Abstract
A second cytadhesin-like protein, MGC2, was identified in the avian respiratory pathogen Mycoplasma gallisepticum. The 912-nucleotide mgc2 gene encodes a 32.6-kDa protein with 40.9 and 31.4% identity with the M. pneumoniae P30 and M. genitalium P32 cytadhesins, respectively. Functional studies with reverse transcription-PCR, immunoblotting, double-sided immunogold labeling, and attachment inhibition assays demonstrated homology to the human mycoplasmal P30 and P32 cytadhesins. These findings suggest that there is a family of cytadhesin genes conserved among pathogenic mycoplasmas infecting widely divergent hosts.
The genus Mycoplasma is unique among prokaryotes because the member species lack cell walls, use UGA to encode tryptophan, and utilize cholesterol in their cell membranes (40). They have been described as minimal cells due to their unusually small size and streamlined genetics, with the smallest recorded genomes of self-replicating organisms (15, 21, 35). Pathogenic mycoplasmas are highly specialized, successfully exploiting respiratory and urinogenital tract niches in a wide variety of vertebrates. Mycoplasma gallisepticum is a significant pathogen of domestic poultry and wild birds. In chickens, the agent produces chronic respiratory disease, while ovaduct infection causes lowered egg production and allows transovarial transmission (49).
M. gallisepticum shares similar pathogenic mechanisms with two human mycoplasmas, Mycoplasma pneumoniae and Mycoplasma genitalium (10, 30). These mycoplasmas exhibit a flask-shaped morphology characterized by a unipolar terminal organelle, or bleb, that is involved in mucosal attachment and gliding motility (7, 25, 46). The tip organelle of M. pneumoniae has been extensively studied and functions through the interactions of cytadhesins, cytadhesin accessory proteins, and elements of a primitive intracellular cytoskeleton (26).
We previously reported the sequence and characterization of MGC1, the M. gallisepticum homolog of M. pneumoniae P1, M. genitalium MgPa, and the Mycoplasma pirum cytadhesins (24, 45). A second cytadhesin has been identified in both M. pneumoniae and M. genitalium. Attenuated class II variants of M. pneumoniae are hemadsorption negative and lack a 30-kDa protein designated P30 (4, 5, 27). Revertants expressing P30 reacquire virulence (28). P30 was also found to be membrane associated and localized on the tip organelle (4, 5). Furthermore, a monoclonal antibody raised against P30 blocked M. pneumoniae attachment, suggesting its importance in cytadherence (36). Additional analysis of P30 mutants has suggested the importance of a repeated proline-rich amino acid domain in cytadherence, virulence, and postinfection autoimmunity (12, 31). The genes encoding P30 and the M. genitalium homolog, P32, have been sequenced and mapped (11, 15, 22, 41).
Cloning procedures and DNA sequencing.
The MGC1 gene is located within plasmid pMG25, which contains an 8.3-kb fragment of the M. gallisepticum genome cloned into Bluescript vector KSII (24). The 1.3-kb region upstream from mgc1 was subcloned from pMG25 into KSII as two PstI fragments and sequenced with Sequenase version 2.0 (U.S. Biochemical, Cleveland, Ohio) according to the manufacturer’s instructions. We designed specific primers to sequence the two subclones and the PstI junction in pMG25. Oligonucleotide primers were purchased from Ransom Hill (La Jolla, Calif.). The nucleotide sequence was analyzed with the Sequence Analysis Software Package of the Genetics Computer Group (13).
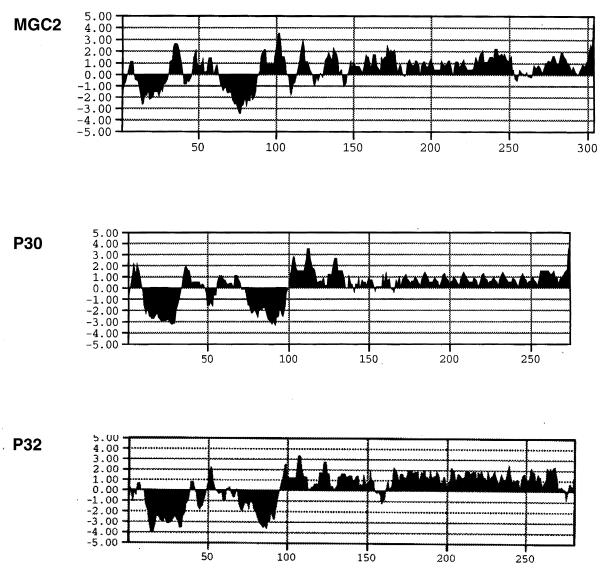
An open reading frame (ORF), 912 nucleotides (nt) long and with a G+C content of 44.8%, was identified and designated mgc2. The deduced MGC2 protein contains 304 amino acids (aa), with a predicted molecular mass of 32,700 Da. There is a single tryptophan, encoded by TGA, at amino acid position 62. At the amino terminus there is a positively charged 10-aa region followed by a 20- to 21-aa hydrophobic core (Leu11-Leu30), suggestive of a signal sequence (Fig. 1 and 2). Based on von Heijne’s −3, −1 rule for identifying signal sequence cleavage sites (47), a possible signal sequence cleavage site exists after Thr31 or Ser32. A Kyte-Doolittle hydrophilicity plot of MGC2 (averaged over a 7-aa window) shows a second highly hydrophobic region, consistent with the presence of a transmembrane domain, from Phe63 to Ala89. The carboxy-terminal two-thirds of the protein (aa 102 to 304) is rich in proline (20%) and glycine (15%) residues.
FIG. 1.
Comparison of the deduced amino acid sequences of MGC2, M. pneumoniae P30 (10), and M. genitalium P32 (15, 36) with the GAP alignment program from the Sequence Analysis Software Package of the Genetics Computer Group (13). Residues conserved in all three proteins are shaded in black, while sequences shared by two of the three mycoplasmas are shaded in gray; dashes represent gaps.
FIG. 2.
Amino acid hydrophilicity plots of MGC2, M. pneumoniae P30, and M. genitalium P32 made with the Kyte-Doolittle algorithm averaged over a 7-aa window (MacVector version 4.1; IBI Kodak, New Haven, Conn.). Positive values indicate increased hydrophilicity, while negative numbers represent increased hydrophobicity. The ordinate values represent the amino acid residues.
Comparison of MGC2 to the M. pneumoniae cytadhesin P30 and to the P32 sequences of M. genitalium showed the deduced MGC2 sequence to be 40.9% identical to that of P30 and 31.4% identical to that of P32. In addition there are 30 shared proline residues, including consensus tryptophan residues at amino acid positions 73 in P30, 68 in P32, and 62 in MGC2. The hydrophilicity plots of the three proteins are nearly superimposable (Fig. 2). The carboxy end of MGC2 (aa 185 to 304), like those of P30 and P32, has the characteristics of cytoskeletal matrix proteins, such as collagen, elastin, vitronectin, and keratin. The carboxy end of the molecule is distinguished by two identical overlapping 24-aa sequences, located from Met185 to Pro208 and from Met206 to Pro230 (Fig. 1). These repeats share the residues Met206, Pro207, and Pro208. Overlapping sequence repeats are also characteristic of P30 and P32 (11, 12, 41). Within the repeated regions (aa 185 to 229 of MGC2, 177 to 254 of P30, and 163 to 260 of P32) the proteins share the following repeated amino acid motifs: Arg-Pro-Gly-Phe, Arg-Pro-Gly, and Pro-Gly.
Transcriptional analysis and organization of the cytadhesin operons.
The transcription initiation site of mgc1, the first gene of an approximately 8-kb operon, is located within the mgc2 coding region (24). Immediately upstream from mgc2 is a 67-nt A+T-rich (83.6%) region. Although two Escherichia coli consensus −10 promoter sequences were found 38 (TATTAT) and 100 nt (TATAAT) from the ATG initiation codon of mgc2, and a possible −35 TTGAAA promoter sequence is found 112 nt from the start site, no consensus Shine-Delgarno sequence was located. Primer extension was unable to identify a transcriptional start site for mgc2 within the region, and Northern blot analysis suggests that the gene is part of a larger transcript (data not shown). Further characterization of the complex regulatory relationship between these two M. gallisepticum cytadhesin-containing operons is currently under way.
The complete genome sequences of M. genitalium (580 kb) and M. pneumoniae (816 kb) have been completed (15, 22). Both genomes contain six segments in which the order of orthologous genes is conserved. However, within the respective genomes, these segments are arranged differently (23). The regions bordering the M. pneumoniae segments have one or more repetitive sequences (RepMP1, RepMP2/3, RepMP4, and RepMP5), and relics of these sequences, with the exception of RepMP1, were found between segments in the M. genitalium genome. It was concluded that reorganization of M. genitalium took place by translocations of the segments through homologous recombination in regions between the repetitive elements (23). The three operons encoding cytadhesin and cytadhesin accessory genes found in M. genitalium and M. pneumoniae are similarly ordered and transcribed in the same direction but map to different segments.
In contrast, the proximity of mgc2 and mgc1 and the presence of the mgc1 transcriptional start site within mgc2 indicate a different genomic organization in M. gallisepticum, since the human homologs for these genes are widely separated and located within different segments of the M. pneumoniae and M. genitalium genomes. We recently completed sequencing the ORF downstream from mgc1 and found it to show 26 and 25% deduced amino acid identity with M. pneumoniae ORF6 and M. genitalium ORF192, respectively, genes found immediately downstream from cytadhesin-encoding P1 and MgPa. At this time it is not known if complete sets of homologous cytadhesin operons are present in M. gallisepticum, organized in one or more genomic regions that might represent an ancestral cytadhesin operon organization. It will be of interest to obtain the complete M. gallisepticum genome sequence, which would allow comparative analysis of a more distantly related pathogenic mycoplasma, infecting another vertebrate class, with the two closely related human pathogens. The presence or absence of related genome segments and cytadhesin operons shared by the three mycoplasmas could provide evolutionary insight into the origins of these organisms.
RT-PCR.
To further investigate mgc2 functionality, reverse transcription-PCR (RT-PCR) was performed. Log-phase M. gallisepticum S6 cells were pelleted by centrifugation at 10,000 × g for 30 min. The cell pellet was extracted with Trizol reagent (Gibco BRL, Gaithersburg, Md.). The aqueous layer, containing RNA, was layered on a 4 M cesium chloride cushion and centrifuged at 90,000 × g for 16 h at 17°C. The RNA pellet was extracted with phenol-chloroform-isoamyl alcohol (29:28:1), ethanol precipitated, and resuspended in diethyl pyrocarbonate-treated water. All RNA samples were subsequently treated with RNase-free DNase (Promega Biotech, Madison, Wis.) for 30 min at 37°C and then phenol-chloroform-isoamyl alcohol extracted as described above. One microgram of M. gallisepticum RNA was RNase treated for use as an RT-PCR negative control. Reverse transcription was conducted with a GeneAmp RNA PCR kit (Perkin-Elmer, Foster City, Calif.).
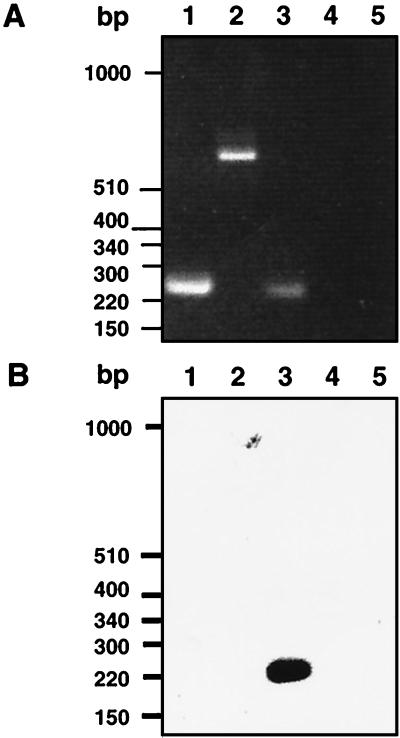
Two mgc2 primers, located at nucleotide positions 144 to 174 and 359 to 341, were used in the PCR. PCR products were electrophoresed on a 1.5% agarose gel and transferred to a nylon membrane for Southern analysis. The nylon blot was incubated in a solution of 1% sodium dodecyl sulfate (SDS), 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate [pH 7.0]), 5× Denhardt’s solution (1× Denhardt’s solution is 0.02% Ficoll, 0.2% polyvinyl pyrrolidone, 0.02% bovine serum albumin), and 200 μg of salmon sperm DNA/ml for 2 h at 50°C. A 32P-labeled mgc2 oligonucleotide probe corresponding to nt 198 to 235 was added, and incubation at 50°C was continued for 12 h. The membrane was washed twice in 0.2× SSC–0.1% SDS for 10 min at room temperature, twice in 0.2× SSC–0.1% SDS for 30 min at 50°C, and once in 0.1× SSC for 30 min at 50°C. As controls for the analysis, we used previously described primers to amplify by RT-PCR a portion of the M. gallisepticum elongation factor (tuf) gene (20). As an additional control, primers at nucleotide positions 2346 to 2372 and 2940 to 2916 (24) were used to RT-PCR amplify a portion of the mgc1 gene. RT-PCR with total M. gallisepticum RNA confirmed the transcription of mgc2. An mgc2-specific RT-PCR product of the predicted size, 211 bp, was identified (Fig. 3A). The controls, portions of the M. gallisepticum tuf and mgc1 transcripts, were amplified by RT-PCR (210 and 580 bp, respectively). No PCR products were observed in samples treated with RNase. Southern analysis was done with a 32P-labeled probe corresponding to nt 198 to 215 of mgc2 (Fig. 3B). The mgc2 primer hybridized only to the mgc2 RT-PCR product.
FIG. 3.
RT-PCR of M. gallisepticum RNA. (A) Agarose gel of RT-PCR results. (B) Gel transferred to a nylon membrane and hybridized with a 32P-labeled oligonucleotide from within mgc2. Lanes 1, M. gallisepticum tuf gene; lanes 2, mgc1; lanes 3, mgc2; lanes 4, RNase-treated mgc2; lanes 5, RNA template without reverse transcriptase. The photographed gel and nylon membrane blot were scanned on an Apple Color I scanner, cropped with Photoshop (Adobe Systems Inc., Mountain View, Calif.), and enlarged and labeled with QuarkXPress (Quark, Inc., Denver, Colo.).
Site-directed mutagenesis and expression cloning of MGC2.
In order to further characterize the mgc2 gene product, we prepared recombinant MGC2 antigen for use in antiserum production. This antiserum was used in immunoprecipitation, Western blotting, immunoelectron microscopy, and attachment inhibition assays. This required site-directed mutagenesis to alter a single TGA (tryptophan codon for amino acid position 62) in the MGC2 gene to a TGG codon with the Altered Sites II site-directed mutagenesis kit (Promega) and the mutagenesis primer 5′ CCCGAACCTTGGTTTTACCA 3′ (the altered codon is underlined). Mutants containing the altered tryptophan codon were confirmed by sequence analysis. MGC2 was overexpressed in E. coli SG13009(pRep4) cells as a six histidine-maltose binding protein-MGC2 fusion protein (Qiagen, Chatsworth, Calif.). The fusion protein was identified as a 75,000-Da band on a 12% SDS polyacrylamide gel after purification on a Talon nickel affinity chromatography column (Clonetech, Palo Alto, Calif.).
Production of anti-MGC2 antiserum and MGC2 detection.
The portion of the SDS polyacrylamide gel corresponding to the migration position of the fusion protein was excised and homogenized with an equal volume of Freund’s complete adjuvant. New Zealand White rabbits were injected subcutaneously with the preparation as previously described (24).
Immunoprecipitations were performed as described by Krause and Baseman (29). Adherent M. gallisepticum S6 cultures, grown in 75-cm2 polystyrene tissue culture flasks containing Frey broth, were washed with phosphate-buffered saline (PBS; 120 mM NaCl, 2.7 mM KCl, 10 mM Na2PO4, pH 7.4). Adherent cells were then disrupted in solubilization buffer (35 mM Tris [pH 8.2], 0.25 M NaCl, 1.6% deoxycholate, 0.1% SDS, and 1 mM phenylmethylsulfonyl fluoride). Cellular debris was removed by centrifugation at 60,000 × g for 30 min. Rabbit anti-MGC2-specific antibody (20 μl) was added to 200 μl of solubilized M. gallisepticum cells and incubated on ice for 1 h. Twenty microliters of Protein G Plus-Protein A agarose (Calbiochem, La Jolla, Calif.) was added to the mixture, and incubation was continued at 4°C for 1 h with gentle mixing. The immunoprecipitate was collected by centrifugation at 17,000 × g for 1 min and washed three times by repeated centrifugation in solubilization buffer. Following the last centrifugation, the precipitate was resuspended in a solution of 3% mercaptoethanol, 3% SDS, 0.3% bromophenol blue, and 10% glycerol loading buffer, boiled for 5 min, and centrifuged for 5 min at 17,000 × g. The supernatant was loaded onto an SDS–10% polyacrylamide gel. After electrophoretic separation, the proteins were transferred to nitrocellulose for Western blot analysis as previously described (24). A 1:100 dilution of rabbit anti-MGC2 antiserum and a 1:2,000 dilution of goat anti-rabbit alkaline phosphatase-conjugated antibody (Bio-Rad, Richmond, Calif.) were used in the analysis.
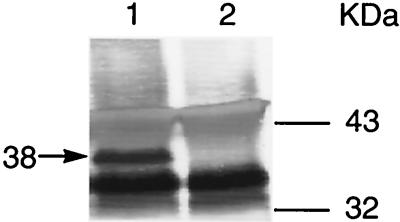
The polyvalent rabbit antiserum detected MGC2 from early-log-phase M. gallisepticum cultures (Fig. 4). The rabbit anti-MGC2 antiserum precipitated a 38-kDa protein (Fig. 4, lane 1), while the preinoculation rabbit serum control did not detect this protein (Fig. 4, lane 2). Additional bands found in the immunoblot with both preinoculation and anti-MGC2 serum treatments were expected, since alkaline phosphatase-labeled goat anti-rabbit immunoglobulin G reacts with the heavy and light chains of rabbit anti-MGC2 antibodies. In addition, both rabbit and goat antibodies could react in immunoblots via their respective Fc fragments with proteins G (30 to 35 kDa) and A (42 kDa). Because of the importance of insuring the specificity of rabbit anti-MGC2 for subsequent experiments, we infected chickens with M. gallisepticum to obtain preinoculation and 4- and 6-week postinfection serum samples. These sera were individually reacted with solubilized M. gallisepticum cells, immunoprecipitated, and subjected to Western blotting as described above. All the chickens from the 4-week (n = 8) and 6-week (n = 8) bleedings showed the 38-kDa band in immunoblots developed with rabbit anti-MGC2 and conjugated goat anti-rabbit antisera. None of the preinoculation and uninfected control sera (n = 8) precipitated the 38-kDa band (data not shown).
FIG. 4.
Identification of MGC2. Solubilized M. gallisepticum cells were immunoprecipitated with rabbit anti-MGC2 antiserum and subjected to Western blot analysis (lane 1); preinoculation rabbit serum was used as an assay control (lane 2). Molecular mass markers and the mass of MGC2 (arrow) are indicated. The Western blot was scanned on an Apple Color I scanner, cropped with Photoshop (Adobe Systems Inc., Mountain View, Calif.), and enlarged and labeled with QuarkXPress (Quark, Inc., Denver, Colo.).
The difference between the predicted (32-kDa) size of MGC2 and the observed (38-kDa) size was also observed in Western blotting assays with solubilized M. gallisepticum cells (data not shown). Several M. pneumoniae proteins are known to have observed molecular masses higher than their predicted masses, as determined from the primary amino acid sequences. Anomalous migrations of M. pneumoniae P30, HMW1, HMW3, P65, and P200 proteins have been reported (5, 14, 37–39). One explanation for these observations is the proline-rich repeated amino acid regions found in these proteins. Prolines provide rigidity and extend the structures of proteins (48). Computer analysis of MGC2 showed that the proline-rich carboxy end of the molecule had homology with collagen, a proline-rich molecule whose beta chain exhibits lower electrophoretic mobility and higher molecular weight than predicted (16).
The results of immunoblotting, taken together with the detection of an RNA transcript by RT-PCR, indicate that mgc2 functions to encode an immunogenic protein.
Immunoelectron microscopy.
To determine the cellular topology of MGC2, double-sided immunogold labeling was conducted with thin sections of M. gallisepticum cells. M. gallisepticum cells were grown to early log phase in Frey broth (pH 7.0), fixed in equal volumes of Frey broth and 2% paraformaldehyde in 0.2 M sodium cacodylate buffer (pH 7.2), and harvested by centrifugation. The resulting pellet was resuspended in 2% low-melting-point agarose, rinsed in buffer (twice for 15 min each time), dehydrated in a graded series of ethanol, and embedded in HM20 resin (Electron Microscopy Sciences, Fort Washington, Pa.) according to the manufacturer’s instructions. The double-sided labeling technique was based on the procedures described for lectin probes and actin (6, 9). Ultrathin sections were examined and photographed at ×20,000 magnification with a Zeiss CEM 902 transmission electron microscope.
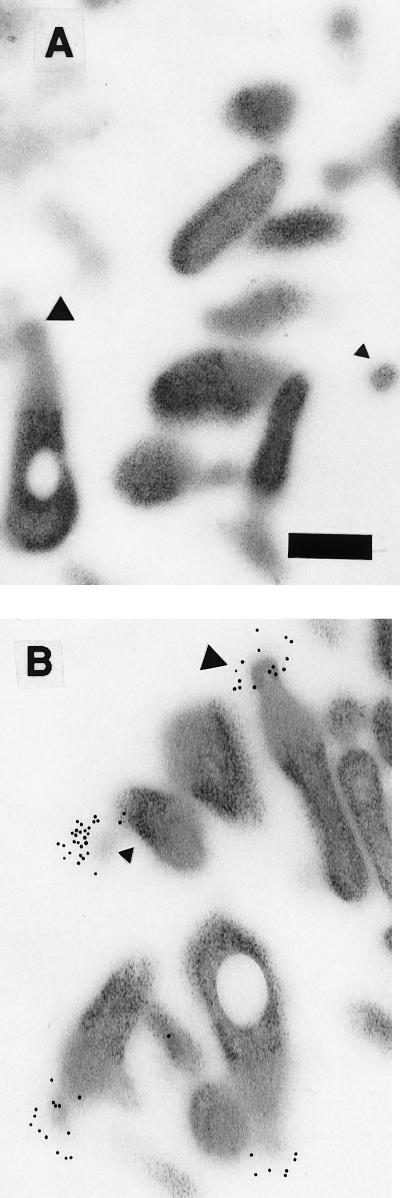
Flask-shaped cells, cut through different longitudinal planes, were clearly visible in the electron micrographs (Fig. 5). Various cross sections through the bleb were also visible. Immunogold labeling was localized on the terminal bleb organelle in sections treated with anti-MGC2 antiserum (Fig. 5B). Gold particles were not visible in the preimmune serum-treated sections (Fig. 5A). As is typical of the flask-shaped pathogenic mycoplasmas, M. gallisepticum attachment to host cells is mediated by the terminal bleb organelle, which permits an intimate association with the host cell membrane (46). In M. pneumoniae and M. genitalium, cytadhesin proteins are concentrated on this structure (26). We demonstrated through immunogold labeling that MGC2 appears to be highly concentrated on the tip of the terminal bleb. In similar experiments, MGC1 was found to be more widely distributed across the bleb (data not shown).
FIG. 5.
Electron micrographs showing double-sided immunogold labeling of M. gallisepticum MGC2 cytadhesin on sectioned M. gallisepticum cells. (A) Sections treated with rabbit preinoculation serum prior to incubation with 10-nm-diameter colloidal-gold-labeled goat anti-rabbit antiserum. (B) Sections reacted with rabbit anti-MGC2 antiserum. Secondary antibodies containing gold particles are distributed on the bleb organelle. The bleb regions in more longitudinal sections are visible (large triangles), while blebs in different planes of cross section are marked (small triangles). Transmission electron microscopy at ×20,000 on a Zeiss transmission electron microscope. Bar, 250 nm.
Based on primary amino acid sequence and electron microscopic labeling, it would appear that MGC2, like P30 and P32, is membrane associated, surface exposed, and localized on the bleb attachment organelle.
Attachment inhibition studies.
Preliminary experiments were conducted to evaluate the use of chicken embryo fibroblast (CEF) cells in subsequent attachment inhibition assays. Erythrocytes were not used because M. gallisepticum has separate hemagglutinating gene products, distinct from those for MGC1 and MGC2 (3, 20, 32–34). CEF monolayers were treated with neuraminidase at 2.5, 5, and 10 mU for 1 h at 37°C before the application of radioactivity-labeled M. gallisepticum cells. After vigorous washing, M. gallisepticum cell-associated radioactivity was significantly (P < 0.05) lower than that of untreated CEF cells, indicating that the sialic acid glycoprotein receptors found on erythrocytes are present on CEFs (2, 18, 19). Neuraminidase treatments reduced binding by 55 to 65% at M. gallisepticum concentrations ranging from 5.5 × 107 to 7.3 × 107 CFU, a result similar to the 58% reduction of M. gallisepticum binding after neuraminidase treatment of the MRC-5 human lung fibroblast cell line (17).
The involvement of MGC2 in attachment to host cell receptors was investigated with the previously described rabbit anti-MGC2 antiserum. Mixtures of antiserum and radiolabeled M. gallisepticum cells were added to CEF cells. After incubation, reduced CEF-associated radioactivity was used to quantitate attachment inhibition.
M. gallisepticum S6 cells, grown to mid-log phase in Frey broth (pH 6.7), were pelleted at 10,000 × g for 2 min, washed once in PBS, and resuspended in Hanks’ balanced salt solution (Gibco BRL, Grand Island, N.Y.) containing 10% methionine-free porcine serum. Prior to being radiolabeled, the cultures were filtered through an 0.8-μm-pore-size membrane to remove clumped cells. M. gallisepticum cells were then labeled with 40 μCi of [35S]methionine (Amersham, Arlington Heights, Ill.) per 108 CFU for 3 h at 37°C with shaking. Radiolabeled cells were washed three times with Hanks’ balanced salt solution and resuspended in M199 medium (Gibco).
CEFs were prepared as previously described (42) and cultured on 96-well tissue culture plates (Corning Costar Corporation, Cambridge, Mass.). The CEFs, grown to confluence in M199 medium containing 5% fetal calf serum, were then washed twice with PBS. Initial titrations of labeled cells and CEFs were used to optimize conditions. The attachment inhibition assay used a 1:10 anti-MGC2 antibody dilution and, as a control, a 1:10 dilution of MGC2 preinoculation rabbit serum, reacted with eight labeled mycoplasma concentrations ranging from 5 × 106 to 40 × 106 CFU. Antibody and mycoplasmas in 200-μl aliquots were incubated for 1 h at 37°C with gentle mixing, added to the washed CEFs, and incubated for an additional 1 h at 37°C. An antibody-free control was also reacted with labeled mycoplasmas diluted only with M199. Following incubation, CEFs with attached M. gallisepticum cells were rigorously washed three times with PBS. Each mycoplasma-and-serum treatment was replicated three times. Individual wells were separated and placed in ScintiVerse (Fisher Scientific, Fair Lawn, N.J.) and counted to determine cell-bound radioactivity. After subtraction of background radioactivity, cell-bound counts were analyzed by two-way analysis of variance, with pairwise comparisons determined by the method of Tukey (43). We also determined that rabbit anti-MGC2 was not mycoplasmacidal or inhibitory of metabolic processes required for attachment, since undiluted antibody did not produce growth or metabolic inhibition by the previously described assays (8, 44).
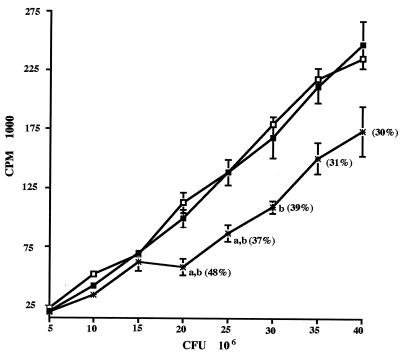
The preinoculation serum and labeled cells without antibody showed a near-linear increase in attachment with increasing numbers of M. gallisepticum CFU (Fig. 6). Pairwise treatment analysis determined that there was significant attachment inhibition (P < 0.01) of anti-MGC2 antibody-treated mycoplasmas at concentrations ranging from 20 × 106 to 30 × 106 CFU, which resulted in 37 to 48% inhibition. The lack of complete inhibition of attachment is likely due to the presence of one or more additional cytadhesins. In parallel attachment inhibition assays conducted under the assay conditions described above, rabbit anti-MGC1 antiserum caused 52 to 60% attachment inhibition compared to controls (P < 0.0001) with M. gallisepticum cells at 5 × 106 to 40 × 106 CFU (data not shown).
FIG. 6.
Antiserum raised against MGC2 blocks [35S]methionine-labeled M. gallisepticum attachment. Increasing numbers of radiolabeled M. gallisepticum cells were pretreated with buffer alone (□), rabbit preinoculation serum diluted 1:10 (▪), or rabbit anti-MGC2 antiserum diluted 1:10 (∗). The values represent means (± standard errors) of three replicate treatments. a, Significant (P < 0.01) difference from the corresponding treatment without antibody; b, significant difference (P < 0.01) from corresponding treatments reacted with preinoculation rabbit serum. The numbers in parentheses indicate the percent inhibition from the preinoculation serum {100 − [(average counts per minute of anti-MGC2 serum treatment/counts per minute of preinoculation serum treatment) × 100]}.
The products of the M. gallisepticum pMGA gene family, unique surface-exposed protein hemagglutinins distinct from MGC1 and MGC2, are putative cytadhesins (3, 20, 32–34). Homologs of the pMGA genes are not found in M. pneumoniae or M. genitalium. In different M. gallisepticum strains, the number of complete pMGA genes that show varying degrees of deduced amino acid homology ranges from 32 to 70 (33, 34). RT-PCR was used to detected four simultaneously expressed pMGA transcripts in the M. gallisepticum S6 strain (20). Recently, the proteins involved in high-frequency phase variation of hemadsorption positive (HA+) and HA− phenotypes were identified (1). The HA+ phenotypes expressed proteins p30, p48, p50, and p80, while p72 was only expressed in the HA− phenotype. The p69 protein, expressed in both HA+ and HA− phenotypes, was the only protein identified as the product of a member of the pMGA family of genes in Western blotting. It has been suggested that the HA phase variation may be important in immune evasion (1, 33). Taken together, these data point to a complex regulation of HA. The relationship of proteins involved in HA and hemagglutination with MGC1 and MGC2 cytadhesins remains unclear, since antisera directed at hemagglutinating and hemadsorbing proteins have not been used to examine attachment inhibition in cells other than erythrocytes. To address this issue, we examined the hemagglutination inhibition (HI) properties of rabbit antisera specific for MGC1 and MGC2 by using chicken erythrocytes in the routine HI diagnostic test (49). These high-titered antisera react in immunoblots at 1:1,000 (MGC1) and 1:2,500 (MGC2) dilutions. However, no HI activity was shown by these sera, suggesting that they do not bind to M. gallisepticum hemagglutinins.
In summary, the data presented in this study provides compelling evidence that MGC2 is the homolog of M. pneumoniae P30 and M. genitalium P32, including 40.9 and 31.4% amino acid homology between MGC2 and the P30 and P32 proteins, respectively. The MGC2 protein has a predicted mass of 32.7 kDa and migrates at an apparent size of 38 kDa. By double-sided immunogold labeling, MGC2, like P30 and P32, is localized on the terminal bleb. Finally, antiserum directed at MGC2 inhibited attachment to CEF cells. The initial attachment of M. gallisepticum to host tissues appears to involve sets of genes closely related to those found in the human mycoplasma pathogens. The fact that mycoplasma pathogens from widely divergent hosts utilize homologous cytadhesins suggests the importance of these membrane proteins to successful exploitation of the host mucosal niche.
Nucleotide sequence accession number.
The nucleotide sequence discussed in this paper was submitted to GenBank under accession no. U23842.
Acknowledgments
This work was supported by USDA NRI CGP grant 93-03408 to J.E.D.
We thank Robin Morgan for her critical reading of the manuscript.
Footnotes
Paper no. 1645 in the Journal Series of the Delaware Agricultural Experiment Station.
REFERENCES
- 1.Athamna A, Rosengarten R, Levisohn S, Kahane I, Yogev D. Adherence of Mycoplasma gallisepticum involves variable surface membrane proteins. Infect Immun. 1997;65:2468–2471. doi: 10.1128/iai.65.6.2468-2471.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banai M, Kahane I, Razin S, Bredt W. Adherence of Mycoplasma gallisepticum to human erythrocytes. Infect Immun. 1978;21:365–372. doi: 10.1128/iai.21.2.365-372.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baseggio N, Glew M D, Markham P F, Whithear K G, Browning G F. Size and genomic location of the pMGA multigene family of Mycoplasma gallisepticum. Microbiology. 1996;142:1429–1435. doi: 10.1099/13500872-142-6-1429. [DOI] [PubMed] [Google Scholar]
- 4.Baseman J B, Drouillard D L, Keith D K, Morrison-Plummer J. Role of Mycoplasma pneumoniae adhesin P1 and accessory proteins in cytadsorption. In: Mergenhagen S E, Rosan B, editors. Molecular basis of oral microbial adhesion. Washington, D.C: American Society for Microbiology; 1985. pp. 18–23. [Google Scholar]
- 5.Baseman J B, Morrison-Plummer D L, Drouillard D L, Puleo-Scheppke B, Tyron V V, Holt S C. Identification of a 32-kilodalton protein of Mycoplasma pneumoniae associated with hemadsorption. Isr J Med Sci. 1987;23:474–479. [PubMed] [Google Scholar]
- 6.Bourett T M, Howard R J. Enhanced labelling of concanavalin A binding sites in fungal endomembranes using a double-sided, indirect method. Mycol Res. 1994;98:769–775. [Google Scholar]
- 7.Carson J L, Hu P-C, Collier A M. Cell structural and functional elements. In: Maniloff J, McElhaney R N, Finch L R, Baseman J B, editors. Mycoplasmas: molecular biology and pathogenesis. Washington, D.C: American Society for Microbiology; 1992. pp. 63–72. [Google Scholar]
- 8.Clyde W C., Jr . Growth inhibition tests. In: Razin S, Tully J G, editors. Methods in mycoplasmology. Vol. 1. New York, N.Y: Academic Press; 1983. pp. 405–410. [Google Scholar]
- 9.Czymmek K J, Bourett J B, Howard R J. Immunolocalization of tubulin and actin in thick-sectioned fungal hyphae after freeze-substitution and metacrylate de-embedment. J Microsc. 1996;181:153–161. [Google Scholar]
- 10.Dallo S F, Baseman J B. Cross-hybridization between the cytadhesin genes of Mycoplasma pneumoniae and Mycoplasma genitalium and genomic DNA of Mycoplasma gallisepticum. Microb Pathog. 1990;8:371–375. doi: 10.1016/0882-4010(90)90096-9. [DOI] [PubMed] [Google Scholar]
- 11.Dallo S F, Chavoya A, Baseman J B. Characterization of the gene for a 30-kilodalton adhesin-related protein of Mycoplasma pneumoniae. Infect Immun. 1990;58:4163–4165. doi: 10.1128/iai.58.12.4163-4165.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dallo S F, Lazzelli A L, Chavoya A, Reddy S P, Baseman J B. Biofunctional domains of the Mycoplasma pneumoniae P30 adhesin. Infect Immun. 1996;64:2595–2601. doi: 10.1128/iai.64.7.2595-2601.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dirksen L B, Proft T, Hilbert H, Plagens H, Herrmann R, Krause D C. Sequence analysis and characterization of the hmw gene cluster of Mycoplasma pneumoniae. Gene. 1996;171:19–25. doi: 10.1016/0378-1119(96)00050-9. [DOI] [PubMed] [Google Scholar]
- 15.Fraser C M, Gocayne J D, White O, Adams M D, Clayton R A, Fleischmann R D, Bult C J, Kerlavage A R, Sutton G, Kelly J M, Fritchman J L, Weidman J F, Small K V, Sandusky M, Fuhrmann J, Nguyen D, Utterback T R, Saudek D M, Phillips C A, Merrick M, Tomb J, Dougherty B A, Bott K F, Hu P, Lucier T S, Peterson S N, Smith H O, Hutchison III C A, Venter J C. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270:395–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 16.Furthmayr H, Timpl R. Characterization of collagen peptides by sodium dodecylsulfate-polyacrylamide electrophoresis. Anal Biochem. 1971;41:510–516. doi: 10.1016/0003-2697(71)90173-4. [DOI] [PubMed] [Google Scholar]
- 17.Geary S J, Gabridge M G, Intres R, Draper D L, Gladd M. Identification of Mycoplasma binding proteins utilizing a 100 kilodalton fibroblast receptor. J Recept Res. 1990;9:465–478. doi: 10.3109/10799898909066071. [DOI] [PubMed] [Google Scholar]
- 18.Gessner B, Lewis T. Sialic acid binding sites: role in hemagglutination by Mycoplasma gallisepticum. Science. 1965;151:353–361. doi: 10.1126/science.151.3710.590. [DOI] [PubMed] [Google Scholar]
- 19.Glasgow L R, Hill R L. Interaction of Mycoplasma gallisepticum with sialyl glycoproteins. Infect Immun. 1980;30:353–361. doi: 10.1128/iai.30.2.353-361.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glew M D, Markham P F, Browning G F, Walker I D. Expression studies on four members of the pMGA multigene family in Mycoplasma gallisepticum S6. Microbiology. 1995;141:3005–3014. doi: 10.1099/13500872-141-11-3005. [DOI] [PubMed] [Google Scholar]
- 21.Herrmann R. Genome structure and organization. In: Maniloff J, McElhaney R N, Finch L R, Baseman J B, editors. Mycoplasmas: molecular biology and pathogenesis. Washington, D.C: American Society for Microbiology; 1992. pp. 157–168. [Google Scholar]
- 22.Himmelreich R, Hilbert H, Plagens H, Pirkl E, Li B-C, Herrmann R. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 1996;24:4420–4449. doi: 10.1093/nar/24.22.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Himmelreich R, Plagens H, Hilbert H, Reiner B, Herrmann R. Comparative analysis of the genomes of the bacteria Mycoplasma pneumoniae and Mycoplasma genitalium. Nucleic Acids Res. 1997;25:701–712. doi: 10.1093/nar/25.4.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keeler C L, Jr, Hnatow L L, Whetzel P, Dohms J E. Cloning and characterization of a putative cytadhesin gene (mgc1) from Mycoplasma gallisepticum. Infect Immun. 1996;64:1541–1547. doi: 10.1128/iai.64.5.1541-1547.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirchhoff H. Motility. In: Maniloff J, McElhaney R N, Finch L R, Baseman J B, editors. Mycoplasmas: molecular biology and pathogenesis. Washington, D.C: American Society for Microbiology; 1992. pp. 289–306. [Google Scholar]
- 26.Krause D C. Mycoplasma pneumoniae cytadherence: unraveling the tie that binds. Mol Microbiol. 1996;20:247–253. doi: 10.1111/j.1365-2958.1996.tb02613.x. [DOI] [PubMed] [Google Scholar]
- 27.Krause D C, Leith D K, Wilson R M, Baseman J B. Identification of Mycoplasma pneumoniae proteins associated with hemadsorption and virulence. Infect Immun. 1982;35:809–817. doi: 10.1128/iai.35.3.809-817.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krause D C, Leith D K, Baseman J B. Reacquisition of specific proteins confers virulence in Mycoplasma pneumoniae. Infect Immun. 1983;39:830–836. doi: 10.1128/iai.39.2.830-836.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krause D C, Baseman J B. Inhibition of Mycoplasma pneumoniae hemadsorption and adherence to respiratory epithelium by antibodies to a membrane protein. Infect Immun. 1983;39:1180–1186. doi: 10.1128/iai.39.3.1180-1186.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krause D C, Taylor-Robinson D. Mycoplasmas which infect humans. In: Maniloff J, McElhaney R N, Finch L R, Baseman J B, editors. Mycoplasmas: molecular biology and pathogenesis. Washington, D.C: American Society for Microbiology; 1992. pp. 417–444. [Google Scholar]
- 31.Layh-Schmitt G, Hilbert H, Pirkl E. A spontaneous hemadsorption-negative mutant of Mycoplasma pneumoniae exhibits a truncated adhesin-related 30-kilodalton protein and lacks the cytadherence-accessory protein HMW1. J Bacteriol. 1995;177:843–846. doi: 10.1128/jb.177.3.843-846.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Markham P F, Glew M D, Brandon M R, Walker I D, Whithear K G. Characterization of a major hemagglutinin protein from Mycoplasma gallisepticum. Infect Immun. 1992;60:3885–3891. doi: 10.1128/iai.60.9.3885-3891.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Markham P F, Glew M D, Whithear K G, Walker I D. Molecular cloning of a member of the gene family that encodes pMGA, a hemagglutinin of Mycoplasma gallisepticum. Infect Immun. 1993;61:903–909. doi: 10.1128/iai.61.3.903-909.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Markham P F, Glew M D, Sykes J E, Bowden T R, Pollocks T D, Browning G F, Whithear K G, Walker I D. The organization of the multigene family which encodes the major cell surface protein, pMGA, of Mycoplasma gallisepticum. FEBS Lett. 1994;325:347–352. doi: 10.1016/0014-5793(94)00991-0. [DOI] [PubMed] [Google Scholar]
- 35.Morowitz H J. The completeness of molecular biology. Isr J Med Sci. 1985;20:750–753. [PubMed] [Google Scholar]
- 36.Morrison-Plummer J, Leith D K, Baseman J B. Biological effects of anti-lipid and anti-protein monoclonal antibodies of Mycoplasma pneumoniae. Infect Immun. 1986;53:398–403. doi: 10.1128/iai.53.2.398-403.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogle K F, Lee K K, Krause D C. Nucleotide sequence analysis reveals novel features of the phase-variable cytadherence accessory protein HMW3 of Mycoplasma pneumoniae. Infect Immun. 1992;60:1633–1641. doi: 10.1128/iai.60.4.1633-1641.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Proft T, Hilbert H, Layh-Schmitt G, Herrmann R. The proline-rich P65 protein of Mycoplasma pneumoniae is a component of the Triton X-100-insoluble fraction and exhibits size polymorphism in the strains M129 and FH. J Bacteriol. 1995;177:3370–3378. doi: 10.1128/jb.177.12.3370-3378.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Proft T, Hilbert H, Plagens H, Herrmann R. The P200 protein of Mycoplasma pneumoniae shows common features with the cytadherence-associated proteins HMW1 and HMW3. Gene. 1996;171:79–82. doi: 10.1016/0378-1119(96)00014-5. [DOI] [PubMed] [Google Scholar]
- 40.Razin S. Characteristics of the mycoplasmas as a group. In: Razin S, Tully J G, editors. Methods in mycoplasmology. Vol. 1. New York, N.Y: Academic Press; 1983. pp. 3–7. [Google Scholar]
- 41.Reddy S P, Rasmussen W G, Baseman J B. Molecular cloning and characterization of an adherence-related operon of Mycoplasma genitalium. J Bacteriol. 1995;177:5943–5951. doi: 10.1128/jb.177.20.5943-5951.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosenberger J K, Klopp S, Krauss W C. Characterization of Newcastle disease viruses isolated from migrating waterfowl in the Atlantic Flyway. Avian Dis. 1975;19:142–149. [PubMed] [Google Scholar]
- 43.SAS Institute, Inc. SAS/STAT user’s guide, version 6. 4th ed. Vol. 2. Cary, N.C: SAS Institute, Inc.; 1989. p. 846. [Google Scholar]
- 44.Taylor-Robinson D. Metabolism inhibition tests. In: Razin S, Tully J G, editors. Methods in mycoplasmology. Vol. 1. New York, N.Y: Academic Press; 1983. pp. 411–417. [Google Scholar]
- 45.Tham T N, Ferris S, Bahraoui E, Montagnier L, Blanchard A. Molecular characterization of the P1-like adhesin gene from Mycoplasma pirum. J Bacteriol. 1994;176:781–788. doi: 10.1128/jb.176.3.781-788.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Uppal P K, Chu H P. Attachment of Mycoplasma gallisepticum to the tracheal epithelium of fowls. Res Vet Sci. 1977;22:259–260. [PubMed] [Google Scholar]
- 47.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Williamson M P. The structure and function of proline-rich regions in proteins. Biochem J. 1994;297:249–260. doi: 10.1042/bj2970249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoder H W., Jr . Mycoplasma gallisepticum infection. In: Calnek B W, Beard C W, Barnes H J, Reid W M, Yoder H W Jr, editors. Diseases of poultry. 9th ed. Ames: Iowa State University Press; 1991. pp. 198–212. [Google Scholar]