatic cancers. BMC Cancer. 2007;7:2. doi: 10.1186/1471-2407-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan L, Wang Y, Wang ZZ, Rong YT, Chen LL, Li Q, Liu T, Chen YH, Li YD, Huang ZH, Peng J. Cell motility and spreading promoted by CEACAM6 through cyclin D1/CDK4 in human pancreatic carcinoma. Oncol Rep. 2016;35:418–426. doi: 10.3892/or.2015.4338. [DOI] [PubMed] [Google Scholar]
- 27.Kurlinkus B, Ger M, Kaupinis A, Jasiunas E, Valius M, Sileikis A. CEACAM6's role as a chemoresistance and prognostic biomarker for pancreatic cancer: A comparison of CEACAM6's diagnostic and prognostic capabilities with those of CA19-9 and CEA. Life (Basel) 2021;11:542. doi: 10.3390/life11060542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pandey R, Zhou M, Islam S, Chen B, Barker NK, Langlais P, Srivastava A, Luo M, Cooke LS, Weterings E, Mahadevan D. Carcinoembryonic antigen cell adhesion molecule 6 (CEACAM6) in pancreatic ductal adenocarcinoma (PDA): An integrative analysis of a novel therapeutic target. Sci Rep. 2019;9:18347. doi: 10.1038/s41598-019-54545-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ilantzis C, DeMarte L, Screaton RA, Stanners CP. Deregulated expression of the human tumor marker CEA and CEA family member CEACAM6 disrupts tissue architecture and blocks colonocyte differentiation. Neoplasia. 2002;4:151–163. doi: 10.1038/sj.neo.7900201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao B, Wang P, Gu L, Liu J. Use of four genes in exosomes as biomarkers for the identification of lung adenocarcinoma and lung squamous cell carcinoma. Oncol Lett. 2021;21:249. doi: 10.3892/ol.2021.12510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Du H, Li Y, Sun R, Yuan Y, Sun S, Zhang Y. CEACAM6 promotes cisplatin resistance in lung adenocarcinoma and is regulated by microRNA-146a and microRNA-26a. Thorac Cancer. 2020;11:2473–2482. doi: 10.1111/1759-7714.13558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim EY, Cha YJ, Jeong S, Chang YS. Overexpression of CEACAM6 activates Src-FAK signaling and inhibits anoikis, through homophilic interactions in lung adenocarcinomas. Transl Oncol. 2022;20:101402. doi: 10.1016/j.tranon.2022.101402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Son SM, Yun J, Lee SH, Han HS, Lim YH, Woo CG, Lee HC, Song HG, Gu YM, Lee HJ, Lee OJ. Therapeutic effect of pHLIP-mediated CEACAM6 gene silencing in lung adenocarcinoma. Sci Rep. 2019;9:11607. doi: 10.1038/s41598-019-48104-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hong KP, Shin MH, Yoon S, Ji GY, Moon YR, Lee OJ, Choi SY, Lee YM, Koo JH, Lee HC, et al. Therapeutic effect of anti CEACAM6 monoclonal antibody against lung adenocarcinoma by enhancing anoikis sensitivity. Biomaterials. 2015;67:32–41. doi: 10.1016/j.biomaterials.2015.07.012. [DOI] [PubMed] [Google Scholar]
- 35.Liu Y, Chudgar N, Mastrogiacomo B, He D, Lankadasari MB, Bapat S, Jones GD, Sanchez-Vega F, Tan KS, Schultz N, et al. A germline SNP in BRMS1 predisposes patients with lung adenocarcinoma to metastasis and can be ameliorated by targeting c-fos. Sci Transl Med. 2022;14:eabo1050. doi: 10.1126/scitranslmed.abo1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y, Polyak D, Lamsam L, Connolly ID, Johnson E, Khoeur LK, Andersen S, Granucci M, Stanley G, Liu B, et al. Comprehensive RNA analysis of CSF reveals a role for CEACAM6 in lung cancer leptomeningeal metastases. NPJ Precis Oncol. 2021;5:90. doi: 10.1038/s41698-021-00228-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X, Tang X, Gu J, Sun Z, Yang S, Mu Y, Guan M, Chen K, Liu W, Ruan H, Xu J. CEACAM6 serves as a biomarker for leptomeningeal metastasis in lung adenocarcinoma. Cancer Med. 2023;12:4521–4529. doi: 10.1002/cam4.5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park W, Chawla A, O'Reilly EM. Pancreatic cancer: A review. JAMA. 2021;326:851–862. doi: 10.1001/jama.2021.13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duxbury MS, Matros E, Clancy T, Bailey G, Doff M, Zinner MJ, Ashley SW, Maitra A, Redston M, Whang EE. CEACAM6 is a novel biomarker in pancreatic adenocarcinoma and PanIN lesions. Ann Surg. 2005;241:491–496. doi: 10.1097/01.sla.0000154455.86404.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duxbury MS, Ito H, Ashley SW, Whang EE. CEACAM6 as a novel target for indirect type 1 immunotoxin-based therapy in pancreatic adenocarcinoma. Biochem Biophys Res Commun. 2004;317:837–843. doi: 10.1016/j.bbrc.2004.03.128. [DOI] [PubMed] [Google Scholar]
- 41.Duxbury MS, Ito H, Zinner MJ, Ashley SW, Whang EE. CEACAM6 gene silencing impairs anoikis resistance and in vivo metastatic ability of pancreatic adenocarcinoma cells. Oncogene. 2004;23:465–473. doi: 10.1038/sj.onc.1207036. [DOI] [PubMed] [Google Scholar]
- 42.Long H, Li Q, Wang Y, Li Q, Liu T, Peng J. Effective combination gene therapy using CEACAM6-shRNA and the fusion suicide gene yCDglyTK for pancreatic carcinoma in vitro. Exp Ther Med. 2013;5:155–161. doi: 10.3892/etm.2012.774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen J, Li Q, An Y, Lv N, Xue X, Wei J, Jiang K, Wu J, Gao W, Qian Z, et al. CEACAM6 induces epithelial-mesenchymal transition and mediates invasion and metastasis in pancreatic cancer. Int J Oncol. 2013;43:877–885. doi: 10.3892/ijo.2013.2015. [DOI] [PubMed] [Google Scholar]
- 44.Yuan L, Zhao J, Zhao S, Dong T, Dong R, Liu D, Ma E, Li Y. ASPER-29 suppresses the metastasis of pancreatic cancer cells by dual inhibition of cathepsin-L and cathepsin-S. Chem Biol Interact. 2022;353:109811. doi: 10.1016/j.cbi.2022.109811. [DOI] [PubMed] [Google Scholar]
- 45.Duxbury MS, Ito H, Ashley SW, Whang EE. CEACAM6 cross-linking induces caveolin-1-dependent, Src-mediated focal adhesion kinase phosphorylation in BxPC3 pancreatic adenocarcinoma cells. J Biol Chem. 2004;279:23176–23182. doi: 10.1074/jbc.M402051200. [DOI] [PubMed] [Google Scholar]
- 46.Duxbury MS, Ito H, Benoit E, Ashley SW, Whang EE. CEACAM6 is a determinant of pancreatic adenocarcinoma cellular invasiveness. Br J Cancer. 2004;91:1384–1390. doi: 10.1038/sj.bjc.6602113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, Gangeswaran R, Zhao X, Wang P, Tysome J, Bhakta V, Yuan M, Chikkanna-Gowda CP, Jiang G, Gao D, et al. CEACAM6 attenuates adenovirus infection by antagonizing viral trafficking in cancer cells. J Clin Invest. 2009;119:1604–1615. doi: 10.1172/JCI37905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duxbury MS, Ito H, Benoit E, Zinner MJ, Ashley SW, Whang EE. Overexpression of CEACAM6 promotes insulin-like growth factor I-induced pancreatic adenocarcinoma cellular invasiveness. Oncogene. 2004;23:5834–5842. doi: 10.1038/sj.onc.1207775. [DOI] [PubMed] [Google Scholar]
- 49.Schölzel S, Zimmermann W, Schwarzkopf G, Grunert F, Rogaczewski B, Thompson J. Carcinoembryonic antigen family members CEACAM6 and CEACAM7 are differentially expressed in normal tissues and oppositely deregulated in hyperplastic colorectal polyps and early adenomas. Am J Pathol. 2000;156:595–605. doi: 10.1016/S0002-9440(10)64764-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jantscheff P, Terracciano L, Lowy A, Glatz-Krieger K, Grunert F, Micheel B, Brümmer J, Laffer U, Metzger U, Herrmann R, Rochlitz C. Expression of CEACAM6 in resectable colorectal cancer: A factor of independent prognostic significance. J Clin Oncol. 2003;21:3638–3646. doi: 10.1200/JCO.2003.55.135. [DOI] [PubMed] [Google Scholar]
- 51.Yang T, Wang H, Li M, Yang L, Han Y, Liu C, Zhang B, Wu M, Wang G, Zhang Z, et al. CD151 promotes colorectal cancer progression by a crosstalk involving CEACAM6, LGR5 and Wnt signaling via TGFβ1. Int J Biol Sci. 2021;17:848–860. doi: 10.7150/ijbs.53657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang J, Xiang H, Lu Y, Wu T. Role and clinical significance of TGF-β1 and TGF-βR1 in malignant tumors (Review) Int J Mol Med. 2021;47:55. doi: 10.3892/ijmm.2021.4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu Y, Xia T, Jin C, Gu D, Yu J, Shi W, Zhang KE, Zhang L, Ye J, Li L. FOXP3 and CEACAM6 expression and T cell infiltration in the occurrence and development of colon cancer. Oncol Lett. 2016;11:3693–3701. doi: 10.3892/ol.2016.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jin C, Liu Y, Zhu J, Xia T, Zhang B, Fei Y, Ma B, Ye J, Chen W. Recombinant Salmonella-based CEACAM6 and 4-1BBL vaccine enhances T-cell immunity and inhibits the development of colorectal cancer in rats: In vivo effects of vaccine containing 4-1BBL and CEACAM6. Oncol Rep. 2015;33:2837–2844. doi: 10.3892/or.2015.3901. [DOI] [PubMed] [Google Scholar]
- 55.Chi F, Chen L, Jin X, He G, Liu Z, Han S. CKAP2L, transcriptionally inhibited by FOXP3, promotes breast carcinogenesis through the AKT/mTOR pathway. Exp Cell Res. 2022;412:113035. doi: 10.1016/j.yexcr.2022.113035. [DOI] [PubMed] [Google Scholar]
- 56.Gong Z, Jia H, Xue L, Li D, Zeng X, Wei M, Liu Z, Tong MCF, Chen GG. The emerging role of transcription factor FOXP3 in thyroid cancer. Rev Endocr Metab Disord. 2022;23:421–429. doi: 10.1007/s11154-021-09684-8. [DOI] [PubMed] [Google Scholar]
- 57.Rodia MT, Ugolini G, Mattei G, Montroni I, Zattoni D, Ghignone F, Veronese G, Marisi G, Lauriola M, Strippoli P, Solmi R. Systematic large-scale meta-analysis identifies a panel of two mRNAs as blood biomarkers for colorectal cancer detection. Oncotarget. 2016;7:30295–30306. doi: 10.18632/oncotarget.8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rodia MT, Solmi R, Pasini F, Nardi E, Mattei G, Ugolini G, Ricciardiello L, Strippoli P, Miglio R, Lauriola M. LGALS4, CEACAM6, TSPAN8, and COL1A2: Blood markers for colorectal cancer-validation in a cohort of subjects with positive fecal immunochemical test result. Clin Colorectal Cancer. 2018;17:e217–e228. doi: 10.1016/j.clcc.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 59.Ferlizza E, Solmi R, Miglio R, Nardi E, Mattei G, Sgarzi M, Lauriola M. Colorectal cancer screening: Assessment of CEACAM6, LGALS4, TSPAN8 and COL1A2 as blood markers in faecal immunochemical test negative subjects. J Adv Res. 2020;24:99–107. doi: 10.1016/j.jare.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ferlizza E, Solmi R, Sgarzi M, Ricciardiello L, Lauriola M. The roadmap of colorectal cancer screening. Cancers (Basel) 2021;13:1101. doi: 10.3390/cancers13051101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roy RK, Hoppe MM, Srivastava S, Samanta A, Sharma N, Tan KT, Yang H, Voon DC, Pang B, The M, et al. CEACAM6 is upregulated by Helicobacter pylori CagA and is a biomarker for early gastric cancer. Oncotarget. 2016;7:55290–55301. doi: 10.18632/oncotarget.10528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Deng X, Liu P, Zhao Y, Wang Q. Expression profiling of CEACAM6 associated with the tumorigenesis and progression in gastric adenocarcinoma. Genet Mol Res. 2014;13:7686–7697. doi: 10.4238/2014.September.26.6. [DOI] [PubMed] [Google Scholar]
- 63.Ru GQ, Han Y, Wang W, Chen Y, Wang HJ, Xu WJ, Ma J, Ye M, Chen X, He XL, et al. CEACAM6 is a prognostic biomarker and potential therapeutic target for gastric carcinoma. Oncotarget. 2017;8:83673–83683. doi: 10.18632/oncotarget.19415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.An F, Zheng C, Zhang G, Zhou L, Wu Y, Hou Z, Zhou Z, Chen K, Zhan Q. Carcinoembryonic antigen related cell adhesion molecule 6 promotes carcinogenesis of gastric cancer and anti-CEACAM6 fluorescent probe can diagnose the precancerous lesions. Front Oncol. 2021;11:643669. doi: 10.3389/fonc.2021.643669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Y, Zang M, Li J, Ji J, Zhang J, Liu X, Qu Y, Su L, Li C, Yu Y, et al. CEACAM6 promotes tumor migration, invasion, and metastasis in gastric cancer. Acta Biochim Biophys Sin (Shanghai) 2014;46:283–290. doi: 10.1093/abbs/gmu001. [DOI] [PubMed] [Google Scholar]
- 66.Zang M, Zhang B, Zhang Y, Li J, Su L, Zhu Z, Gu Q, Liu B, Yan M. CEACAM6 promotes gastric cancer invasion and metastasis by inducing epithelial-mesenchymal transition via PI3K/AKT signaling pathway. PLoS One. 2014;9:e112908. doi: 10.1371/journal.pone.0112908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nieto MA, Sargent MG, Wilkinson DG, Cooke J. Control of cell behavior during vertebrate development by Slug, a zinc finger gene. Science. 1994;264:835–839. doi: 10.1126/science.7513443. [DOI] [PubMed] [Google Scholar]
- 68.Ros MA, Sefton M, Nieto MA. Slug, a zinc finger gene previously implicated in the early patterning of the mesoderm and the neural crest, is also involved in chick limb development. Development. 1997;124:1821–1829. doi: 10.1242/dev.124.9.1821. [DOI] [PubMed] [Google Scholar]
- 69.Cohen ME, Yin M, Paznekas WA, Schertzer M, Wood S, Jabs EW. Human SLUG gene organization, expression, and chromosome map location on 8q. Genomics. 1998;51:468–471. doi: 10.1006/geno.1998.5367. [DOI] [PubMed] [Google Scholar]
- 70.Savagner P. Leaving the neighborhood: Molecular mechanisms involved during epithelial-mesenchymal transition. Bioessays. 2001;23:912–923. doi: 10.1002/bies.1132. [DOI] [PubMed] [Google Scholar]
- 71.Nieto MA. The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- 72.Zang M, Zhang Y, Zhang B, Hu L, Li J, Fan Z, Wang H, Su L, Zhu Z, Li C, et al. CEACAM6 promotes tumor angiogenesis and vasculogenic mimicry in gastric cancer via FAK signaling. Biochim Biophys Acta. 2015;1852:1020–1028. doi: 10.1016/j.bbadis.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 73.Han SU, Kwak TH, Her KH, Cho YH, Choi C, Lee HJ, Hong S, Park YS, Kim YS, Kim TA, Kim SJ. CEACAM5 and CEACAM6 are major target genes for Smad3-mediated TGF-beta signaling. Oncogene. 2008;27:675–683. doi: 10.1038/sj.onc.1210686. [DOI] [PubMed] [Google Scholar]
- 74.Chu C, Liu X, Zhao Z, Shi Z. Circ_0008035 promotes the progression of gastric cancer via the regulation of miR-1256/CEACAM6 axis. Cell Cycle. 2022;21:1091–1102. doi: 10.1080/15384101.2022.2041354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen SY, Zhang RG, Duan GC. Pathogenic mechanisms of the oncoprotein CagA in H. pylori-induced gastric cancer (Review) Oncol Rep. 2016;36:3087–3094. doi: 10.3892/or.2016.5145. [DOI] [PubMed] [Google Scholar]
- 76.Xia R, Zhang B, Wang X, Jia Q. Pathogenic interactions between Helicobacter pylori adhesion protein HopQ and human cell surface adhesion molecules CEACAMs in gastric epithelial cells. Iran J Basic Med Sci. 2019;22:710–715. doi: 10.22038/ijbms.2019.34237.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Behrens IK, Busch B, Ishikawa-Ankerhold H, Palamides P, Shively JE, Stanners C, Chan C, Leung N, Gray-Owen S, Haas R. The HopQ-CEACAM interaction controls CagA translocation, phosphorylation, and phagocytosis of Helicobacter pylori in neutrophils. mBio. 2020;11:e03256–19. doi: 10.1128/mBio.03256-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Toh J, Hoppe MM, Thakur T, Yang H, Tan KT, Pang B, Ho S, Roy R, Ho KY, Yeoh KG, et al. Profiling of gastric cancer cell-surface markers to achieve tumour-normal discrimination. BMJ Open Gastroenterol. 2020;7:e000452. doi: 10.1136/bmjgast-2020-000452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhao ZS, Li L, Wang HJ, Wang YY. Expression and prognostic significance of CEACAM6, ITGB1, and CYR61 in peripheral blood of patients with gastric cancer. J Surg Oncol. 2011;104:525–529. doi: 10.1002/jso.21984. [DOI] [PubMed] [Google Scholar]
- 80.Tsang JY, Kwok YK, Chan KW, Ni YB, Chow WN, Lau KF, Shao MM, Chan SK, Tan PH, Tse GM. Expression and clinical significance of carcinoembryonic antigen-related cell adhesion molecule 6 in breast cancers. Breast Cancer Res Treat. 2013;142:311–322. doi: 10.1007/s10549-013-2756-y. [DOI] [PubMed] [Google Scholar]
- 81.Balk-Møller E, Kim J, Hopkinson B, Timmermans-Wielenga V, Petersen OW, Villadsen R. A marker of endocrine receptor-positive cells, CEACAM6, is shared by two major classes of breast cancer: Luminal and HER2-enriched. Am J Pathol. 2014;184:1198–1208. doi: 10.1016/j.ajpath.2013.12.013. [DOI] [PubMed] [Google Scholar]
- 82.Zhao SJ, Zhao HD, Li J, Zhang H, Gao DT, Wang Q. CD151 promotes breast cancer metastasis by activating TGF-β1/Smad signaling pathway. Eur Rev Med Pharmacol Sci. 2018;22:7314–7322. doi: 10.26355/eurrev_201811_16268. [DOI] [PubMed] [Google Scholar]
- 83.Wang SE. The functional crosstalk between HER2 tyrosine kinase and TGF-β signaling in breast cancer malignancy. J Signal Transduct. 2011;2011:804236. doi: 10.1155/2011/804236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Scott DJ, Parkes AT, Ponchel F, Cummings M, Poola I, Speirs V. Changes in expression of steroid receptors, their downstream target genes and their associated co-regulators during the sequential acquisition of tamoxifen resistance in vitro. Int J Oncol. 2007;31:557–565. doi: 10.3892/ijo.31.3.557. [DOI] [PubMed] [Google Scholar]
- 85.Lewis-Wambi JS, Cunliffe HE, Kim HR, Willis AL, Jordan VC. Overexpression of CEACAM6 promotes migration and invasion of oestrogen-deprived breast cancer cells. Eur J Cancer. 2008;44:1770–1779. doi: 10.1016/j.ejca.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Khoury T, Kanehira K, Wang D, Ademuyiwa F, Mojica W, Cheney R, Morrison C, Conroy J, Nowak N, Liu S. Breast carcinoma with amplified HER2: A gene expression signature specific for trastuzumab resistance and poor prognosis. Mod Pathol. 2010;23:1364–1378. doi: 10.1038/modpathol.2010.125. [DOI] [PubMed] [Google Scholar]
- 87.Ihnen M, Kilic E, Köhler N, Löning T, Witzel I, Hagel C, Höller S, Kersten JF, Müller V, Jänicke F, et al. Protein expression analysis of ALCAM and CEACAM6 in breast cancer metastases reveals significantly increased ALCAM expression in metastases of the skin. J Clin Pathol. 2011;64:146–152. doi: 10.1136/jcp.2010.082602. [DOI] [PubMed] [Google Scholar]
- 88.Koh EY, You JE, Jung SH, Kim PH. Biological functions and identification of novel biomarker expressed on the surface of breast cancer-derived cancer stem cells via proteomic analysis. Mol Cells. 2020;43:384–396. doi: 10.14348/molcells.2020.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gong L, Liu X, Jiao L, Yang X, Lemoff A, Liu X. CK2-mediated phosphorylation of SUZ12 promotes PRC2 function by stabilizing enzyme active site. Nat Commun. 2022;13:6781. doi: 10.1038/s41467-022-34431-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ieta K, Tanaka F, Utsunomiya T, Kuwano H, Mori M. CEACAM6 gene expression in intrahepatic cholangiocarcinoma. Br J Cancer. 2006;95:532–540. doi: 10.1038/sj.bjc.6603276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rose JB, Correa-Gallego C, Li Y, Nelson J, Alseidi A, Helton WS, Allen PJ, D'Angelica MI, DeMatteo RP, Fong Y, et al. The role of biliary carcinoembryonic antigen-related cellular adhesion molecule 6 (CEACAM6) as a biomarker in cholangiocarcinoma. PLoS One. 2016;11:e0150195. doi: 10.1371/journal.pone.0150195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tian C, Zhang B, Ge C. Effect of CEACAM6 silencing on the biological behavior of human gallbladder cancer cells. Oncol Lett. 2020;20:2677–2688. doi: 10.3892/ol.2020.11806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Litkouhi B, Litkouhi B, Fleming E, Welch WR, Berkowitz RS, Birrer MJ, Mok SC. Overexpression of CEACAM6 in borderline and invasive mucinous ovarian neoplasms. Gynecol Oncol. 2008;109:234–239. doi: 10.1016/j.ygyno.2008.01.031. [DOI] [PubMed] [Google Scholar]
- 94.du Manoir S, Delpech H, Orsetti B, Jacot W, Pirot N, Noel J, Colombo PE, Sardet C, Theillet C. In high-grade ovarian carcinoma, platinum-sensitive tumor recurrence and acquired-resistance derive from quiescent residual cancer cells that overexpress CRYAB, CEACAM6, and SOX2. J Pathol. 2022;257:367–378. doi: 10.1002/path.5896. [DOI] [PubMed] [Google Scholar]
- 95.Ye Y, Wang J, Liang F, Song P, Yan X, Wu S, Huang X, Han P. Identification of key genes for HNSCC from public databases using bioinformatics analysis. Cancer Cell Int. 2021;21:549. doi: 10.1186/s12935-021-02254-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bednarek K, Kostrzewska-Poczekaj M, Szaumkessel M, Kiwerska K, Paczkowska J, Byzia E, Ustaszewski A, Janiszewska J, Bartochowska A, Grenman R, et al. Downregulation of CEACAM6 gene expression in laryngeal squamous cell carcinoma is an effect of DNA hypermethylation and correlates with disease progression. Am J Cancer Res. 2018;8:1249–1261. [PMC free article] [PubMed] [Google Scholar]
- 97.Cameron S, de Long LM, Hazar-Rethinam M, Topkas E, Endo-Munoz L, Cumming A, Gannon O, Guminski A, Saunders N. Focal overexpression of CEACAM6 contributes to enhanced tumourigenesis in head and neck cancer via suppression of apoptosis. Mol Cancer. 2012;11:74. doi: 10.1186/1476-4598-11-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang Z, Luo C, Wang H, Yan X, Liu W, Meng Z. CEACAM6 is associated with osteosarcoma metastasis and facilitates epithelial-mesenchymal transition in osteosarcoma cells. Onco Targets Ther. 2018;11:3159–3166. doi: 10.2147/OTT.S161807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lasa A, Serrano E, Carricondo M, Carnicer MJ, Brunet S, Badell I, Sierra J, Aventín A, Nomdedéu JF. High expression of CEACAM6 and CEACAM8 mRNA in acute lymphoblastic leukemias. Ann Hematol. 2008;87:205–211. doi: 10.1007/s00277-007-0388-1. [DOI] [PubMed] [Google Scholar]
- 100.Kanderová V, Hrusák O, Kalina T. Aberrantly expressed CEACAM6 is involved in the signaling leading to apoptosis of acute lymphoblastic leukemia cells. Exp Hematol. 2010;38:653–660.e1. doi: 10.1016/j.exphem.2010.03.018. [DOI] [PubMed] [Google Scholar]
- 101.Steiner N, Hajek R, Nachbaur D, Borjan B, Sevcikova S, Göbel G, Gunsilius E. Levels of CEACAM6 in peripheral blood are elevated in patients with plasma cell disorders: A potential new diagnostic marker and a new therapeutic target? Dis Markers. 2019;2019:1806034. doi: 10.1155/2019/1806034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhu R, Ge J, Ma J, Zheng J. Carcinoembryonic antigen related cell adhesion molecule 6 promotes the proliferation and migration of renal cancer cells through the ERK/AKT signaling pathway. Transl Androl Urol. 2019;8:457–466. doi: 10.21037/tau.2019.09.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Igami K, Uchiumi T, Shiota M, Ueda S, Tsukahara S, Akimoto M, Eto M, Kang D. Extracellular vesicles expressing CEACAM proteins in the urine of bladder cancer patients. Cancer Sci. 2022;113:3120–3133. doi: 10.1111/cas.15438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wu SJ, Arundhathi A, Wang HC, Chen CY, Cheng TM, Yuan SF, Wang YM. Migration and invasion of NSCLC suppressed by the downregulation of Src/focal adhesion kinase using single, double and tetra domain anti-CEACAM6 antibodies. Transl Oncol. 2021;14:101057. doi: 10.1016/j.tranon.2021.101057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Youssef A, Haskali MB, Gorringe KL. The protein landscape of mucinous ovarian cancer: Towards a theranostic. Cancers (Basel) 2021;13:5596. doi: 10.3390/cancers13225596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Han ZW, Lyv ZW, Cui B, Wang YY, Cheng JT, Zhang Y, Cai WQ, Zhou Y, Ma ZW, Wang XW, et al. The old CEACAMs find their new role in tumor immunotherapy. Invest New Drugs. 2020;38:1888–1898. doi: 10.1007/s10637-020-00955-w. [DOI] [PubMed] [Google Scholar]
- 107.Johnson B, Mahadevan D. Emerging role and targeting of carcinoembryonic antigen-related cell adhesion molecule 6 (CEACAM6) in human malignancies. Clin Cancer Drugs. 2015;2:100–111. doi: 10.2174/2212697X02666150602215823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Burgos M, Cavero-Redondo I, Álvarez-Bueno C, Galán-Moya EM, Pandiella A, Amir E, Ocaña A. Prognostic value of the immune target CEACAM6 in cancer: A meta-analysis. Ther Adv Med Oncol. 2022;14:17588359211072621. doi: 10.1177/17588359211072621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Schäfer D, Tomiuk S, Küster LN, Rawashdeh WA, Henze J, Tischler-Höhle G, Agorku DJ, Brauner J, Linnartz C, Lock D, et al. Identification of CD318, TSPAN8 and CD66c as target candidates for CAR T cell based immunotherapy of pancreatic adenocarcinoma. Nat Commun. 2021;12:1453. doi: 10.1038/s41467-021-21774-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Witzens-Harig M, Hose D, Jünger S, Pfirschke C, Khandelwal N, Umansky L, Seckinger A, Conrad H, Brackertz B, Rème T, et al. Tumor cells in multiple myeloma patients inhibit myeloma-reactive T cells through carcinoembryonic antigen-related cell adhesion molecule-6. Blood. 2013;121:4493–4503. doi: 10.1182/blood-2012-05-429415. [DOI] [PubMed] [Google Scholar]
- 111.Jin C, Duan X, Liu Y, Zhu J, Zhang K, Zhang Y, Xia T, Fei Y, Ye J. T cell immunity induced by a bivalent Salmonella-based CEACAM6 and 4-1BBL vaccines in a rat colorectal cancer model. Oncol Lett. 2017;13:3753–3759. doi: 10.3892/ol.2017.5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Schettini F, Barbao P, Brasó-Maristany F, Galván P, Martínez D, Paré L, De Placido S, Prat A, Guedan S. Identification of cell surface targets for CAR-T cell therapies and antibody-drug conjugates in breast cancer. ESMO Open. 2021;6:100102. doi: 10.1016/j.esmoop.2021.100102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zeligs KP, Morelli MP, David JM, Neuman M, Hernandez L, Hewitt S, Ozaki M, Osei-Tutu A, Anderson D, Andresson T, et al. Evaluation of the anti-tumor activity of the humanized monoclonal antibody NEO-201 in preclinical models of ovarian cancer. Front Oncol. 2020;10:805. doi: 10.3389/fonc.2020.00805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cheng TM, Murad YM, Chang CC, Yang MC, Baral TN, Cowan A, Tseng SH, Wong A, Mackenzie R, Shieh DB, Zhang J. Single domain antibody against carcinoembryonic antigen-related cell adhesion molecule 6 (CEACAM6) inhibits proliferation, migration, invasion and angiogenesis of pancreatic cancer cells. Eur J Cancer. 2014;50:713–721. doi: 10.1016/j.ejca.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 115.Riley CJ, Engelhardt KP, Saldanha JW, Qi W, Cooke LS, Zhu Y, Narayan ST, Shakalya K, Croce KD, Georgiev IG, et al. Design and activity of a murine and humanized anti-CEACAM6 single-chain variable fragment in the treatment of pancreatic cancer. Cancer Res. 2009;69:1933–1940. doi: 10.1158/0008-5472.CAN-08-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Niu G, Murad YM, Gao H, Hu S, Guo N, Jacobson O, Nguyen TD, Zhang J, Chen X. Molecular targeting of CEACAM6 using antibody probes of different sizes. J Control Release. 2012;161:18–24. doi: 10.1016/j.jconrel.2012.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lee H, Jang Y, Park S, Jang H, Park EJ, Kim HJ, Kim H. Development and evaluation of a CEACAM6-targeting theranostic nanomedicine for photoacoustic-based diagnosis and chemotherapy of metastatic cancer. Theranostics. 2018;8:4247–4261. doi: 10.7150/thno.25131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zou J, Chen S, Li Y, Zeng L, Lian G, Li J, Chen S, Huang K, Chen Y. Nanoparticles modified by triple single chain antibodies for MRI examination and targeted therapy in pancreatic cancer. Nanoscale. 2020;12:4473–4490. doi: 10.1039/c9nr04976b. [DOI] [PubMed] [Google Scholar]
- 119.Ordoñez C, Screaton RA, Ilantzis C, Stanners CP. Human carcinoembryonic antigen functions as a general inhibitor of anoikis. Cancer Res. 2000;60:3419–3424. [PubMed] [Google Scholar]
- 120.Chuang PC, Lu CW, Tsai CC, Tseng SH, Su WH. MicroRNA-128 confers anti-endothelial adhesion and anti-migration properties to counteract highly metastatic cervical cancer cells' migration in a parallel-plate flow chamber. Int J Mol Sci. 2020;22:215. doi: 10.3390/ijms22010215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Blumenthal RD, Hansen HJ, Goldenberg DM. Inhibition of adhesion, invasion, and metastasis by antibodies targeting CEACAM6 (NCA-90) and CEACAM5 (carcinoembryonic antigen) Cancer Res. 2005;65:8809–8817. doi: 10.1158/0008-5472.CAN-05-0420. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.