Abstract
The HIV-1 Envelope (Env) glycoprotein facilitates host cell fusion through a complex series of receptor-induced structural changes. Although remarkable progress has been made in understanding the structures of various Env conformations, microsecond timescale dynamics have not been studied experimentally. Here, we used time-resolved, temperature-jump small-angle x-ray scattering to monitor structural rearrangements in an HIV-1 Env SOSIP ectodomain construct with microsecond precision. In two distinct Env variants, we detected a transition that correlated with known Env structure rearrangements with a time constant in the hundreds of microseconds range. A previously unknown structural transition was also observed, which occurred with a time constant below 10 μs, and involved an order-to-disorder transition in the trimer apex. Using this information, we engineered an Env SOSIP construct that locks the trimer in the prefusion closed state by connecting adjacent protomers via disulfides. Our findings show that the microsecond timescale structural dynamics play an essential role in controlling the Env conformation with impacts on vaccine design.
Fast movements in the HIV-1 Envelope protein control access to antibody epitopes with impacts on vaccine design.
INTRODUCTION
The HIV-1 Envelope (Env) glycoprotein mediates viral attachment and entry into host cells (1, 2). This is accomplished through a complex series of receptor CD4 and co-receptor CCR5/CXCR4-induced Env structural transitions that ultimately result in virion-host cell membrane fusion and host cell infection (3–5). The Env ectodomain is composed of three hetero-protomers, each consisting of a receptor-binding gp120 domain and fusion machinery containing the gp41 domain (Fig. 1A) (1–4, 6). In the prefusion closed state, the gp120 domains surround the gp41 domains, thus sequestering the fusion machinery before CD4 receptor and CCR5/CXCR4 co-receptor binding (4). Receptor CD4 engagement induces internal rearrangements and rotation of the gp120 domains to expose the gp41 fusion elements (3, 4). Env ectodomain structures determined using x-ray crystallography and cryo–electron microscopy (cryo-EM) have identified key structural elements within these domains that control these movements (2, 7–9). Interprotomer contacts between gp120 subunits at the viral membrane distant trimer apex and contact between each gp120 with a trimeric gp41 three-helix bundle characterize the closed state (Fig. 1B, left) (9). These stabilizing inter- and intraprotomer contacts are disrupted in the CD4-induced, open Env structure, resulting in an open-state configuration (Fig. 1B, right) (9). Although these studies have provided detailed end-state configurations for this important transition, less is known about the structural intermediates the Env visits along this opening path.
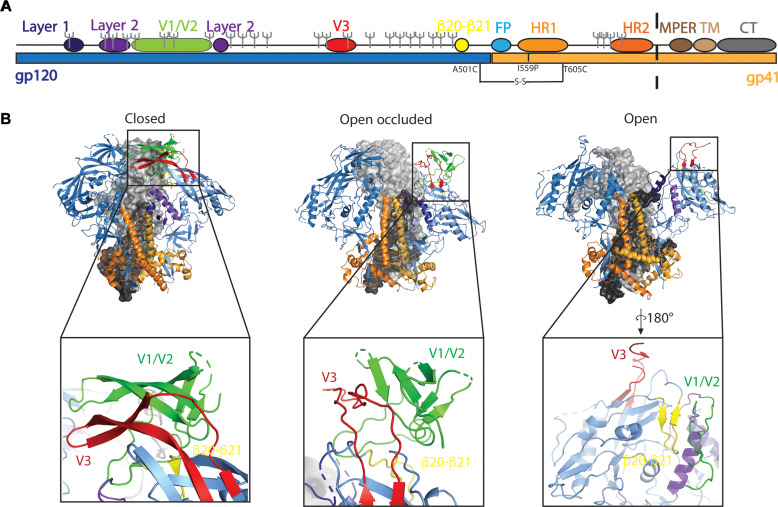
Fig. 1. The HIV-1 Env glycoprotein is structurally dynamic.
(A) Linear sequence of the HIV-1 Env with gp120 in blue and gp41 in light orange identifying structural elements layer 1 (navy), layer 2 (purple), variable domains 1 and 2 (V1/V2; green), variable domain 3 (V3; red), β20-β21 (yellow), fusion peptide (FP; cyan), heptad repeat 1 (HR1; orange), heptad repeat 2 (HR2; dark orange), membrane-proximal external region (MPER; brown), transmembrane domain (TM; tan), and the cytoplasmic tail (CT; gray) allosteric elements. CH505 transmitted founder virus glycosylation sites are denoted by gray forks. The black dashed line represents the location at which the SOSIP ectodomains are truncated. (B) Structures of a prefusion closed Env trimer (PDB ID 6UDA, left), an open-occluded Env trimer (PDB ID 5VN8, middle), and an open Env trimer (PDB ID 5VN3, right) from the side viewpoint. Allosteric elements of the gp120 and gp41 domains are colored according to (A). Insets show zoomed-in views of V1/V2, V3, and β20-β21 elements in each conformation.
Receptor-induced movement from the prefusion, closed state results in the elimination of apex interprotomer contacts between two sequence variable loop–containing segments, V1/V2 and V3 (4, 10–12). The final state for an Env ectodomain bound to CD4 and a co-receptor mimicking antibody consisted of rotated gp120s, an exposed, flexible V3 loop, and a dynamic, conformationally displaced V1/V2 (12, 13). Another structure determined with an Env ectodomain interacting with a broadly neutralizing antibody, b12, showed a similar rotation of gp120 but no major rearrangements in V1/V2 or V3 (14). Several additional antibody-bound structures with these gp120 rotated, V1/V2 occluded (open-occluded) configurations (Fig. 1B, middle) have been identified, suggesting that this state may be an intermediate in the closed to open transition (14–16). Single-molecule Förster resonance energy transfer (smFRET) studies (17–20), double electron-electron resonance (DEER) experiments (21), and hydrogen-deuterium exchange (HDX) mass spectrometry experiments (22–25) indicate that Env ectodomain is structurally dynamic in the absence of receptor. Correlation between these observations and known structures remains challenging, and observable timescales of these measurements limit interrogation of microsecond timescale structural transitions that may control these movements.
Knowledge of Env structure and dynamics has nevertheless played an important role in HIV-1 vaccine immunogen design. Preventing transitions from the prefusion, closed configuration is thought to be essential for selecting functional improbable broadly neutralizing antibody mutations required for neutralization induction by vaccination (2, 26, 27). The Env glycoprotein is the primary target for these design efforts involving both full-length membrane-associated gp160 and truncated soluble gp140 constructs. The soluble ectodomains are often stabilized using an intraprotomer gp120 to gp41 linking disulfide (SOS) mutation and Ile559➔Pro (IP) mutation to prevent gp120 shedding and movements from the prefusion state, respectively (2, 27). Additional stabilization for these SOSIP immunogens has proven essential to ensure that gp41 fusion elements are not exposed (7, 26–39). These include structure-based design modifications within the allosteric components of gp120 to gp41 intra- and interprotomer contacts toward the trimer base. Despite this additional stabilization, vaccination can still induce poorly neutralizing epitope targeting antibodies (15, 40). While smFRET (17–20) and HDX (22–25) experiments have characterized Env dynamics on the milliseconds to seconds and seconds to hours timescales, respectively, microsecond timescale structural movements have not been studied experimentally. An improved understanding of the Env opening pathway would provide useful information to guide the design of greater closed-state Env trimer stability.
Here, to probe Env opening on microsecond timescales, we performed time-resolved, temperature-jump (TR, T-Jump) small-angle x-ray scattering (SAXS) experiments using a CH505 variant (41) and a CH848 variant (42) Env ectodomain SOSIP trimers containing a stabilizing disulfide bond between positions 201C and 433C (DS). The CH505 variant data revealed a rapidly forming structural intermediate with a time constant of ~7 μs that moved to a distinct structural state with a time constant of ~466 μs. The CH848 variant time constants were ~2 and ~200 μs, indicating that, though more thermostable and resistant to open-state targeting antibody binding, CH848 transition rates are enhanced compared to the CH505 variant. Structure-based modeling of these data suggested that the structural states correspond to the breaking of interprotomer gp120 contacts at the trimer apex followed by gp120 domain rotation outward, away from the central axis. This led us to develop an interprotomer Env apex stapled construct that was designed to eliminate access to the first, rapidly forming intermediate. Structures of this stabilized construct complexed with receptor binding site–targeting antibodies revealed a prefusion closed Env state, with one antibody, b12, showing marked shifts in its angles of approach compared to antibody binding to unstabilized Env. These results show that knowledge of transient intermediate structural states can guide vaccine immunogen design.
RESULTS
SAXS captures conformational transitions in HIV-1 Env glycoproteins
TR, T-Jump SAXS experiments are referred to as pump-probe experiments. In the pumping stage, the system equilibrium is perturbed, in this case by rapid heating of the water surrounding the sample by an infrared (IR) laser. The probe stage occurs at a time delay relative to the pump stage and acts as a readout for the state of the system as the system moves to its new equilibrium (fig. S5A). In this study, we used SAXS as a probe to investigate fast timescale structure rearrangements. Successful application of this method requires a construct that can readily undergo conformational transitions with the applied T-Jump and where the conformational change can be detected by SAXS measurement. Therefore, we sought first to identify an Env construct suitable for TR, T-Jump SAXS experiments. We used biolayer interferometry (BLI) to measure conformation-specific antibody binding to two SOSIP-stabilized Env gp140 ectodomains from BG505 (43) and a CH505 transmitted founder virus–derived Env chimera (CH505; CH505-derived gp120 and a BG505-derived gp41 needed to stabilize the protein for expression) (41). Both BG505 and CH505 were isolated from people living with HIV (41, 43). Binding was tested to antibodies PGT145 (Env closed state–specific, trimer apex targeting) (44), 17b (Env open state–specific, co-receptor binding site targeting) (45), and 19b (Env open state–specific, linear V3 tip epitope targeting) (46). While CD4-induced Envs in the open conformation bind 17b and 19b efficiently, Envs from certain HIV-1 isolates access these open conformations and bind 17b and 19b even in the absence of CD4 (17–20). PGT145 binds a quaternary epitope contributed by all three protomers, and its binding confirms the presence of a closed trimer (17, 19). Both CH505 and BG505 SOSIP bound PGT145, confirming the presence of closed trimers in both Env preparations (Fig. 2A, right). CH505 SOSIP interacted with both 17b (Fig. 2A, left) and 19b (Fig. 2A, middle), while BG505 did not, indicating that CH505 SOSIP was more structurally labile and may, therefore, more readily respond to perturbation in the T-Jump experiments.
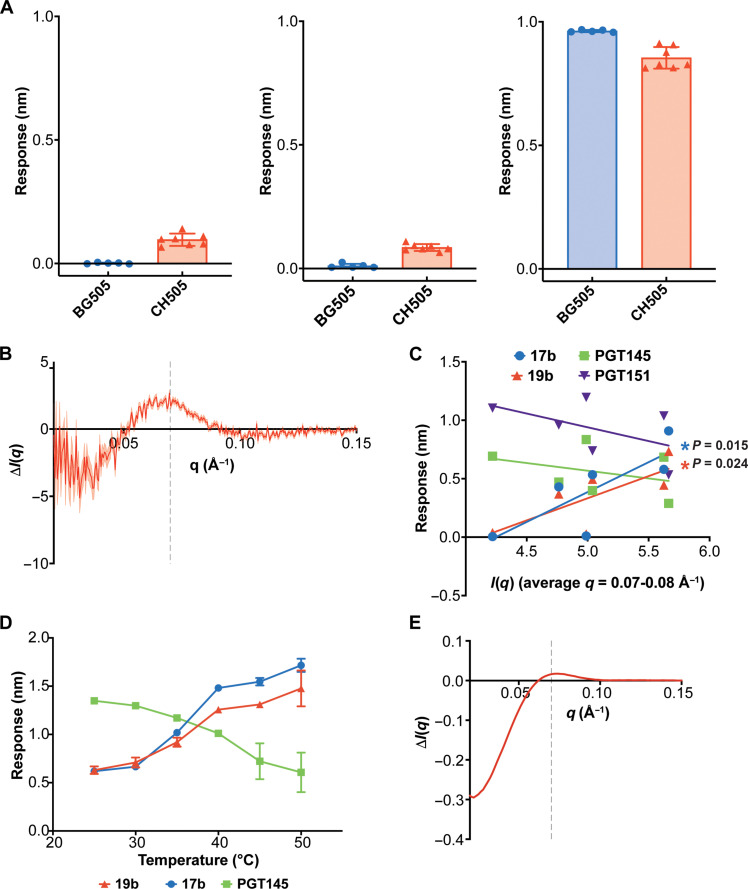
Fig. 2. Static SAXS profiles capture HIV-1 Env conformation.
(A) BLI binding responses for 17b (left), 19b (middle), and PGT145 (right) for BG505 SOSIP (blue circles) and CH505 SOSIP (red triangles). Error bars indicate the SD from the arithmetic mean aggregated from five and seven independently produced BG505 and CH505 SOSIP lots, respectively. (B) Static SAXS scattering difference curve representing the intensity differences between the BG505 and CH505 curves. Propagated SE represented by the shaded region. The scattering difference feature (q) peak at ~0.07 Å−1 is indicated by the gray dashed line. (C) SAXS scattering intensity averages between 0.07 and 0.08 Å−1 versus BLI binding responses for 17b (blue circles), 19b (red triangles), PGT145 (green squares), and PGT151, (purple inverted triangles) determined for a panel of SOSIP Envs. (D) CH505 Env SOSIP temperature series binding responses for interactions with 17b (blue circles), 19b (red triangles), and PGT145 (green squares). The error bars indicate the SD from the arithmetic mean (N = 3). (E) SAXS scattering difference curves calculated by subtracting the CH505 Env SOSIP SAXS scattering profile at 25°C from the CH505 Env SOSIP scattering profile at 50°C. The scattering difference feature peak is indicated at q = 0.07 Å−1 by the gray dashed line.
SAXS data for proteins collected in solution involve isotropic scattering due to molecular movement. As a result, the scattering curves in the lower angle range report low-resolution features related to protein size and overall shape. Identifying specific curve features that correlate with differences in specific structural features can aid in assigning the observed transitions to structural changes. Owing to the marked differences exhibited by CH505 and BG505 SOSIPs in their binding to conformation-specific antibodies (Fig. 2A), we hypothesized that a clear differentiating feature would be found between their respective scattering profiles. We measured static SAXS profiles for CH505 and BG505 SOSIPs (fig. S1A) and calculated difference profiles by subtracting the BG505 Env SOSIP scattering curve from the CH505 Env SOSIP scattering curve (Fig. 2B). The difference profiles show a noticeable feature with a peak at a scattering vector (q) value of 0.07 Å−1 with a downward trend between 0.01 and 0.05 Å−1 followed by a rise between 0.00 and 0.01 Å−1 (Fig. 2B). We next asked whether this SAXS feature correlates with conformation-specific antibody binding for a panel of CH505-derived Env SOSIPs, including the CH505 transmitted founder, CH505 variants isolated at weeks 53, 78, and 100 after infection (47), a 4.1-stabilized (48) CH505 transmitted founder, and the BG505 Env SOSIP. We measured SAXS profiles for each Env SOSIP (figs. S1A and S2) and antibody binding for each to 17b, 19b, PGT145, and the trimer-specific, gp120-gp41 interface binding antibody PGT151 via BLI (fig. S1C). Linear regression of the average intensity at the feature (0.07 to 0.08 Å−1) versus BLI response for each antibody revealed significant correlation between the SAXS and binding data for 17b and 19b (P = 0.015 and 0.024, respectively; Fig. 2C). Binding of PGT145 and PGT151 was not significantly correlated with the SAXS data, although a clear negative trend was observed, consistent with differences in closed- and open-state populations (P = 0.127 and 0.270, respectively; Fig. 2C). The lack of significant correlation suggested that an additional factor affects binding that is not captured in the SAXS profile. As PGT145 and PGT151, but not 17b and 19b, must navigate the glycan shield, differences in variant glycan shields likely affected the correlation. Together, these results show that SAXS effectively reports known differences in structural state between SOSIPs.
We next extracted component curves from the set of static SAXS curves using singular value decomposition (SVD). This method splits the data into matrices of component curves, or left vectors, singular values, and contributions, or right vectors, that, when multiplied, return the original curves (49). This acts to remove noise from the data and to separate signals into distinct components that may correlate with physical properties of interest (here, differences in open-state propensity). The input for this analysis here is a matrix of SAXS intensities (fig. S1A), and the output is a series of vectors that separate the signal. This effectively removes noise from the signal and can separate contributions to the signal that correspond to different processes. It is important to note that SVD does not necessarily separate the signal into physically meaningful vectors. The first left vector was composed of a negative peak between 0.02 and 0.05 Å−1 with a minor positive peak at 0.07 Å−1 (fig. S1B). The second left vector showed features matching an inversion of the BG505 versus CH505 difference curve, consistent with this vector describing open or open-like state propensity (fig. S1B). The first left vector is consistent with a reduction in particle density or size. These results show that Env constructs that differ in their propensity to access open states exhibit differing SAXS profiles that, at the 0.07 Å−1 peak, report on the propensity of the SOSIP to occupy open or open-like states.
HIV-1 Env glycoprotein conformational transitions can be induced by increasing temperature
The TR, T-Jump SAXS experiments perturb the conformational equilibrium via a T-Jump initiated by a rapid IR laser pulse. It is therefore essential that the transition of interest occurs within the temperature range sampled in the experiment. We selected the CH505 SOSIP for these experiments due to its observed conformational lability and its ability to access the Env open state even in the absence of CD4 induction. To determine the effects of increasing temperature on the conformational dynamics of the CH505 SOSIP Env, we measured 17b, 19b, and PGT145 binding at 25°, 30°, 35°, 40°, 45°, and 50°C. As expected, elevated temperatures increased 17b and 19b binding while reducing PGT145 binding, demonstrating an increase in the population of open or open-like states at higher temperatures (Fig. 2D).
We next asked whether SAXS would capture structural changes in the CH505 SOSIP Env associated with increased temperature. We measured static SAXS scattering profiles for the CH505 SOSIP at 25°, 35°, 40°, 44°, and 50°C (fig. S3). The difference profiles displayed the same features as observed in the difference profiles for CH505 and BG505 SOSIPs at 0.07 Å−1 (Fig. 2E). Unlike the static difference profile between CH505 and BG505 at 25°C (Fig. 2B), the curve displayed a downward trend at scattering vector values <0.03 Å−1 (Fig. 2E). We performed an SVD of the static temperature series dataset to examine the component curves that define the signal. The first and second components in this dataset mirror those of the static SAXS SOSIP panel (fig. S3H). This suggests that the CH505 conformation is temperature sensitive, with higher temperatures leading to increased openness. Together, these results demonstrate that the conformational transitions in CH505 SOSIP are sensitive to temperature and can be measured using SAXS.
TR, T-Jump SAXS reveals time-dependent changes in CH505 SOSIP SAXS profiles
We next examined the CH505 SOSIP response to rapid system heating using TR, T-Jump SAXS. We first optimized the initial equilibrium temperature of our system. On the basis of our previous temperature-dependent binding experiments, we initiated T-Jumps from 40°, 42°, 44°, and 46°C with a probe delay of 10 μs and 1 ms and found that the difference signal in the region of interest (0.02 to 0.10 Å−1) was maximal at 44°C. We then determined the extent of laser-induced heating of the system when jumping from 44°C. Linear regression analysis of the temperature-dependent static SAXS profiles second SVD right vector (50) indicated that our system T-Jump was ~6°C, leading to a final perturbed system temperature of ~50°C (fig. S7). Next, to probe the CH505 SOSIP conformation at different times after heating, we measured scattering at post-laser excitation delay times of 500 ns, 1.5 μs, 3 μs, 5 μs, 10 μs, 50 μs, 100 μs, 500 μs, 1 ms, 10 ms, and 100 ms (Fig. 3A and fig. S5). The scattering difference curve measured at 500 ns shows a prominent negative peak between 0.02 and 0.05 Å−1 (fig. S5E). This is suggestive of a process that occurs faster than our measurement dead time. This feature becomes increasingly prominent at greater time delays up to 1 ms with a marked increase between 0.02 and 0.05 Å−1 between 1.5 and 3 μs (Fig. 3A). A second signal at q = 0.07 Å−1 becomes more prominent as the delay time is increased (Fig. 3A). These results are consistent with the static SAXS SVD analysis of the SOSIP panel, indicating that the transitions correspond to similar structural differences (fig. S1B). At longer time delays of 10 and 100 ms, a period over which the system temperature is returning to its initial state, these difference features begin to shift toward zero (Fig. 3A), indicating that the Env relaxes back to its initial equilibrium state distribution. Together, these results demonstrate that the CH505 Env conformation exhibits reversible structural transitions on the microsecond timescale. We next asked whether a more stable Env variant would show these transitions. We measured TR, T-Jump SAXS profiles for a CH848 (42) Env variant SOSIP trimer that included a disulfide bond designed to block movements to the Env open state. Unlike the CH505 SOSIP trimer, the CH848 variant shows no binding to the open-state targeting antibody 17b and markedly weaker binding to 19b, suggesting that the CH848 trimer accesses the open state less readily (fig. S1C). Thermal denaturation experiments also indicate that the CH848 SOSIP trimer is more stable than the CH505 SOSIP trimer, with denaturation inflection temperatures of 73.6° and 68.9°C, respectively. TR, T-Jump SAXS data were collected at time delays of 5 μs, 10 μs, 50 μs, 100 μs, 250 μs, 500 μs, 750 μs, 1 ms, 10 ms, and 100 ms, revealing SAXS differences over the same range as observed in CH505 (Fig. 3B and fig. S6). Therefore, despite increased stability and reduced open-state antibody binding relative to the CH505 trimer, these results show that CH848 undergoes similar structural transitions at the measured timescales.
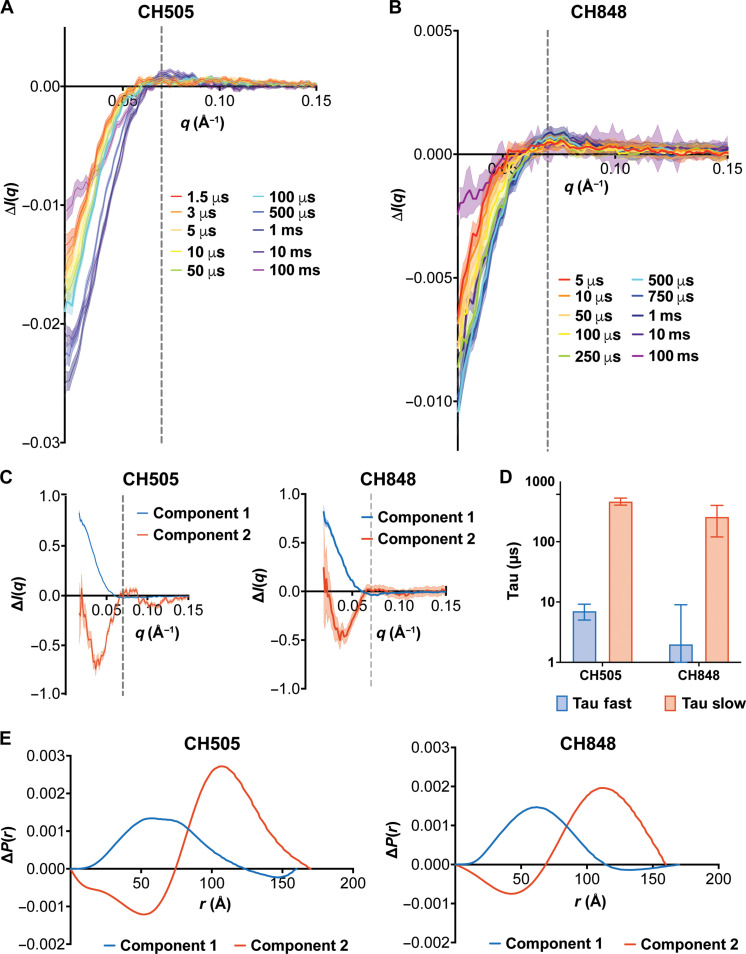
Fig. 3. TR, T-Jump SAXS of HIV-1 Env reveals two structural transitions.
(A) CH505 Env SOSIP TR, T-Jump SAXS scattering difference curves for 1.5-μs (red), 3-μs (orange), 5-μs (light orange), 10-μs (yellow), 50-μs (green), 100-μs (cyan), 500-μs (blue), 1-ms (indigo), 10-ms (violet), and 100-ms (magenta) time delays. (B) CH848 SOSIP Env TR, T-Jump SAXS scattering difference curves for 5-μs (red), 10-μs (orange), 50-μs (light orange), 100-μs (yellow), 250-μs (green), 500-μs (cyan), 750-μs (blue), 1-ms (indigo), 10-ms (violet), and 100-ms (magenta) time delays. In (A) and (B), the gray dashed line at q = 0.07 Å−1 indicates the scattering difference feature peak. Shaded regions indicate the SE from the arithmetic mean replicate measures. (C) Deconvoluted TR, T-Jump SAXS components 1 (blue) and component 2 (red) from the REGALS decomposition of SAXS difference curves for CH505 Env SOSIP (left) and CH848 Env SOSIP (right). The gray dashed line at q = 0.07 Å−1 indicates the location of the feature peak. (D) Fast (blue) and slow (red) time constants from double exponential fits of SVD component 1 for CH505 and CH848 time-resolved data. (E) REGALS pair distance distribution differences for components 1 (blue) and 2 (red) for CH505 (left) and CH848 (right) Env SOSIPs.
Env transitions through a rapidly forming intermediate
We next determined the number of distinct structural transitions occurring in the CH505 and CH848 trimer TR, T-Jump SAXS data. We first extracted component curves using SVD. Here, we focused on difference curves between 1.5 μs and 1 ms, as this time range reports on structural transitions before cooling of the sample at longer timescales. The results showed that the same two components observed in the static SAXS data for the CH505 and BG505 SOSIP panel (fig. S1B) were present in the CH505 (fig. S8B) and CH848 data (fig. S9B). The CH505 right vector first component transitions occurred at both shorter and longer time delays, while the second component transition occurred only at longer time delays (fig. S8D). Both the left vectors (fig. S8C) and right vectors (fig. S8E) for SVD components 3 to 8 fluctuated randomly about zero, indicating that these components did not contribute substantially to the TR SAXS signal. An SVD analysis of the CH848 data revealed that the same two components but only single transitions in the first and second components were discernible by visual inspection. We next fit kinetic models to the first two right vectors. A double exponential fit to the CH505 first component yielded time constants of 7 ± 2 μs and 466 ± 63 μs (Fig. 3D and table S4). A single exponential fit to the second right vector yielded a time constant of 789 ± 183 μs (Fig. 3D and table S4). We then fit a double exponential model to the CH505 area under the curve (AUC) between 0.02 and 0.1 Å−1. The results were consistent with the two-transition model of the first SVD component fit time constants of 7 ± 2 μs and 502 ± 40 μs (fig. S8H and table S4). Single exponential fits to the CH848 first and second right vectors yielded time constants of 213 ± 69 μs and 110 ± 15 μs, respectively, indicating that the structural rearrangements occurred at similar timescales (Fig. 3D, fig. S9D, and table S4). Calculations based on the AUCs for CH848 revealed a single transition with a time constant of 197 ± 55 μs, consistent with the SVD timescales (fig. S9H and table S4).
The CH505 first component changes on two timescales. The slower component one transition coincides with the second component transition, indicating that these transitions occur simultaneously. It is important to note that these component one reductions in scattering intensity at each timescale do not necessarily correspond to the same structural change. The CH848 single exponential kinetic model yielded a time constant similar to that of the single exponential kinetic model for the second component (table S4). The AUC single exponential kinetic model fit for CH848 was consistent with both SVD component fits (table S4). These results indicated that this CH848 transition is most similar to the second CH505 transition where components one and two change together. Fitting of a double exponential kinetic model to the CH848 component one yielded time constants of 2 ± 7 μs and 260 ± 140 μs (Fig. 3D). The large error for the fast time constant suggests that this model describes the data poorly. However, the slower transition in this model is consistent with that of the single exponential fits to component one, component two, and the AUC results. This suggests that the double-exponential model effectively captures the slow process. The poor fit is likely due to the absence of data points in the relevant timescales in the range of 1 to 5 μs. The double exponential fit supports a model of the CH848 results in which a transition matching the CH505 fast time constant occurs more quickly. Additional data at timescales between 1 and 5 μs would be needed to robustly evaluate this model. Nevertheless, the fast-forming intermediate observed in CH505 likely occurs in CH848. These results indicate that both SOSIPs display rapid transitions to open or open-like states and indicate that the two SOSIPs transition with differing time constants.
SVD does not necessarily decompose data into physically meaningful components (51). A recently developed algorithm, termed REGALS, deconvolves SAXS datasets by applying experimentally determined restraints to SVD deconvolutions so that the components are physically realistic (51). We performed a REGALS deconvolution on TR, T-Jump SAXS difference profiles to identify individual component scattering profiles in the CH505 and CH848 SOSIP time-resolved SAXS data (Fig. 3C and figs. S8, F and G, and S9, F and G). We included two components as identified in both the SVD and AUC analyses with minimum dimensions of 154 and 160 Å for CH505 and CH848, respectively, and maximum dimensions of 173 and 162 Å for CH505 and CH848, respectively. These dimensions were determined based on the dimensions of Env structure models (figs. S10A and S11A) and iterative model optimization. Fitting of the data yielded overall χ2 values of ~1.0 and 0.124 for CH505 (fig. S8F) and CH848 (fig. S9F), respectively. Each of the REGALS components (Fig. 3C) displayed similarities to their respective SVD components (figs. S8B and S9B) despite being mirrored about the x axis.
In addition to splitting the scattering difference curves into distinct components, REGALS returns pair distance distribution differences describing component particle atom-atom distance differences (Fig. 3E). The distance distribution difference for component 2 in both the CH505 and CH848 results indicated that structural changes occur over a relatively large range up to ~150 Å. The CH505 and CH848 component 2 ΔP(r) distributions showed marked reductions in pairwise distances between 0 and 75 Å and concomitant increases between 75 and 150 Å. The largest distance changes for component 1 were over a smaller range with a mean of ~60 Å for CH505 and CH848 (Fig. 3E). The results for CH505 showed that the component 2 concentration began to increase at longer time delays, while component 1 decreased at both early and long time delays (fig. S8G). The CH848 results showed that component 1 increased at longer time delays, while component 2 decreased at longer time delays (fig. S9G). These results are consistent with the changing contributions of the components determined by the SVD right vector analysis (figs. S8D and fig. S9D). This REGALS analysis, together with the SVD analysis, is consistent with a model consisting of two distinct structural changes in CH505 and CH848, with distance distribution information useful for structure-based modeling.
The slower transition involves the formation of an open-occluded state
We next asked how the transitions and states observed in CH505 and CH848 SOSIPs here relate to known HIV-1 Env ectodomain structures and dynamics. Previous smFRET studies have shown that an asymmetric intermediate connects the closed and open configurations (17–20). Transitions from the ground state to this asymmetric state and from this state to the open state may occur within the millisecond dead time of the measurement (20), which would be consistent with the processes measured here (Fig. 3D and table S4). The asymmetric state is characterized by a single protomer open state with two protomers in a distinct conformation. On the basis of the correlation between open-state targeting antibody binding and static SAXS at increasing temperature (Fig. 1C), the first transition observed in the TR, T-Jump SAXS dataset corresponds to transitions that are initiated from a closed state. Two distinct structural states of SOSIP Envs have been observed previously including open-occluded states in which each gp120 domain is rotated outward from the primary trimer axis without major rearrangements in V1/V2 or V3 (Fig. 1B, middle), and an open state in which the gp120 domains are rotated outward and V1/V2 and V3 have rearranged (Fig. 1B, right). We first asked whether our measures could effectively distinguish between theoretical SAXS difference curves calculated from a prefusion closed-state model and either an open-occluded state or an open-state model. An SVD analysis of the closed, open-occluded, and open model theoretical SAXS curves (figs. S10C and S11C) indicated two primary signals consistent with the components extracted from the TR, T-Jump experimental difference curves (figs. S8A and S9A). The SAXS difference curve for a transition from closed to open occluded resembles the SVD and REGALS component 2 (Fig. 3C), with a prominent negative and positive peak at low scattering angles (Fig. 4A and fig. S12B). The P(r) function difference curves were like the REGALS component P(r) difference curves, albeit inverted across the distance axis (Fig. 4A and fig. S12B). The closed to open SAXS difference curve was dominated by a single positive peak that did not match the SAXS features (Fig. 4B). The open-occluded versus open SAXS and P(r) function difference curves did not show similarities to any of the SVD or REGALS curves (Fig. 4C). This indicates that transitions between these two states did not occur or were not resolved in the dataset. These modeling results indicate that the second component transition involves movement to a state similar to known open-occluded state structures where gp120 domains have rotated outward from the trimer central axis without substantial rearrangements in the V1/V2 and V3 regions.
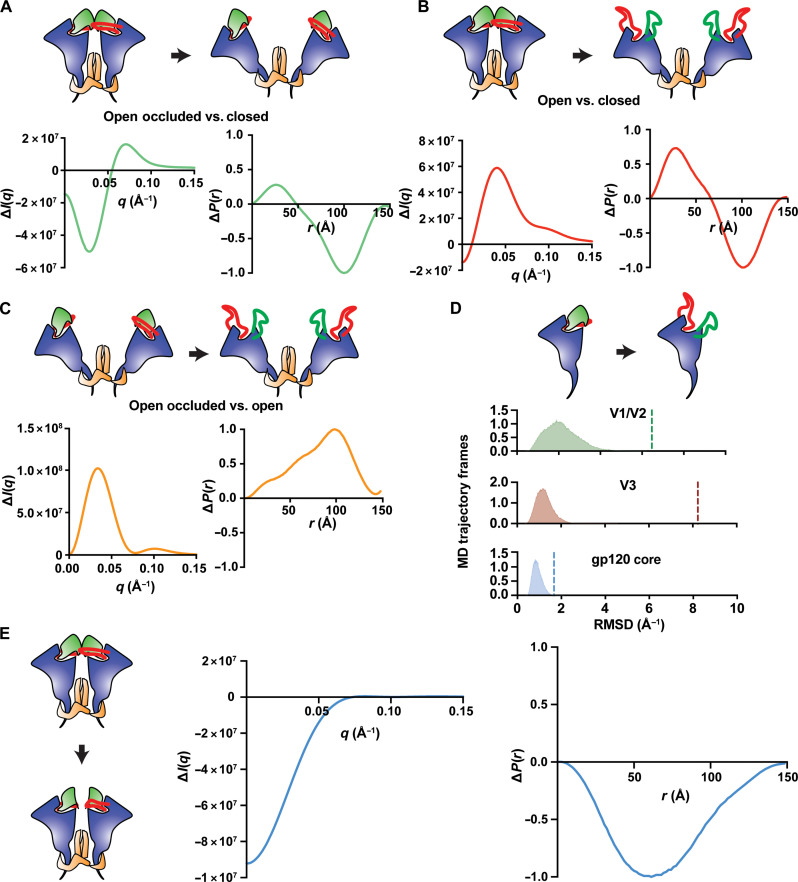
Fig. 4. Fast and slow processes correspond to two distinct structure transitions.
(A to E) Cartoon diagrams represent distinct Env structural states. Relevant regions of variability include V1/V2 (green), V3 (red), gp120 (blue), and gp41 (orange). (A) Theoretical SAXS difference curve (bottom, left) and the theoretical pair distance distribution difference curve (bottom, right) for the closed to open-occluded transition. (B) Theoretical scattering difference curves (bottom, left) and pair distance distribution difference curve (bottom, right) for the closed to open conformational transition. (C) Theoretical SAXS scattering difference curve (bottom, left) and pair distance distribution difference curve (bottom, left) for the open-occluded to open transition. (D) Cartoon depiction of the V1/V2 and V3 rearrangement in a gp120 monomer (top). The RMSD (bottom) for V1/V2 (green), V3 (red), and gp120 core (blue) β sheet α carbons for an aggregated 250 5-μs MD simulations, each containing a total of 1000 frames. The dashed lines represent the RMSD for the given domain between the closed and open conformations. (E) Theoretical SAXS scattering difference curve (middle) and the pair distance distribution difference curve (right) for the apex order-to-disorder transition.
We next examined the structural transition involved in the rapidly forming intermediate. The first REGALS component P(r) difference function involves a substantial loss of scattering centers within a range of 0 to 100 Å but without concomitant increases in more distant scattering centers. This is consistent with a loss of scattering density in the Env. The HIV-1 Env is a heavily glycosylated protein with a dense network of conformationally dynamic interactions (52–54). We, therefore, asked whether changes in the glycan shield could explain the two SVD and REGALS components. An SVD analysis based on theoretical SAXS profiles for nonglycosylated and mannose-9 (Man9) glycosylated Env closed, open-occluded, and open models indicated that the overall features between the two are similar with shifts in the scattering vector position of the peaks that are related to differences in particle size between the two sets (fig. S10D). The nonglycosylated models display the SAXS features of interest, suggesting that the glycans alone do not give rise to the observed difference signal.
The fast-forming intermediate involves an order-to-disorder transition in the trimer apex
We next asked whether the TR, T-Jump SAXS intermediate state observed in both the CH505 and CH848 SOSIP trimers involves rearrangements in the gp120 V1/V2 and V3 elements. The V1/V2 element packs against V3 and the gp120 outer domain in the closed state, which both contact the adjacent protomer (Fig. 1B, left and middle). In the open and open-occluded states, the gp120s rotate outward from the trimer central axis (Fig. 1B, middle and right). In the open state, the V3 loop transitions from an ordered loop packed against V1/V2 and the gp120 outer domain to a disordered state (Fig. 1B, right). In addition, the V1/V2 region becomes dislodged from the outer domain (Fig. 1B, right). Movements in V1/V2 result in strand switching from a closed-state three–β sheet structure to an open-state four–β sheet structure in β20-β21, residues 418 to 445, that is referred to as the bridging sheet (Fig. 1B, right). Each of these rearrangements results in substantial differences in structure and may be preceded by structural intermediates. Isolated gp120 domains in solution are known to expose the bridging sheet epitope, which is targeted by the CCR5 binding site monoclonal antibody (mAb), 17b, as well as V3 loop targeting antibodies such as mAbs 19b, F39F, and 3074 (55). This contrasts with stabilized SOSIP trimers, which do not show binding to these mAbs (7, 26–32, 35, 38, 39, 56, 57). We used molecular dynamics (MD) simulation to monitor the structural plasticity of the closed-state gp120 domain V1/V2 and V3. For this, we extracted a gp120 domain from a closed-state CH505 SOSIP trimer. This smaller system allowed us to (i) reach longer simulated timescales due to the reduction in the number of atoms in the system, and (ii) observe enhanced gp120 dynamics in the V1/V2 and V3 region due to the absence of interprotomer contacts. We collected a total of 250 independent 5-μs MD simulation trajectories run at a system temperature of 50°C to examine the V1/V2 and V3 conformation. Open-state V1/V2 and V3 conformations have root mean square deviations (RMSDs) of 6.5 and 8.2 Å relative to the closed state, respectively (Fig. 4D). The average V1/V2 and V3 RMSDs relative to the initial closed structure are 2.1 ± 0.8 Å and 1.3 ± 0.4 Å, respectively, compared to the conformationally invariant gp120 outer domain β sheet average of 0.9 ± 0.2 Å (Fig. 4D). The maximum RMSDs observed for each of these elements were 6.1, 4.6, and 2.1 Å, for V1/V2, V3, and β20-β21, respectively. Substantial V1/V2 displacement and strand switching in β20-β21 were not observed in any of the simulations.
Although the V1/V2 and V3 elements did not show transitions consistent with movement to an open or open-like state, an order-to-disorder transition in the loops that form interprotomer apex contacts in the closed-state trimer was observed. This led us to ask whether an order-to-disorder transition in the apex could explain the change in scattering observed in the fast-forming intermediate. The loss of scattering density suggested by the REGALS component 1 P(r) difference function is consistent with an order-to-disorder transition. This is due to a reduced density differential for a packed protein fold and a well-hydrated, disordered chain. We, therefore, asked whether a structure missing the gp120-to-gp120 contacting loops could give rise to the observed scattering difference curves and ΔP(r) function. The difference profile for both the scattering and P(r) functions calculated for models missing residues 160 to 171 and 306 to 317 matched the observed REGALS component 1 (Fig. 4E and fig. S12A). This is reflected as a reduction in scattering density and likely corresponds to a transition to a disordered state for this portion of the trimer. Together, these results are consistent with a model of a T-Jump transition that eliminates contacts at the trimer apex and gp120 rotation to an open-occluded state.
Interprotomer disulfide bonds stabilize the closed Env trimer
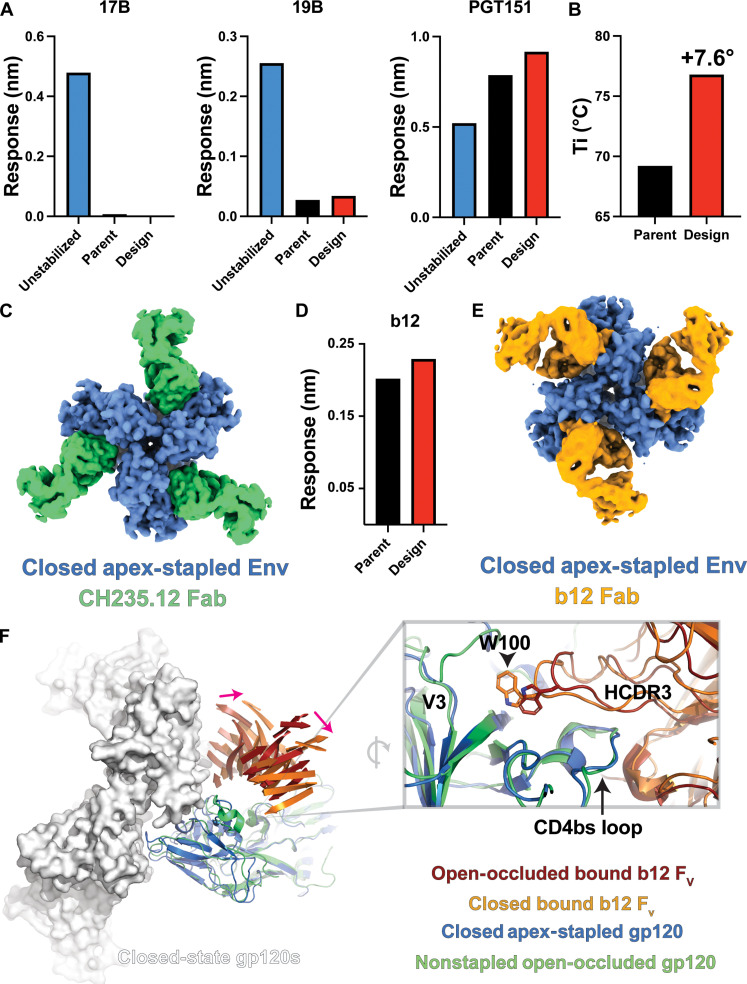
The TR, T-Jump SAXS results indicate that even in Env SOSIPs that do not show substantial open-state targeting antibody binding, movements to a more open configuration can still occur. A recent macaque immunization study found that, despite immunogen stabilization in a prefusion closed conformation, antibodies targeting open-occluded states were induced, consistent with our observations (15). Our modeling of the SAXS data suggests that changes in gp120-to-gp120 apex contacts play an important role in controlling gp120 rotation. Thus, blocking rearrangements in the apex is likely necessary to prevent the Env from transitioning to the open-occluded conformation. We, therefore, used a combination of in silico tools to identify possible interprotomer apex stabilizing mutations. From this analysis, we identified a V127C-D167C disulfide that stapled V1/V2 contact regions between gp120 protomers in a CH505 SOSIP trimer design parent (58). This design was further stabilized using the previously reported F14 (39) and SOSIP 2P (59) mutations to improve expression and folding in addition to a CD4bs antibody binding enhancing N197D glycan deletion mutation (60). A closed Env trimer conformation was identified by negative-stain electron microscopy (NSEM). However, SDS–polyacrylamide gel electrophoresis (SDS-PAGE) analysis and binding to 19b indicated that the Env SOSIP preparation included a population with improperly formed interprotomer disulfides that exposed the V3 epitope (fig. S13). Single-particle cryo-EM analysis of a CH505 apex-stapled SOSIP design bound to CH235.12 yielded two distinct populations, including a closed state and another state lacking well-defined density in the V1/V2 apical region (Fig. 5C and fig. S14B). To eliminate the open state, we further purified the Env SOSIP sample by negative selection using the V3 loop targeting 3074 antibody (61). This resulted in the elimination of the non-interprotomer–linked bands in SDS-PAGE (fig. S13B), reduction of 19b binding (Fig. 5A and fig. S13A), and enhancement of trimer-specific, gp120/gp41 interface targeting PGT151 binding (Fig. 5A and fig. S13A). The apex-stapled design displayed a marked increase in thermal denaturation inflection temperature (Ti + 7.6°C; Fig. 5B and fig. S13D). The closed-state configuration of the interprotomer disulfide stapled design displayed lower resolution density at the interprotomer apex loop contacts suggestive of conformational variability at this site (fig. S14). Density consistent with the formation of a disulfide between the protomers was nevertheless visible at a lowered map contour level (fig. S17A). Together, these results show that disulfide linkages between protomers at the trimer apex can effectively stabilize the trimer.
Fig. 5. Interprotomer disulfide bonds stabilize the closed Env trimer.
(A) Binding responses for a CH505 SOSIP, CH505 parent SOSIP containing previously identified stabilizing mutations, and the interprotomer disulfide-stapled CH505 SOSIP design interacting with the co-receptor binding 17b, V3 loop binding 19b, and the closed-state apex-interactive, trimer-specific PGT145 mAbs. (B) Differential fluorescence thermal denaturation inflection point temperature for the parent CH505 SOSIP and the interprotomer disulfide-stapled CH505 SOSIP. (C) Gaussian filtered map of the interprotomer disulfide-stapled CH505 SOSIP design bound to the CH235.12 Fab. (D) Binding responses for PGT151 captured stabilized CH505 parent SOSIP and interprotomer disulfide-stapled CH505 SOSIP design interacting with the b12 Fab. (E) Gaussian filtered map of the interprotomer disulfide-stapled CH505 SOSIP design bound to the b12 Fab. (F) Left: Structure comparison between the closed-state b12-bound interprotomer disulfide-stapled CH505 SOSIP design and the open-occluded state b12-bound B41 isolate SOSIP. A single b12-bound gp120 domain from B41 is aligned to a closed-state CH505 design gp120 to highlight the shift in the b12 Fv position (only β sheets shown for clarity; pink arrows indicate shift direction). Two additional gp120 domains are shown as surfaces to highlight the potential for b12 clashes. Right: Alignment of the b12-bound apex-stapled CH505 SOSIP design and B41 isolate gp120 domains highlighting differences in the HCDR3 position.
We next asked whether open-occluded state antibody binding was eliminated in the interprotomer disulfide-linked trimers. A previous structural study showed that the CD4bs b12 antibody interacted with the B41 isolate Env ectodomain in an open-occluded conformation (14). Another study showed that the capture of a BG505 isolate Env ectodomain with PGT145 eliminated b12 binding, presumably by locking the trimer apex in the close configuration (15). The apex-stapled CH505 SOSIP design binding with the b12 antibody Fab by BLI was like that of the non–apex-stapled construct (Fig. 5D). Examination of the b12-bound structures of each by NSEM indicated that, in the non–apex-stapled construct, apex contacts were broken, consistent with an open-occluded state, while the apex contacts in the apex-stapled construct remained intact (fig. S15). A previous study found that capture of a trimer by PGT145 eliminated b12 binding, likely through PGT145-mediated stabilization of the closed state (15). The ability for b12 to bind in the CH505 design is likely due to the elimination of the N197 glycosylation site via the introduced N197D mutation, which would eliminate steric hindrance from the glycan. Model fitting of b12 into the cryo-EM density of the apex-stapled CH505 SOSIP showed that a substantial shift in its angle of approach permitted interaction of b12 with this apex-stapled construct, which was fixed in the closed configuration and unable to access the open-occluded conformation preferred by b12 (fig. S15). We obtained a 4.5-Å resolution cryo-EM reconstruction of the b12-bound apex-stapled construct to examine these changes in greater detail (Fig. 5 and fig. S16). The map showed that the prefusion, closed configuration is retained with b12 interaction and that the b12 angle of approach is substantially shifted compared to a previously determined open-occluded state b12-bound B41 isolate Env SOSIP (Fig. 5F) (14). This results in a shift of the HCDR3, bringing residue W100 closer to the gp120 V3 loop (Fig. 5F and fig. S16). The overall fold of the gp120 domain is similar, with a gp120 RMSD of ~1.3 Å relative to a previously determined CH505 SOSIP structure bound to the CH235 unmutated common ancestor (UCA) antibody Fab (fig. S17B) (58). The shifted position appears to be facilitated through rearrangements in the β20-β21 and b12 contact with the V1/V2 region in the apex-stapled construct relative to the B41 isolate bound, open-occluded state structure (Fig. 5F and fig. S17C). Comparison of the CH235.12 and b12 CH505 apex staple design to a CH505 structure bound to the CH235 UCA antibody Fab indicated that the gp120 domains were shifted inward, toward the primary trimer axis (fig. S17C). This suggests that the disulfide causes closer interactions between the gp120 domains. These results show that disulfide linkages at the trimer apex between protomers can effectively eliminate movements to an open-occluded, intermediate state.
DISCUSSION
Here, we show that the CH505 and CH848 DS Env SOSIP conformations transitioned to open-occluded–like states on the microsecond timescale. The CH505 results showed that a rapidly forming intermediate precedes transitions to an open-occluded–like state. Uncertainty in the model fits to the CH848 data could not unequivocally establish that this rapidly forming intermediate occurred in the CH848 DS SOSIP. Model comparisons are nevertheless supportive of a two-state CH848 transition in which the intermediate transitions and the open-occluded transitions are more rapid compared to CH505. The gp120 MD simulations show that the closed-state fold from which they were initiated is stable on the timescale of the fast-forming intermediate apart from the interprotomer contacting gp120 loops. These loops showed an order-to-disorder transition in the simulations. This led us to test a model for early intermediate formation involving loss of scattering density at the apex loop contacts. We found that this model is consistent with both our SVD and REGALS analysis, suggesting that these loops undergo an ordered to disordered transition. The second slower transition is consistent with scattering profile differences between the CH505 and BG505 SOSIP Env panel and open-state antibody binding. Thus, this state likely corresponds with an Env in which gp120 is rotated outward, giving rise to a greater distance between the gp120 apex residues. It is not clear from the scattering data here, however, to what extent the V1/V2 and V3 rearrangements have occurred or the extent of gp120 rotation.
Most stabilizing designs focus on disabling allosteric rearrangements coupled with V1/V2 rearrangement and improving gp120 to gp41 interactions (7, 18, 20, 25, 48, 52, 53, 62–69). The induction of antibodies that target the open-occluded state of Env by vaccination with prefusion stabilized trimers and the presence of an intermediate consistent with this state here suggest that stabilization between apex contacts is needed to eliminate shifts in gp120 domain position. As blocking transitions out of the closed state is essential for inducing neutralizing antibodies by vaccination, we used this information to design an Env SOSIP with a disulfide staple at the apex to eliminate this disorder transition. This effect was confirmed by the closed state–bound configuration of the b12-neutralizing antibody. It is important to note that the position of this disulfide may not be suitable for differing Env variants. A previous HDX experiment demonstrated that hyperstabilized designs differed in their ability to limit Env dynamics depending on the variant in which the stabilization mutations were added (25). Several variants showed little response, while one variant showed markedly reduced dynamics. The ability of the DS stabilized CH848 variant to access the same states as the less stable CH505 variant is consistent with these results, suggesting that stabilization mutations do not necessarily block major rearrangements in gp120 rotation. Unlike the mutations examined by HDX, the apex staple involves a chemical linkage that, unless broken, does not allow separation of the gp120 domains. Nevertheless, it is not certain that all Env variants can be locked via mutation at these specific sites. Several potential sites for interprotomer gp120 linkage are available that may increase the extensibility of this stabilization approach should these mutations fail to form disulfides or act to limit protein expression.
When interpreting low-resolution scattering results, it is important to consider alternative explanations of the data. The Env glycan shield likely plays a major role in determining the density of the particle solvent shell and, in combination with the possibility of differential glycan interactions, complicates differentiating transitions that involve protein and those that may involve only glycan. We find that modeling the trimer in different combinations of closed and open states here with and without glycans yields nearly identical difference curve features. This suggests that the early transitions we observe are protein related. The solvent shell of nonglycosylated proteins responds to the T-Jump within ~250 ns (62). The relaxation timescale for individual glycan motions is typically under 1 μs at a given temperature (70), which together suggests that any changes in the glycan in response to the T-Jump would likely occur well below our observed intermediate timescale. These considerations together led us to conclude that a substantial portion of time-dependent scattering differentials we observe are associated with protein movements.
It is also important to consider that, although the Env SOSIPs used here were extensively purified and characterized, we cannot entirely rule out the possibility that multiple, yet unidentified, folded species exist in our samples. This too could lead to two different transition rates that, rather than sequential movements, are related to relative transition propensities between two different folded states. In the case of our apex-stapled design, a differing folded state population was readily identified in our cryo-EM three-dimensional (3D) classification. No such state has been identified in any of our related SOSIP constructs (39, 58, 71, 72), nor were substantial lot-to-lot quality control variations observed that would indicate fold instability. Furthermore, a different fold would likely result in a distinct thermal denaturation profile, so a discernible bi- or multimodal denaturation profile would be expected. This was not observed for our samples here. We, therefore, conclude that the transitions we observe are indeed related to sequential events of a single-fold species. Last, it is difficult to robustly quantify the extent of gp120 rotation and the degree of conformational dynamics in the end state observed here. Additional experiments are needed to compare SAXS measurements for trimers locked in known conformational states with other structural data.
The findings from this study show that the transition from a closed state to one in which the gp120 domains rotate outward away from the timer central axis involves sequential movements. Rather than gp120 rotation occurring simultaneously with apex contact breakage, the gp120 apex loop contacts first become disordered. Our results suggest that this occurs without complete V1/V2 rearrangements that result in the formation of the bridging sheet. This is consistent with previous studies showing that V3 loop exposure can occur without exposure of the bridging sheet (10). The transition to a more open configuration occurred at a nearly two orders of magnitude longer timescale compared to the order-to-disorder transition in the CH505 Env SOSIP. With contacts at the apex broken, control of this transition must involve the remaining gp120 to gp41 contacts. At equilibrium, this sequential ordering of each transition creates a kinetic switch where gp120 rotation is controlled by the joint probability of both the order-to-disorder apex transition and breakage of the remaining gp120 to gp41 contacts occurring. Changes in the relative probabilities of these transitions likely explain the reduction in CH848 time constants and may explain differences in open-state probabilities between different Env variants. In turn, these differences in microsecond timescale transition rates could determine sensitivities to receptor-induced conformational transitions.
The relationship between the transitions we observe here and the native, full-length gp160 HIV-1 Env is an open question. Native Env trimers are membrane-embedded, which may alter the conformational transitions between Env structural states and their kinetics relative to the transitions measured here in the soluble gp140 construct. Membrane-embedded gp160 Env is known to transition between multiple structural states similar to the soluble SOSIP gp140 constructs (2, 17, 18, 73) and also shares structural similarity with gp140 SOSIP structures (74, 75). Thus, the results presented here are likely relevant for gp160 Env. The specific relationship between previously observed states is nevertheless unknown owing to the absence of the transmembrane domain and the addition of trimer stabilizing SOSIP mutations in these constructs. These are important questions as any susceptibility to movement beyond a prefusion closed state could be exploited by maturing antibodies in the context of vaccination. These states are rare in tier 2 viruses prior to CD4 receptor binding. The impacts on vaccination outcome are twofold: Poorly neutralizing antibodies targeting open-occluded states are more likely to be induced and antibodies with broad neutralizing potential can acquire mutations favoring these states rather than mutations that are critical to improving neutralization function, thus limiting their development toward breadth. The apex-stapled design presented here prevents the movement of gp120, thus eliminating these possibilities. Consistent with impacts on antibody interactions, the NSEM and cryo-EM structures of b12 revealed marked changes in the antibody angle of approach with the apex staple compared to a nonstapled construct.
In summary, these results show that transient intermediate states observable only on a microsecond timescale play an essential role in controlling HIV-1 Env conformation. Blocking these early transitions is likely an important consideration in vaccine development efforts to ensure that maturing antibodies remain on track to develop neutralization breadth.
MATERIALS AND METHODS
Recombinant HIV-1 Env SOSIP production
A chimeric CH505 transmitted founder (CH505) envelope SOSIP, containing the CH505 gp120 and truncated BG505 gp41 domains, a 4.1-stabilized CH505TF SOSIP, CH505 SOSIPs isolated from the CH505 person living with HIV at weeks 53, 78, and 100 after infection, a DS-stabilized (76) CH848 day 949 10.17 envelope SOSIP (42), and an N332 BG505 envelope SOSIP (27) were expressed in Freestyle 293-F cells (Thermo Fisher Scientific, catalog no. R79007). Before transfection, cells were diluted in Freestyle 293 Expression Medium (catalog no. 12338018) to 1.25 × 106 cells/ml at a volume of 950 ml. Plasmid DNAs expressing the envelope SOSIP and furin were cotransfected at a 4:1 ratio (650 and 150 μg per transfection liter, respectively) and incubated with 293fectin transfection reagent (Thermo Fisher Scientific, catalog no. 12347019) in Opti-MEM I Reduced Serum Medium (Thermo Fisher Scientific, catalog no. 31985062) to allow for complex formation. The diluted mixture was added to the cell culture, which was incubated at 37°C, 9% CO2 on a shaker at 120 rpm for 6 days. On day 6, the cell supernatant was harvested by centrifuging the cell culture at 4000g for 45 min. The supernatant was filtered with a 0.45-μm polyethersulfone (PES) filter and concentrated to approximately 100 ml using a Vivaflow 200 cross-flow cassette (Sartorius, catalog no. VF20P2).
Envelope SOSIPs were purified using a PGT145 affinity chromatography column equilibrated in 15 mM Hepes and 150 mM NaCl (pH 7.1). A PGT145 Ab affinity column was made by coupling PGT145 mAbs to CNBr-activated Sepharose 4B (catalog no. 170430-01, GE Biosciences) and packed into a Tricorn column (GE Healthcare). The supernatant was applied over the column at 2 ml/min using an AKTA go chromatography system (Cytiva) followed by three-column volume wash steps with 15 mM Hepes and 150 mM NaCl. Protein was eluted off the column using 3 M MgCl2 and diluted in 15 mM Hepes and 150 mM NaCl buffer. The protein sample was buffer-exchanged into 15 mM Hepes and 150 mM NaCl by ultrafiltration using a 100-kDa molecular weight cutoff Amicon Ultra-15 Centrifugal Filter Unit (Millipore Aldrich, catalog no. UFC9010) and concentrated to <0.5 ml for size exclusion chromatography. Size exclusion chromatography was performed using a Superose 6 10/300 GL Column (Cytiva) on an AKTA go system in 15 mM Hepes and 150 mM NaCl. Fractions containing trimeric SOSIP were collected.
All lots produced were subjected to quality control including analytical size exclusion chromatography, SDS-PAGE, thermal shift analysis, BLI, and NSEM to assure the presence of well-folded Env trimers. Production lots deviating from mean observed values in each experiment greater than 1 SD were not used in the experiments. For the TR, T-Jump SAXS experiments, all CH505 SOSIP lots were combined. The combined sample was subjected to the same quality control after completion of the TR, T-Jump SAXS experiments to verify sample integrity.
Recombinant antibody and antibody Fab production
Antibodies were expressed in Expi293F cells in the Expi293 Expression System (Thermo Fisher Scientific, catalog no. A1435101). Cells were diluted to 1.25 × 106 cells/ml at a volume of 100 ml. Heavy- and light-chain plasmid DNAs, at a ratio of 1:1 (50 μg each per 100-ml transfection), were mixed in Opti-MEM I Reduced Serum Medium (Thermo Fisher Scientific, catalog no. 31985062) and incubated with Expifectamine 293 transfection reagent (Thermo Fisher Scientific, catalog no. A14525) to create the DNA-reagent complex. The mixture was added to the prepared cell culture, which was transferred to be incubated at 37°C, 8% CO2 on a shaker at 120 rpm for 6 days. On day 6, the cell supernatant was harvested by centrifuging the cell culture at 4500 rpm for 45 min. The supernatant was filtered with a 0.45-μm PES filter.
Each antibody was purified with a Cytiva HiTrap Protein A HP antibody purification column (Cytiva, catalog no. 29048576) equilibrated in 1× phosphate-buffered saline (PBS). The harvested supernatant was applied over the equilibrated column at a flow rate of 0.5 ml/min using an AKTA start chromatography system (Cytiva), followed by three-column volume wash steps of 1× PBS. The antibodies were then eluted off with Pierce immunoglobulin G (IgG) elution buffer (pH 2.8; Thermo Fisher Scientific, catalog no. 21004). The sample buffer was neutralized with tris (pH 9) at 100 μl/ml (Thermo Fisher Scientific, catalog no. J60707). Lots produced were subjected to quality control including SDS-PAGE and thermal shift analysis.
Antibody Fabs were prepared using Lys-C digestion. A digestion mixture of Lys-C enzyme (Thermo Fisher Scientific, catalog no. 90307) and IgG (1:2000 ratio) was combined in 600 μl of 1× PBS for each sample. Each sample was incubated at 37°C for 2 hours. Following the incubation, the digested mAb was removed from the incubator, and a proteinase inhibitor is added to the solution to stop the digestion process. The sample was then purified with a Cytiva HiTrap Protein A HP antibody purification column (Cytiva, catalog no. 29048576), collecting the Fab fragments that flow through. The Fab fragments are then buffer-exchanged into 15 mM Hepes and 150 mM NaCl. Each Fab lot produced was subjected to quality control by SDS-PAGE and thermal shift analysis.
Thermal shift assay
Thermal shift assay was performed using Tycho NT.6 (NanoTemper Technologies). Envelope ectodomain SOSIPs were diluted (0.15 mg ml−1) in 15 mM Hepes buffer with 150 mM NaCl at pH 7.1. Intrinsic fluorescence was recorded at 330 and 350 nm while heating the sample from 35° to 95°C at a rate of 3°C min−1. The ratio of fluorescence (350/330 nm) and the inflection temperatures (Ti) were calculated by Tycho NT.6.
Biolayer interferometry
Sample mAb and Fab binding were obtained using BLI (OctetR4, FortéBio). Antibodies were immobilized on anti-human IgG Fc capture (FortéBio) sensor tips, while Fabs were immobilized on protein A sensor tips, each via immersion in mAb (4.5 μg/ml) in 15 mM Hepes buffer with 150 mM NaCl at pH 7.1 for 300 s followed by washing in 15 mM Hepes buffer with 150 mM NaCl at pH 7.1 for 60 s at 1000 rpm. The sensor tips were then immersed in the SOSIP-containing wells (45 μg/ml) for 180 s. Reported binding corresponds to values and the end of the association phase. Data were evaluated using the Octet Data Analysis software (FortéBio).
Negative-stain electron microscopy
An aliquot at room temperature was diluted to 20 μg/ml with 0.1% (w/v) n-dodecyl β-d-maltoside [50 mM tris (pH 7.4), 150 mM NaCl, and sodium deoxycholate (0.03 mg/ml)]. The sample was immediately applied to a glow-discharged carbon-coated EM grid for 8 to 10 s, then blotted, and stained with uranyl formate (2 g/dl) for 1 min, blotted, and air-dried. Grids were examined on a Philips EM420 electron microscope operating at 120 kV and nominal magnification of ×49,000, and 114 images were collected on a 76-megapixel charge-coupled device camera at 2.4 Å per pixel. Images were analyzed by 2D class averages using standard protocols with Relion 3.0 (77).
TR, T-Jump small-angle x-ray scattering beamline setup
The BioCARS 14-ID-B beamline at the Advanced Photon Source at Argonne National Laboratory was used to conduct TR, T-Jump SAXS and static temperature series SAXS experiments. CH505 protein was concentrated to a final concentration of 5.125 mg/ml in 15 mM Hepes buffer with 150 mM NaCl at pH 7.1, for a total volume of ~4 ml. Two separate solutions of CH848 protein were concentrated in 15 mM Hepes buffer with 150 mM NaCl at pH 7.1 to final concentrations of 1.66 mg/ml in ~3 ml and 1.4 mg/ml in ~2 ml, respectively. The protein sample was syringe-filtered with a 0.22-μm filter and degassed before use. A peristaltic pump–injected protein/buffer solution through a 700-μm quartz capillary mounted on a custom, temperature-controlled aluminum nitride holder. T-Jumps of the sample were initiated using a 7-ns IR laser pulse at 1.443-μm wavelength and 1.1 mJ per pulse to excite the O-H stretch of water. The sample was probed with a 1.6-μs (11-bunch mode) or 3.6-μs (24-bunch mode) pulse of pink x-ray beam with photon energy of 12 keV and bandpass of 300 eV after a variable time delay (fig. S5A). Images were collected using a Rayonix MX340-HS x-ray detector in a scattering vector (q) range of 0 to 2.5 Å−1.
T-Jump calibration
The magnitude of the T-Jump was calibrated as previously described (50). We measured the static SAXS signal of buffer at 25° to 51°C for CH505 buffer data and 30° to 45°C for CH848 buffer data. For each respective dataset, we combined T-Jump results with the static results and performed an SVD decomposition of the data. The second component left vector displayed a profile matching differences in the buffer signal. We therefore performed a linear regression analysis of the second component right vectors for the static temperature data. The magnitude of the T-Jumps was then determined on the basis of the slopes and intercepts from the respective fits.
Static SAXS
Static SAXS scattering profiles of BG505 (43) and the CH505 (41) Env SOSIP panel were collected at the Lawrence Berkeley National Laboratory Advanced Light Source SIBYLS 12.3.1 beamline (63, 64, 78, 79). Samples were shipped overnight at −79°C and stored at −80°C before data collection. A series of concentrations between 0.5 mg/ml and 3 to 5 mg/ml were collected with a total exposure time of 10 s. Data were reduced and analyzed using the ATSAS program suite.
Static SAXS temperature series
Static SAXS temperature series data were measured at the Advanced Photon Source BioCARS 14-ID-B beamline used for TR, T-Jump SAXS, described above. Static SAXS profiles for 15 mM Hepes buffer and CH505 Env sample (5.125 mg/ml) at 25°, 35°, 44°, and 50°C (fig. S3) were collected. For CH848 Env samples, static SAXS profiles for 15 mM Hepes buffer, CH848 (1.66 mg/ml), and CH848 (1.4 mg/ml) were collected at 30°, 35°, 40°, and 44°C (fig. S4). Static SAXS signals were collected using 24 bunches in continuous translation mode at 20 Hz with 3.6 μs per pulse and 197 pulses per image exposure time on the BioCARS 14-ID-B beamline. A total of 50 images were collected for each set for scattering vector range q = 0 to 2.5 Å−1.
CH505 and CH848 time-delay series
TR, T-Jump SAXS data were collected at the Advanced Photon Source BioCARS 14-ID-B beamline. Data were collected for a CH505 Env SOSIP sample (5.125 mg/ml) at several different time delays after sample heating from 44° to ~50°C by IR pulse T-Jump: 500 ns, 1.5 μs, 3.0 μs, 5.0 μs, 10 μs, 50 μs, 100 μs, 500 μs, 1 ms, 10 ms, and 100 ms. Time-resolved SAXS data were collected for CH848 Env SOSIP samples (1.66 and 1.4 mg/ml) at 5 μs,10 μs, 50 μs, 100 μs, 250 μs, 500 μs, 750 μs, 1 ms, 10 ms, and 100 ms. For each time delay measured, TR, T-Jump SAXS was also measured at −10 and −5 μs before heating with IR laser (“laser off”; fig. S5A). To accommodate the large change in timescales and minimize systematic errors due to experimental drift, we measured TR, T-Jump SAXS for buffer and Env in multiple sets, with overlapping time delays between each set. The CH505 Env SOSIP TR, T-Jump SAXS was measured in three sets. CH505 Env set 1 includes −10 μs, −5 μs, 10 μs, 50 μs, 100 μs, 500 μs, and 1 ms measured with 24 bunches in continuous translation mode at 20 Hz with 3.6 μs per pulse and 197 pulses per image (65), collecting 250 images for each time delay. Set 2 for CH505 Env SOSIP was measured with 11 bunches in 20-Hz continuous translation mode with 1.6 μs per pulse and 197 pulses per image (65) and includes −10, −5, 1.5, 3, and 5 μs, with 362 images for each time delay. Two hundred images at 3.6 μs per pulse and 197 pulses per image were collected for CH505 Env set 3, which was measured in 5-Hz step translation mode with 24 bunches and included time delays of −10 μs, −5 μs, 1 ms, 10 ms, and 100 ms. Twenty-five images for the 500-ns time delay were collected separately using a single bunch at 20-Hz continuous translation mode (65) with the T-Jump starting from 42°C. CH848 data were collected in three sets. CH848 Env set 1 includes −10 μs, −5 μs, 5 μs, 10 μs, 50 μs, 100 μs, 500 μs, and 1 ms measured with 24 bunches in continuous translation mode at 20 Hz with 3.6 μs per pulse and 197 pulses per image (65), collecting 100 images for each time delay. Set 2 for CH848 Env SOSIP was measured with 24 bunches in 20-Hz continuous translation mode at 3.6 μs per pulse and 197 pulses per image (65) and includes −10 μs, −5 μs, 5 μs, 10 μs, 50 μs, 100 μs, 250 μs, 500 μs, 750 μs, and 1 ms, with 100 images for each time delay. One hundred images were collected for CH848 Env SOSIP set 3, which was measured in 5-Hz step translation mode with 24 bunches at 3.6 μs per pulse and 197 pulses per image (65) and included time delays of −10 μs, −5 μs, 1 ms, 10 ms, and 100 ms.
TR, T-Jump SAXS profiles of buffer were collected using the same protocol. For CH505 datasets, 100 images for each time delay in buffer set 1, including −10-, −5-, 1.5-, 3.0-, and 5.0-μs time delays, were collected with 11 bunches in 20-Hz continuous translation mode with 1.6 μs per pulse and 197 pulses per image (65). CH505 buffer set 2 was collected with 24 bunches in 20-Hz continuous translation mode with 3.6 μs per pulse and 197 pulses per image (65) and consisted of 50 images each at time delays of −10 μs, −5 μs, 5 μs, 10 μs, 25 μs, 50 μs, 100 μs, 250 μs, 500 μs, and 1 ms. CH505 Hepes set 3 was measured in 24 bunches in 5-Hz step mode with 3.6 μs per pulse and 197 pulses per image (65), collecting 50 images each at −10 μs, −5 μs, 1 ms, 10 ms, and 100 ms time delays. TR SAXS for CH848 buffer was collected in two sets. For CH848 buffer set 1, 25 images were collected for each time delay −10 μs, −5 μs, 5 μs, 10 μs, 50 μs, 100 μs, 250 μs, 500 μs, 750 μs, and 1 ms with 24 bunches in 20-Hz continuous translation mode (65). For CH848 buffer set 2, 25 images were collected for each time delay of −10 μs, −5 μs, 1 ms, 10 ms, and 100 ms with 24 bunches in 5-Hz step mode (65). Both CH848 buffer sets had 3.6 μs per pulse with 197 pulses per image x-ray exposure time.
Data reduction
Scattering intensity (I) was binned as a function of the scattering vector (q, Å−1) and radially averaged to produce isotropic scattering curves of the scattering vector, q, and intensity at q, I(q), with q calculated according to Eq. 1
| (1) |
where θ is one-half the scattering angle and λ is the x-ray wavelength. A mask was applied below 0.02 or 0.025 Å−1 q for CH505 and CH848, respectively, to eliminate the scattering signal near the beam stop for all collected images for static and TR, T-Jump SAXS datasets collected on the BioCARS 14-ID-B beamline. The scattering curves were normalized to the isosbestic point for water, 1.4 to 1.6 Å−1. This data reduction was performed with custom software at the BioCARS beamline.
Data processing
For static SAXS scattering curves, outliers were detected using two iterations of SVD to remove curves with first right vector values exceeding 2.5 SDs from the mean. After outlier removal, the remaining curves from the overlapping time delays in CH848 protein sets 1 and 2 were pooled. Once outliers were removed, average curves for each temperature were calculated for buffer, CH505, and CH848 Env SOSIP samples. The procedure was identical for buffer and Env SOSIPs. The remaining curves were used to determine the average static curves for buffer, CH505, and CH848 Env SOSIP. The average buffer curves were scaled to the average CH505 and CH848 curves in the scattering vector range q = 1.5 to 2.5 Å−1 and q = 0.5 to 1.0 Å−1, respectively, with the scaling factors (s) calculated according to Eq. 2
| (2) |
The scaled average buffer scattering curves were subtracted from the average CH505 and CH848 static scattering curves for each temperature to produce buffer-corrected average static SAXS scattering curves. The CH848 buffer curves were smoothed using the Savitzky-Golay filtering method (66) using a window length of 250 and third-order polynomial.
To identify outliers for TR, T-Jump SAXS curves, two iterations of SVD for both laser on and laser off scattering curves were used to detect scattering curves with first right vector values exceeding 2.5 SDs from the mean. A second iteration of SVD analysis was performed to ensure that any outliers detected in the first SVD iteration did not skew the calculation of the mean. This outlier analysis was carried out for −10- and −5-μs laser-off scattering profiles as well as the scattering profiles for each time delay after heating with the IR laser pulse. Difference curves were then calculated for the remaining scattering curves by subtracting the −10-μs laser-off curve for CH505 and −5-μs laser-off curve for CH848 from each time delay. An iterative χ2 analysis was used to detect difference curves with a χ2 > 1.5 for TR, T-Jump SAXS at each time delay. The same procedure was performed on buffer, CH505, and CH848 Env SOSIPs. Average difference curves for each time delay were calculated from the remaining difference curves. For each time delay difference profile, the corresponding average buffer difference curves were scaled using Eq. 2 to the average CH505 and CH848 Env difference curves and subtracted from the respective average CH505 difference profiles to yield average, buffer-corrected TR, T-Jump SAXS difference curves for CH505 and CH848 Env SOSIPs. These buffer-subtracted curves were used for the remaining TR, T-Jump and static SAXS data analysis.
Average buffer-subtracted CH505 and CH848 scattering profiles were determined using the laser-on scattering curves for CH505 and CH848 Env SOSIPs, respectively, and buffer T-Jump scattering curves. The same procedure was followed for determining these buffer-subtracted T-Jump scattering curves as for the static SAXS curve processing described above. Guinier analysis was performed in ATSAS primus software (67) to determine the particle radius of gyration (Rg) and to inspect for radiation damage and aggregation for both static (figs. S2, A to F, S3, C to F, and S4, C to F, and tables S1 and S2) and TR, T-Jump SAXS (table S3) scattering data. Scattering profile Kratky plots and pair-distance distributions [P(r)] were also assessed for each static (figs. S2, G and H, S3, B and G, and S4, B and G) and TR, T-Jump (figs. S5, C and D, and S6, B and C) scattering profiles in ATSAS.
TR, T-Jump component analysis
An SVD-based analysis of the averaged, buffer-subtracted CH505 and CH848 TR, T-Jump difference curves, and CH505 and CH848 static temperature series scattering curves was conducted to decompose the SAXS signals. The singular values were used to determine that there are two component signals (figs. S8A and S9A). The SVD left vectors can reveal individual feature curves contributing to the SAXS profiles, while the right vectors show the relative contributions of the corresponding left vectors at each time delay or temperature. Individual components from the buffer-subtracted CH505 and CH848 Env SOSIP temperature series static SAXS curves were also identically extracted by SVD.
TR, T-Jump kinetic analysis
Kinetic analysis was carried out using a bootstrapping method using 1000 resamples from the CH505 and CH848 TR, T-Jump difference curves. For each time delay in the CH505 and CH848 TR, T-Jump datasets, 80% of the difference curves remaining after outlier subtraction were selected and averaged. The previously determined average TR, T-Jump buffer curves were subtracted from the respective bootstrapped TR, T-Jump CH505 and CH848 curves to yield to the buffer-corrected, bootstrapped average curves for each time delay. For each bootstrap iteration, SVD and AUC analysis was performed to determine the average SVD left vectors, right vectors, and AUC. The first two right vectors from SVD analysis and the AUC were fit to a single exponential model in Eq. 3 for CH848 SOSIP Env. For CH505 SOSIP Env, the first right vector and AUC were fit to a double exponential in Eq. 4, while the second right vector was fit with a single exponential model (Eq. 3). The number of components to include in the models was determined from the SVD singular values.
| (3) |
| (4) |
Modeling
Atomic models of CH505 and CH848 Env SOSIPs in closed (Closed), closed with no apex contacts (Closed_NoApex), open-occluded (OpenOccluded), and open (Open) conformations were built using Modeller10.1 (68). The templates used for the models included Protein Data Bank (PDB) ID 6UM6 (71) for closed protomers, PDB ID 6CM3 (13) for the open protomers, and PDB ID 7TFN (15) for the open-occluded protomers. Sequence alignments between CH505 or CH848 Env SOSIP and templates were conducted using Clustal Omega in the DNASTAR MegAlign software. Disulfide patches were added to Modeller scripts to ensure the formation of disulfide bonds.
Glycosite (69) was used to predict the possible N-linked glycosylation sites in the CH505 and CH848 Env SOSIPs, and Glycosylator (80) was used to glycosylate each model with Man9 glycans. Three rounds of refinement were performed to remove glycan clashes. All rounds of refinement used 10 iterations, a 0.01 mutation rate, and a population size of 30. The first two rounds of refinement used 10 generations, while the last refinement used 20 generations. The number of individues (80) was decreased by two between each refinement.
The theoretical SAXS profiles for each glycosylated model were calculated using the FoXS server (81). P(r) distributions for these theoretical SAXS scattering profiles were determined in ATSAS primus software (67), and ΔP(r) curves were calculated by subtracting the closed P(r) curve from the Open or OpenOccluded P(r) curve and compared to the experimental and REGALS SAXS profiles and ΔP(r) curves. SVD analysis was performed on the theoretical scattering curves for the Closed, OpenOccluded, and Open models both with Man9 glycans and without glycosylation to extract component signals for transitions between these structures. Guinier analyses and pair distance distribution were performed in ATSAS Primus software (67) on each theoretical SAXS profile to determine Rg (table S5).
Molecular dynamics
The CHARMM-GUI Glycan Modeler (82–87) was used to glycosylate glycosite-predicted (70) glycosylation sites with Man5 glycans and prepared gp120 systems from the CH505 model based on PDB ID 6UDA for simulation. A total of 250, 5-μs trajectories were run for closed gp120 at 50°C. The Amber22 software package with pmemd CUDA implementation (88–90) in the CHARMM36 force field (91–94) and TIP3P water model (95) was used for simulations. A 150-Å octahedral box was used to allow for V1/V2 and V3 dislocation during simulation. Both systems were neutralized with 150 mM NaCl. Ten thousand steps of energy minimization were performed with 1000 steps of steepest descent minimization with restraints (500 kcal/mol·Å) on protein. A second unrestrained energy minimization consisting of 10,000 steps and 1000 steepest descent steps followed. Minimization was followed by 20 ps of heating at constant volume with restraints (10 kcal/mol·Å) applied to protein atoms and a subsequent 5 ns equilibration with no restraints, with a constant temperature of 323.15 K maintained using a Langevin thermostat (96), and pressure of 1 atm was maintained with isotropic position scaling. Two hundred fifty nanoseconds of unrestrained simulations in the NVT ensemble was performed to equilibrate the system, followed by 5 μs of production simulation in the NVT ensemble. Electrostatic interactions were calculated with the Particle Mesh Ewald method (97) with a cutoff of 12 Å and switching distance of 10 Å. The SHAKE algorithm (98) with hydrogen mass repartitioning (99) was used to constrain hydrogen atoms and allow for a 4-fs time step. The cpptraj tool in AmberTools21 (100) was used to determine the RMSD time series of V1/V2 (residues 129 to 133, 135 to 150, 154 to 163, 168 to 177, 181 to 183, and 190 to 193 in HXB2 isolate numbering), V3 (residues 297 to 308 and 311 to 319 in HXB2 isolate numbering), and gp120 core (residues 288 to 291, 280 to 284, 355 to 358, 370 to 374, 377 to 382, 411 to 416, 439 to 452, and 462 to 466 in HXB2 isolate numbering) β sheet α carbons for each replicate of MD simulation. A custom Python script was used to determine the average RMSD and frequency distributions for all replicates aggregated together.
Rational structure-based design
The CH235.UCA-bound CH505.M5.G458Y SOSIP trimer structure (6UDA) was prepared in Maestro (101) using the protein preparation wizard (102) followed by in silico mutagenesis using Schrödinger’s cysteine mutation (103) and residue scanning (104) tools. Residue scanning was first performed for V1/V2 and V3 region interprotomer contact residues. Scores and visual inspection were used in the selection of the prepared construct. The selected design was prepared in a previously reported CH505 isolate SOSIP backbone (76) with the introduction of an expression-enhancing 2P mutation (60), an allosteric machinery disabling set of mutations, F14 (48), and a CD4bs antibody binding enhancing N197D mutation (60).
Cryo-EM sample preparation, data collection, and processing
Purified envelope SOSIP ectodomain preparations were prepared at concentrations of 4 to 5 mg/ml in 15 mM Hepes buffer with 150 mM NaCl at pH 7.1 and mixed with Fab at a 1:5 molar ratio. A total of 2.5 μl of the complex was deposited on a CF-1.2/1.3 grid that had been glow-discharged for 15 s in a PELCO easiGlow glow-discharge cleaning system. After 30-s incubation in >95% humidity, the excess protein was blotted away for 2.5 s before being plunge-frozen in liquid ethane using a Leica EM GP2 plunge freezer (Leica Microsystems). Frozen grids were imaged in a Talos Arctica (Thermo Fisher Scientific) equipped with a K3 detector (Gatan). Individual frames were aligned, dose-weighted, and contrast transfer function (CTF) corrected followed by particle picking, 2D classification, ab initio model generation, heterogeneous refinements, homogeneous 3D refinements, and local resolution calculations in cryoSPARC (Table 1) (105).
Table 1. Cryo-EM data collection and refinement statistics.
| CH505 design + CH235.12 Fab | CH505 design + b12 Fab | |
|---|---|---|
| Data collection | ||
| Microscope | Talos Arctica G3 | Talos Arctica G3 |
| Voltage (kV) | 200 | 200 |
| Electron dose (e-/Å2) | 57 | 58 |
| Detector | Gatan K3 | Gatan K3 |
| Pixel size (Å) | 0.874 | 0.874 |
| Defocus range (μm) | 0.1–2.88 | 0.1–2.48 |
| Magnification | 45,000 | 45,000 |
| Reconstruction | ||
| Software | cryoSPARC | cryoSPARC |
| Particles | 116,771 | 151,789 |
| Symmetry | C3 | C3 |
| Box size (pixel) | 300 | 300 |
| Resolution (Å) (FSC 0.143)* | 4.0 | 4.5 |
| Validation | ||
| Protein residues | 2,415 | 2,439 |
| EMRinger score | 2.5 | 1.24 |
| RMSDs | ||
| Bond lengths (Å) | 0.005 | 0.006 |
| Bond angles (°) | 0.924 | 0.998 |
| Molprobity score | 1.49 | 1.72 |
| Clash score | 2.98 | 5.43 |
| Rotamer outliers (%) | 0.05 | 0.05 |
| Ramachandran | ||
| Favored regions (%) | 94.06 | 93.38 |
| Disallowed regions (%) | 0.34 | 0.38 |
*Resolutions are reported according to the FSC 0.143 gold-standard criterion.
†Statistics are reported for the protein residues within the complex excluding the antibody constant domains.
Cryo-EM structure fitting and analysis
The CH505.M5.G458Y SOSIP structure (6UDA) and either CH235.12 (5F96) or b12 (2NY7) structures were used to fit the cryo-EM maps in ChimeraX (106). Mutations were made in PyMol (107). Coordinates were then fitted using Isolde (108) followed by iterative refinement using Phenix (109) real-space refinement. Structure and map analyses were performed using PyMol, Chimera (110), and ChimeraX (106).
Acknowledgments
We thank L. X. Chen of Northwestern University for providing the heating cell for TR, T-Jump SAXS experiments.
Funding: This project was supported by NIH, National Institute of Allergy and Infectious Diseases, Division of AIDS Consortia for HIV/AIDS Vaccine Development (CHAVD) grants UM1AI144371 (to B.F.H.), R01AI145687 (to P.A.), and U54AI170752 (to R.H. and P.A.) and Translating Duke Health Initiative (to R.H. and P.A.). A portion of this work was conducted at the Advanced Light Source (ALS), a national user facility operated by Lawrence Berkeley National Laboratory on behalf of the US Department of Energy (DOE), Office of Basic Energy Sciences, through the Integrated Diffraction Analysis Technologies (IDAT) program, supported by DOE Office of Biological and Environmental Research. Additional support comes from the NIH project ALS-ENABLE (P30 GM124169) and a High-End Instrumentation Grant S10OD018483. This research also used resources of the Advanced Photon Source, a DOE Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under contract no. DE-AC02-06CH11357. The use of BioCARS was also supported by the National Institute of General Medical Sciences (https://nigms.nih.gov/about/overview/pages/BBCB.aspx) of the NIH (http://nih.gov/) under grant number P41 GM118217. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. TR setup at Sector 14 was funded in part through a collaboration with P. Anfinrud (NIH/NIDDK). A part of this work was conducted at the ALS, a national user facility operated by Lawrence Berkeley National Laboratory on behalf of the DOE, Office of Basic Energy Sciences, through the IDAT program, supported by DOE Office of Biological and Environmental Research. Additional support comes from the NIH project ALS-ENABLE (P30 GM124169) and a High-End Instrumentation Grant S10OD018483.
Author contributions: A.L.B. and R.H. designed, conducted, and interpreted the TR, T-Jump SAXS experiments. A.L.B. and R.H. developed the TR, T-Jump SAXS analysis code and analyzed TR, T-Jump SAXS data. C.S., A.W., Y.B., P.B., and K.O.S. prepared all proteins for this study. C.S., K.A., and S.M.A. collected and analyzed BLI data. R.J.E. and K.M. collected and analyzed NSEM data. R.H. collected the cryo-EM data. R.H. and P.A. analyzed the cryo-EM data. R.H. designed the apex-stapled immunogen. B.F.H. reviewed data and immunogen design plans. All authors read and edited the manuscript.
Competing interests: A patent application covering HIV-1 Env modifications based on this study has been submitted by Duke University. The authors declare no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Cryo-EM reconstructions and atomic models have been deposited in the EMDB and wwPDB with accession codes 8SXI and 8SXJ and EMD-40854 and EMD-40853. The SAXS data, including processed SAXS curves, have been deposited to the SASBDB with accession codes SASDTZ9, SASDU22, SASDU32, SASDU42, SASDTX9, SASDTY9 for the CH505 transmitted founder, CH505 variants isolated at weeks 53, 78, and 100 post-infection, a 4.1 stabilized CH505 transmitted founder, and the BG505 Env SOSIP panel, SASDT29, SASDT39, SASDT49, SASDT59 for the CH505 temperature series static data, SASDT69, SASDT79, SASDT89, SASDT99, SASDTA9, SASDTB9, SASDTC9, SASDTD9, SASDTE9, SASDTF9, SASDTG9 for CH505 T-Jump data, SASDTH9, SASDTJ9, SASDTK9, SASDTL9 for the CH848 temperature series static data, and SASDTM9, SASDTN9, SASDTP9, SASDTQ9, SASDTR9, SASDTS9, SASDTT9, SASDTU9, SASDTV9, SASDTW9 for CH848 T-Jump data. The code used to process and analyze the SAXS data is available on GitLab: https://gitlab.oit.duke.edu/tr_t-jump_saxs/y22-23, doi:10.5281/zenodo.10424314. Filtered molecular dynamics trajectories are available on Zenodo (10.5281/zenodo.10451687).
Supplementary Materials
This PDF file includes:
Figs. S1 to S17
Tables S1 to S5
REFERENCES AND NOTES
- 1.Tran E. E., Borgnia M. J., Kuybeda O., Schauder D. M., Bartesaghi A., Frank G. A., Sapiro G., Milne J. L., Subramaniam S., Structural mechanism of trimeric HIV-1 envelope glycoprotein activation. PLOS Pathog. 8, e1002797 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ward A. B., Wilson I. A., The HIV-1 envelope glycoprotein structure: Nailing down a moving target. Immunol. Rev. 275, 21–32 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shaik M. M., Peng H., Lu J., Rits-Volloch S., Xu C., Liao M., Chen B., Structural basis of coreceptor recognition by HIV-1 envelope spike. Nature 565, 318–323 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwong P. D., Wyatt R., Robinson J., Sweet R. W., Sodroski J., Hendrickson W. A., Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393, 648–659 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harrison S. C., Viral membrane fusion. Nat. Struct. Mol. Biol. 15, 690–698 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weissenhorn W., Dessen A., Harrison S. C., Skehel J. J., Wiley D. C., Atomic structure of the ectodomain from HIV-1 gp41. Nature 387, 426–430 (1997). [DOI] [PubMed] [Google Scholar]
- 7.Torrents de la Pena A., Julien J. P., de Taeye S. W., Garces F., Guttman M., Ozorowski G., Pritchard L. K., Behrens A. J., Go E. P., Burger J. A., Schermer E. E., Sliepen K., Ketas T. J., Pugach P., Yasmeen A., Cottrell C. A., Torres J. L., Vavourakis C. D., van Gils M. J., LaBranche C., Montefiori D. C., Desaire H., Crispin M., Klasse P. J., Lee K. K., Moore J. P., Ward A. B., Wilson I. A., Sanders R. W., Improving the Immunogenicity of Native-like HIV-1 envelope trimers by hyperstabilization. Cell Rep. 20, 1805–1817 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ward A. B., Wilson I. A., Insights into the trimeric HIV-1 envelope glycoprotein structure. Trends Biochem. Sci. 40, 101–107 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bennett A. L., Henderson R., HIV-1 envelope conformation, allostery, and dynamics. Viruses 13, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zolla-Pazner S., Cohen S. S., Boyd D., Kong X. P., Seaman M., Nussenzweig M., Klein F., Overbaugh J., Totrov M., Structure/function studies involving the V3 region of the HIV-1 envelope delineate multiple factors that affect neutralization sensitivity. J. Virol. 90, 636–649 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Powell R. L. R., Totrov M., Itri V., Liu X., Fox A., Zolla-Pazner S., Plasticity and epitope exposure of the HIV-1 envelope trimer. J. Virol. 91, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang H., Cohen A. A., Galimidi R. P., Gristick H. B., Jensen G. J., Bjorkman P. J., Cryo-EM structure of a CD4-bound open HIV-1 envelope trimer reveals structural rearrangements of the gp120 V1V2 loop. Proc. Natl. Acad. Sci. U.S.A. 113, E7151–E7158 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang H., Barnes C. O., Yang Z., Nussenzweig M. C., Bjorkman P. J., Partially open HIV-1 envelope structures exhibit conformational changes relevant for co-receptor binding and fusion. Cell Host Microbe 24, 579–592.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ozorowski G., Pallesen J., de Val N., Lyumkis D., Cottrell C. A., Torres J. L., Copps J., Stanfield R. L., Cupo A., Pugach P., Moore J. P., Wilson I. A., Ward A. B., Open and closed structures reveal allostery and pliability in the HIV-1 envelope spike. Nature 547, 360–363 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Z., Dam K. A., Bridges M. D., Hoffmann M. A. G., DeLaitsch A. T., Gristick H. B., Escolano A., Gautam R., Martin M. A., Nussenzweig M. C., Hubbell W. L., Bjorkman P. J., Neutralizing antibodies induced in immunized macaques recognize the CD4-binding site on an occluded-open HIV-1 envelope trimer. Nat. Commun. 13, 732 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.K. A. Dam, C. Fan, Z. Yang, P. J. Bjorkman, Structural characterization of HIV-1 Env heterotrimers bound to one or two CD4 receptors reveals intermediate Env conformations. bioRxiv 2023.01.27.525985 [Preprint] (2023). 10.1101/2023.01.27.525985. [DOI]
- 17.Munro J. B., Gorman J., Ma X., Zhou Z., Arthos J., Burton D. R., Koff W. C., Courter J. R., Smith A. B. III, Kwong P. D., Blanchard S. C., Mothes W., Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science 346, 759–763 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Munro J. B., Mothes W., Structure and dynamics of the native HIV-1 Env trimer. J. Virol. 89, 5752–5755 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu M., Ma X., Castillo-Menendez L. R., Gorman J., Alsahafi N., Ermel U., Terry D. S., Chambers M., Peng D., Zhang B., Zhou T., Reichard N., Wang K., Grover J. R., Carman B. P., Gardner M. R., Nikic-Spiegel I., Sugawara A., Arthos J., Lemke E. A., Smith A. B. III, Farzan M., Abrams C., Munro J. B., McDermott A. B., Finzi A., Kwong P. D., Blanchard S. C., Sodroski J. G., Mothes W., Associating HIV-1 envelope glycoprotein structures with states on the virus observed by smFRET. Nature 568, 415–419 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma X., Lu M., Gorman J., Terry D. S., Hong X., Zhou Z., Zhao H., Altman R. B., Arthos J., Blanchard S. C., Kwong P. D., Munro J. B., Mothes W., HIV-1 Env trimer opens through an asymmetric intermediate in which individual protomers adopt distinct conformations. eLife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stadtmueller B. M., Bridges M. D., Dam K. M., Lerch M. T., Huey-Tubman K. E., Hubbell W. L., Bjorkman P. J., DEER spectroscopy measurements reveal multiple conformations of HIV-1 SOSIP envelopes that show similarities with envelopes on native virions. Immunity 49, 235–246.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davenport T. M., Guttman M., Guo W., Cleveland B., Kahn M., Hu S. L., Lee K. K., Isolate-specific differences in the conformational dynamics and antigenicity of HIV-1 gp120. J. Virol. 87, 10855–10873 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kong L., Huang C. C., Coales S. J., Molnar K. S., Skinner J., Hamuro Y., Kwong P. D., Local conformational stability of HIV-1 gp120 in unliganded and CD4-bound states as defined by amide hydrogen/deuterium exchange. J. Virol. 84, 10311–10321 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guttman M., Kahn M., Garcia N. K., Hu S. L., Lee K. K., Solution structure, conformational dynamics, and CD4-induced activation in full-length, glycosylated, monomeric HIV gp120. J. Virol. 86, 8750–8764 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hodge E. A., Naika G. S., Kephart S. M., Nguyen A., Zhu R., Benhaim M. A., Guo W., Moore J. P., Hu S. L., Sanders R. W., Lee K. K., Structural dynamics reveal isolate-specific differences at neutralization epitopes on HIV Env. iScience 25, 104449 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Taeye S. W., Ozorowski G., Torrents de la Pena A., Guttman M., Julien J. P., van den Kerkhof T. L., Burger J. A., Pritchard L. K., Pugach P., Yasmeen A., Crampton J., Hu J., Bontjer I., Torres J. L., Arendt H., DeStefano J., Koff W. C., Schuitemaker H., Eggink D., Berkhout B., Dean H., LaBranche C., Crotty S., Crispin M., Montefiori D. C., Klasse P. J., Lee K. K., Moore J. P., Wilson I. A., Ward A. B., Sanders R. W., Immunogenicity of stabilized HIV-1 envelope trimers with reduced exposure of non-neutralizing epitopes. Cell 163, 1702–1715 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanders R. W., Derking R., Cupo A., Julien J. P., Yasmeen A., de Val N., Kim H. J., Blattner C., de la Pena A. T., Korzun J., Golabek M., de Los Reyes K., Ketas T. J., van Gils M. J., King C. R., Wilson I. A., Ward A. B., Klasse P. J., Moore J. P., A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLOS Pathog. 9, e1003618 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guenaga J., Garces F., de Val N., Stanfield R. L., Dubrovskaya V., Higgins B., Carrette B., Ward A. B., Wilson I. A., Wyatt R. T., Glycine substitution at helix-to-coil transitions facilitates the structural determination of a stabilized subtype C HIV envelope glycoprotein. Immunity 46, 792–803.e3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He L., Kumar S., Allen J. D., Huang D., Lin X., Mann C. J., Saye-Francisco K. L., Copps J., Sarkar A., Blizard G. S., Ozorowski G., Sok D., Crispin M., Ward A. B., Nemazee D., Burton D. R., Wilson I. A., Zhu J., HIV-1 vaccine design through minimizing envelope metastability. Sci. Adv. 4, eaau6769 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang P., Gorman J., Geng H., Liu Q., Lin Y., Tsybovsky Y., Go E. P., Dey B., Andine T., Kwon A., Patel M., Gururani D., Uddin F., Guzzo C., Cimbro R., Miao H., McKee K., Chuang G. Y., Martin L., Sironi F., Malnati M. S., Desaire H., Berger E. A., Mascola J. R., Dolan M. A., Kwong P. D., Lusso P., Interdomain stabilization impairs CD4 binding and improves immunogenicity of the HIV-1 envelope trimer. Cell Host Microbe 23, 832–844.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chuang G. Y., Geng H., Pancera M., Xu K., Cheng C., Acharya P., Chambers M., Druz A., Tsybovsky Y., Wanninger T. G., Yang Y., Doria-Rose N. A., Georgiev I. S., Gorman J., Joyce M. G., O'Dell S., Zhou T., McDermott A. B., Mascola J. R., Kwong P. D., Structure-based design of a soluble prefusion-closed HIV-1 Env trimer with reduced CD4 affinity and improved immunogenicity. J. Virol. 91, e02268 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Medina-Ramirez M., Garces F., Escolano A., Skog P., de Taeye S. W., Del Moral-Sanchez I., McGuire A. T., Yasmeen A., Behrens A. J., Ozorowski G., van den Kerkhof T., Freund N. T., Dosenovic P., Hua Y., Gitlin A. D., Cupo A., van der Woude P., Golabek M., Sliepen K., Blane T., Kootstra N., van Breemen M. J., Pritchard L. K., Stanfield R. L., Crispin M., Ward A. B., Stamatatos L., Klasse P. J., Moore J. P., Nemazee D., Nussenzweig M. C., Wilson I. A., Sanders R. W., Design and crystal structure of a native-like HIV-1 envelope trimer that engages multiple broadly neutralizing antibody precursors in vivo. J. Exp. Med. 214, 2573–2590 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steichen J. M., Kulp D. W., Tokatlian T., Escolano A., Dosenovic P., Stanfield R. L., McCoy L. E., Ozorowski G., Hu X., Kalyuzhniy O., Briney B., Schiffner T., Garces F., Freund N. T., Gitlin A. D., Menis S., Georgeson E., Kubitz M., Adachi Y., Jones M., Mutafyan A. A., Yun D. S., Mayer C. T., Ward A. B., Burton D. R., Wilson I. A., Irvine D. J., Nussenzweig M. C., Schief W. R., HIV vaccine design to target germline precursors of glycan-dependent broadly neutralizing antibodies. Immunity 45, 483–496 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kulp D. W., Steichen J. M., Pauthner M., Hu X., Schiffner T., Liguori A., Cottrell C. A., Havenar-Daughton C., Ozorowski G., Georgeson E., Kalyuzhniy O., Willis J. R., Kubitz M., Adachi Y., Reiss S. M., Shin M., de Val N., Ward A. B., Crotty S., Burton D. R., Schief W. R., Structure-based design of native-like HIV-1 envelope trimers to silence non-neutralizing epitopes and eliminate CD4 binding. Nat. Commun. 8, 1655 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang L., Sharma S. K., Cottrell C., Guenaga J., Tran K., Wilson R., Behrens A. J., Crispin M., de Val N., Wyatt R. T., Structure-guided redesign improves NFL HIV Env trimer integrity and identifies an inter-protomer disulfide permitting post-expression cleavage. Front. Immunol. 9, 1631 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharma S. K., de Val N., Bale S., Guenaga J., Tran K., Feng Y., Dubrovskaya V., Ward A. B., Wyatt R. T., Cleavage-independent HIV-1 Env trimers engineered as soluble native spike mimetics for vaccine design. Cell Rep. 11, 539–550 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guenaga J., Dubrovskaya V., de Val N., Sharma S. K., Carrette B., Ward A. B., Wyatt R. T., Structure-guided redesign increases the propensity of HIV Env to generate highly stable soluble trimers. J. Virol. 90, 2806–2817 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sliepen K., Han B. W., Bontjer I., Mooij P., Garces F., Behrens A. J., Rantalainen K., Kumar S., Sarkar A., Brouwer P. J. M., Hua Y., Tolazzi M., Schermer E., Torres J. L., Ozorowski G., van der Woude P., de la Pena A. T., van Breemen M. J., Camacho-Sanchez J. M., Burger J. A., Medina-Ramirez M., Gonzalez N., Alcami J., LaBranche C., Scarlatti G., van Gils M. J., Crispin M., Montefiori D. C., Ward A. B., Koopman G., Moore J. P., Shattock R. J., Bogers W. M., Wilson I. A., Sanders R. W., Structure and immunogenicity of a stabilized HIV-1 envelope trimer based on a group-M consensus sequence. Nat. Commun. 10, 2355 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Henderson R., Lu M., Zhou Y., Mu Z., Parks R., Han Q., Hsu A. L., Carter E., Blanchard S. C., Edwards R. J., Wiehe K., Saunders K. O., Borgnia M. J., Bartesaghi A., Mothes W., Haynes B. F., Acharya P., Munir Alam S., Disruption of the HIV-1 envelope allosteric network blocks CD4-induced rearrangements. Nat. Commun. 11, 520 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bianchi M., Turner H. L., Nogal B., Cottrell C. A., Oyen D., Pauthner M., Bastidas R., Nedellec R., McCoy L. E., Wilson I. A., Burton D. R., Ward A. B., Hangartner L., Electron-microscopy-based epitope mapping defines specificities of polyclonal antibodies elicited during HIV-1 BG505 envelope trimer immunization. Immunity 49, 288–300.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liao H. X., Lynch R., Zhou T., Gao F., Alam S. M., Boyd S. D., Fire A. Z., Roskin K. M., Schramm C. A., Zhang Z., Zhu J., Shapiro L., Program N. C. S., Mullikin J. C., Gnanakaran S., Hraber P., Wiehe K., Kelsoe G., Yang G., Xia S. M., Montefiori D. C., Parks R., Lloyd K. E., Scearce R. M., Soderberg K. A., Cohen M., Kamanga G., Louder M. K., Tran L. M., Chen Y., Cai F., Chen S., Moquin S., Du X., Joyce M. G., Srivatsan S., Zhang B., Zheng A., Shaw G. M., Hahn B. H., Kepler T. B., Korber B. T., Kwong P. D., Mascola J. R., Haynes B. F., Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature 496, 469–476 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bonsignori M., Kreider E. F., Fera D., Meyerhoff R. R., Bradley T., Wiehe K., Alam S. M., Aussedat B., Walkowicz W. E., Hwang K. K., Saunders K. O., Zhang R., Gladden M. A., Monroe A., Kumar A., Xia S. M., Cooper M., Louder M. K., McKee K., Bailer R. T., Pier B. W., Jette C. A., Kelsoe G., Williams W. B., Morris L., Kappes J., Wagh K., Kamanga G., Cohen M. S., Hraber P. T., Montefiori D. C., Trama A., Liao H. X., Kepler T. B., Moody M. A., Gao F., Danishefsky S. J., Mascola J. R., Shaw G. M., Hahn B. H., Harrison S. C., Korber B. T., Haynes B. F., Staged induction of HIV-1 glycan-dependent broadly neutralizing antibodies. Sci. Transl. Med. 9, eaai7514 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu X., Parast A. B., Richardson B. A., Nduati R., John-Stewart G., Mbori-Ngacha D., Rainwater S. M., Overbaugh J., Neutralization escape variants of human immunodeficiency virus type 1 are transmitted from mother to infant. J. Virol. 80, 835–844 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee J. H., Andrabi R., Su C. Y., Yasmeen A., Julien J. P., Kong L., Wu N. C., McBride R., Sok D., Pauthner M., Cottrell C. A., Nieusma T., Blattner C., Paulson J. C., Klasse P. J., Wilson I. A., Burton D. R., Ward A. B., A broadly neutralizing antibody targets the dynamic HIV envelope trimer apex via a long, rigidified, and anionic β-hairpin structure. Immunity 46, 690–702 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Merk A., Subramaniam S., HIV-1 envelope glycoprotein structure. Curr. Opin. Struct. Biol. 23, 268–276 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moore J. P., Trkola A., Korber B., Boots L. J., Kessler J. A. II, McCutchan F. E., Mascola J., Ho D. D., Robinson J., Conley A. J., A human monoclonal antibody to a complex epitope in the V3 region of gp120 of human immunodeficiency virus type 1 has broad reactivity within and outside clade B. J. Virol. 69, 122–130 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lynch R. M., Tran L., Louder M. K., Schmidt S. D., Cohen M., Members C. C. T., Dersimonian R., Euler Z., Gray E. S., Abdool Karim S., Kirchherr J., Montefiori D. C., Sibeko S., Soderberg K., Tomaras G., Yang Z. Y., Nabel G. J., Schuitemaker H., Morris L., Haynes B. F., Mascola J. R., The development of CD4 binding site antibodies during HIV-1 infection. J. Virol. 86, 7588–7595 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saunders K. O., Verkoczy L. K., Jiang C., Zhang J., Parks R., Chen H., Housman M., Bouton-Verville H., Shen X., Trama A. M., Scearce R., Sutherland L., Santra S., Newman A., Eaton A., Xu K., Georgiev I. S., Joyce M. G., Tomaras G. D., Bonsignori M., Reed S. G., Salazar A., Mascola J. R., Moody M. A., Cain D. W., Centlivre M., Zurawski S., Zurawski G., Erickson H. P., Kwong P. D., Alam S. M., Levy Y., Montefiori D. C., Haynes B. F., Vaccine induction of heterologous tier 2 HIV-1 neutralizing antibodies in animal models. Cell Rep. 21, 3681–3690 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmidt M., Rajagopal S., Ren Z., Moffat K., Application of singular value decomposition to the analysis of time-resolved macromolecular x-ray data. Biophys. J. 84, 2112–2129 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thompson M. C., Barad B. A., Wolff A. M., Sun Cho H., Schotte F., Schwarz D. M. C., Anfinrud P., Fraser J. S., Temperature-jump solution x-ray scattering reveals distinct motions in a dynamic enzyme. Nat. Chem. 11, 1058–1066 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meisburger S. P., Xu D., Ando N., REGALS: A general method to deconvolve x-ray scattering data from evolving mixtures. IUCrJ 8, 225–237 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang M., Huang J., Simon R., Wang L. X., MacKerell A. D. Jr., Conformational heterogeneity of the HIV envelope glycan shield. Sci. Rep. 7, 4435 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Berndsen Z. T., Chakraborty S., Wang X., Cottrell C. A., Torres J. L., Diedrich J. K., Lopez C. A., Yates J. R. III, van Gils M. J., Paulson J. C., Gnanakaran S., Ward A. B., Visualization of the HIV-1 Env glycan shield across scales. Proc. Natl. Acad. Sci. U.S.A. 117, 28014–28025 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chakraborty S., Wagh K., Gnanakaran S., Lopez C. A., Development of Martini 2.2 parameters for N-glycans: A case study of the HIV-1 Env glycoprotein dynamics. Glycobiology 31, 787–799 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kovacs J. M., Nkolola J. P., Peng H., Cheung A., Perry J., Miller C. A., Seaman M. S., Barouch D. H., Chen B., HIV-1 envelope trimer elicits more potent neutralizing antibody responses than monomeric gp120. Proc. Natl. Acad. Sci. U.S.A. 109, 12111–12116 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kwon Y. D., Pancera M., Acharya P., Georgiev I. S., Crooks E. T., Gorman J., Joyce M. G., Guttman M., Ma X., Narpala S., Soto C., Terry D. S., Yang Y., Zhou T., Ahlsen G., Bailer R. T., Chambers M., Chuang G. Y., Doria-Rose N. A., Druz A., Hallen M. A., Harned A., Kirys T., Louder M. K., O'Dell S., Ofek G., Osawa K., Prabhakaran M., Sastry M., Stewart-Jones G. B., Stuckey J., Thomas P. V., Tittley T., Williams C., Zhang B., Zhao H., Zhou Z., Donald B. R., Lee L. K., Zolla-Pazner S., Baxa U., Schon A., Freire E., Shapiro L., Lee K. K., Arthos J., Munro J. B., Blanchard S. C., Mothes W., Binley J. M., McDermott A. B., Mascola J. R., Kwong P. D., Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat. Struct. Mol. Biol. 22, 522–531 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu Q., Acharya P., Dolan M. A., Zhang P., Guzzo C., Lu J., Kwon A., Gururani D., Miao H., Bylund T., Chuang G. Y., Druz A., Zhou T., Rice W. J., Wigge C., Carragher B., Potter C. S., Kwong P. D., Lusso P., Quaternary contact in the initial interaction of CD4 with the HIV-1 envelope trimer. Nat. Struct. Mol. Biol. 24, 370–378 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.LaBranche C. C., Henderson R., Hsu A., Behrens S., Chen X., Zhou T., Wiehe K., Saunders K. O., Alam S. M., Bonsignori M., Borgnia M. J., Sattentau Q. J., Eaton A., Greene K., Gao H., Liao H. X., Williams W. B., Peacock J., Tang H., Perez L. G., Edwards R. J., Kepler T. B., Korber B. T., Kwong P. D., Mascola J. R., Acharya P., Haynes B. F., Montefiori D. C., Neutralization-guided design of HIV-1 envelope trimers with high affinity for the unmutated common ancestor of CH235 lineage CD4bs broadly neutralizing antibodies. PLOS Pathog. 15, e1008026 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wrapp D., Mu Z., Thakur B., Janowska K., Ajayi O., Barr M., Parks R., Mansouri K., Edwards R. J., Hahn B. H., Acharya P., Saunders K. O., Haynes B. F., Structure-based stabilization of SOSIP Env enhances recombinant ectodomain durability and yield. J. Virol. 97, e0167322 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Crooks E. T., Tong T., Chakrabarti B., Narayan K., Georgiev I. S., Menis S., Huang X., Kulp D., Osawa K., Muranaka J., Stewart-Jones G., Destefano J., O'Dell S., LaBranche C., Robinson J. E., Montefiori D. C., McKee K., Du S. X., Doria-Rose N., Kwong P. D., Mascola J. R., Zhu P., Schief W. R., Wyatt R. T., Whalen R. G., Binley J. M., Vaccine-elicited tier 2 HIV-1 neutralizing antibodies bind to quaternary epitopes involving glycan-deficient patches proximal to the CD4 binding site. PLOS Pathog. 11, e1004932 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hioe C. E., Wrin T., Seaman M. S., Yu X., Wood B., Self S., Williams C., Gorny M. K., Zolla-Pazner S., Anti-V3 monoclonal antibodies display broad neutralizing activities against multiple HIV-1 subtypes. PLOS ONE 5, e10254 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rimmerman D., Leshchev D., Hsu D. J., Hong J., Kosheleva I., Chen L. X., Direct observation of insulin association dynamics with time-resolved x-ray scattering. J. Phys. Chem. Lett. 8, 4413–4418 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hura G. L., Menon A. L., Hammel M., Rambo R. P., Poole F. L. II, Tsutakawa S. E., Jenney F. E. Jr., Classen S., Frankel K. A., Hopkins R. C., Yang S. J., Scott J. W., Dillard B. D., Adams M. W., Tainer J. A., Robust, high-throughput solution structural analyses by small angle x-ray scattering (SAXS). Nat. Methods 6, 606–612 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Putnam C. D., Hammel M., Hura G. L., Tainer J. A., X-ray solution scattering (SAXS) combined with crystallography and computation: Defining accurate macromolecular structures, conformations and assemblies in solution. Q. Rev. Biophys. 40, 191–285 (2007). [DOI] [PubMed] [Google Scholar]
- 65.Kosheleva I., Henning R., Kim I., Kim S. O., Kusel M., Srajer V., Sample-minimizing co-flow cell for time-resolved pump-probe x-ray solution scattering. J. Synchrotron Radiat. 30, 490–499 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Savitzky A., Golay M. J. E., Smoothing and differentiation of data by simplified least squares procedures. Anal. Chem. 36, 1627–1639 (1964). [Google Scholar]
- 67.Manalastas-Cantos K., Konarev P. V., Hajizadeh N. R., Kikhney A. G., Petoukhov M. V., Molodenskiy D. S., Panjkovich A., Mertens H. D. T., Gruzinov A., Borges C., Jeffries C. M., Svergun D. I., Franke D., ATSAS 3.0: Expanded functionality and new tools for small-angle scattering data analysis. J. Appl. Cryst. 54, 343–355 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sali A., Blundell T. L., Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 (1993). [DOI] [PubMed] [Google Scholar]
- 69.Zhang M., Gaschen B., Blay W., Foley B., Haigwood N., Kuiken C., Korber B., Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology 14, 1229–1246 (2004). [DOI] [PubMed] [Google Scholar]
- 70.Guttman M., Weinkam P., Sali A., Lee K. K., All-atom ensemble modeling to analyze small-angle x-ray scattering of glycosylated proteins. Structure 21, 321–331 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Saunders K. O., Wiehe K., Tian M., Acharya P., Bradley T., Alam S. M., Go E. P., Scearce R., Sutherland L., Henderson R., Hsu A. L., Borgnia M. J., Chen H., Lu X., Wu N. R., Watts B., Jiang C., Easterhoff D., Cheng H. L., McGovern K., Waddicor P., Chapdelaine-Williams A., Eaton A., Zhang J., Rountree W., Verkoczy L., Tomai M., Lewis M. G., Desaire H. R., Edwards R. J., Cain D. W., Bonsignori M., Montefiori D., Alt F. W., Haynes B. F., Targeted selection of HIV-specific antibody mutations by engineering B cell maturation. Science 366, eaay7199 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.R. Henderson, Y. Zhou, V. Stalls, K. Wiehe, K. O. Saunders, K. Wagh, K. Anasti, M. Barr, R. Parks, S. M. Alam, B. Korber, B. F. Haynes, A. Bartesaghi, P. Acharya, Structural basis for breadth development in a HIV-1 neutralizing antibody. bioRxiv 2022.09.14.507935 [Preprint] (2022). 10.1101/2022.09.14.507935. [DOI]
- 73.Pugach P., Ozorowski G., Cupo A., Ringe R., Yasmeen A., de Val N., Derking R., Kim H. J., Korzun J., Golabek M., de Los Reyes K., Ketas T. J., Julien J. P., Burton D. R., Wilson I. A., Sanders R. W., Klasse P. J., Ward A. B., Moore J. P., A native-like SOSIP.664 trimer based on an HIV-1 subtype B env gene. J. Virol. 89, 3380–3395 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Torrents de la Pena A., Rantalainen K., Cottrell C. A., Allen J. D., van Gils M. J., Torres J. L., Crispin M., Sanders R. W., Ward A. B., Similarities and differences between native HIV-1 envelope glycoprotein trimers and stabilized soluble trimer mimetics. PLOS Pathog. 15, e1007920 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pan J., Peng H., Chen B., Harrison S. C., Cryo-EM structure of full-length HIV-1 Env bound with the Fab of antibody PG16. J. Mol. Biol. 432, 1158–1168 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Joyce M. G., Georgiev I. S., Yang Y., Druz A., Geng H., Chuang G. Y., Kwon Y. D., Pancera M., Rawi R., Sastry M., Stewart-Jones G. B. E., Zheng A., Zhou T., Choe M., Van Galen J. G., Chen R. E., Lees C. R., Narpala S., Chambers M., Tsybovsky Y., Baxa U., McDermott A. B., Mascola J. R., Kwong P. D., Soluble prefusion closed DS-SOSIP.664-Env trimers of diverse HIV-1 strains. Cell Rep. 21, 2992–3002 (2017). [DOI] [PubMed] [Google Scholar]
- 77.Zivanov J., Nakane T., Forsberg B. O., Kimanius D., Hagen W. J., Lindahl E., Scheres S. H., New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dyer K. N., Hammel M., Rambo R. P., Tsutakawa S. E., Rodic I., Classen S., Tainer J. A., Hura G. L., High-throughput SAXS for the characterization of biomolecules in solution: A practical approach. Methods Mol. Biol. 1091, 245–258 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Classen S., Hura G. L., Holton J. M., Rambo R. P., Rodic I., McGuire P. J., Dyer K., Hammel M., Meigs G., Frankel K. A., Tainer J. A., Implementation and performance of SIBYLS: A dual endstation small-angle x-ray scattering and macromolecular crystallography beamline at the advanced light source. J. Appl. Cryst. 46, 1–13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lemmin T., Soto C., Glycosylator: A Python framework for the rapid modeling of glycans. BMC Bioinformatics 20, 513 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schneidman-Duhovny D., Hammel M., Tainer J. A., Sali A., FoXS, FoXSDock and MultiFoXS: Single-state and multi-state structural modeling of proteins and their complexes based on SAXS profiles. Nucleic Acids Res. 44, W424–W429 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jo S., Kim T., Iyer V. G., Im W., CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008). [DOI] [PubMed] [Google Scholar]
- 83.Jo S., Song K. C., Desaire H., MacKerell A. D. Jr., Im W., Glycan Reader: Automated sugar identification and simulation preparation for carbohydrates and glycoproteins. J. Comput. Chem. 32, 3135–3141 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Park S. J., Lee J., Patel D. S., Ma H., Lee H. S., Jo S., Im W., Glycan Reader is improved to recognize most sugar types and chemical modifications in the Protein Data Bank. Bioinformatics 33, 3051–3057 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Park S. J., Lee J., Qi Y., Kern N. R., Lee H. S., Jo S., Joung I., Joo K., Lee J., Im W., CHARMM-GUI Glycan Modeler for modeling and simulation of carbohydrates and glycoconjugates. Glycobiology 29, 320–331 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brooks B. R., Brooks C. L. III, Mackerell A. D. Jr., Nilsson L., Petrella R. J., Roux B., Won Y., Archontis G., Bartels C., Boresch S., Caflisch A., Caves L., Cui Q., Dinner A. R., Feig M., Fischer S., Gao J., Hodoscek M., Im W., Kuczera K., Lazaridis T., Ma J., Ovchinnikov V., Paci E., Pastor R. W., Post C. B., Pu J. Z., Schaefer M., Tidor B., Venable R. M., Woodcock H. L., Wu X., Yang W., York D. M., Karplus M., CHARMM: The biomolecular simulation program. J. Comput. Chem. 30, 1545–1614 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee J., Cheng X., Swails J. M., Yeom M. S., Eastman P. K., Lemkul J. A., Wei S., Buckner J., Jeong J. C., Qi Y., Jo S., Pande V. S., Case D. A., Brooks C. L. III, MacKerell A. D. Jr., Klauda J. B., Im W., CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 12, 405–413 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gotz A. W., Williamson M. J., Xu D., Poole D., Le Grand S., Walker R. C., Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1. Generalized Born. J. Chem. Theory Comput. 8, 1542–1555 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Salomon-Ferrer R., Gotz A. W., Poole D., Le Grand S., Walker R. C., Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh Ewald. J. Chem. Theory Comput. 9, 3878–3888 (2013). [DOI] [PubMed] [Google Scholar]
- 90.Le Grand S., Gotz A. W., Walker R. C., SPFP: Speed without compromise—A mixed precision model for GPU accelerated molecular dynamics simulations. Comput. Phys. Commun. 184, 374–380 (2013). [Google Scholar]
- 91.Best R. B., Zhu X., Shim J., Lopes P. E., Mittal J., Feig M., Mackerell A. D. Jr., Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone ϕ, ψ and side-chain χ1 and χ2 dihedral angles. J. Chem. Theory Comput. 8, 3257–3273 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Guvench O., Mallajosyula S. S., Raman E. P., Hatcher E., Vanommeslaeghe K., Foster T. J., Jamison F. W. II, Mackerell A. D. Jr., CHARMM additive all-atom force field for carbohydrate derivatives and its utility in polysaccharide and carbohydrate-protein modeling. J Chem Theory Comput 7, 3162–3180 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mallajosyula S. S., Guvench O., Hatcher E., Mackerell A. D. Jr., CHARMM additive all-atom force field for phosphate and sulfate linked to carbohydrates. J. Chem. Theory Comput. 8, 759–776 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Raman E. P., Guvench O., MacKerell A. D. Jr., CHARMM additive all-atom force field for glycosidic linkages in carbohydrates involving furanoses. J. Phys. Chem. B 114, 12981–12994 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L., Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983). [Google Scholar]
- 96.Loncharich R. J., Brooks B. R., Pastor R. W., Langevin dynamics of peptides: The frictional dependence of isomerization rates of N-acetylalanyl-N′-methylamide. Biopolymers 32, 523–535 (1992). [DOI] [PubMed] [Google Scholar]
- 97.Essmann U., Perera L., Berkowitz M. L., Darden T., Lee H., Pedersen L. G., A smooth particle mesh Ewald method. J. Chem. Phys. 103, 8577–8593 (1995). [Google Scholar]
- 98.Ryckaert J. P., Ciccotti G., Berendsen H. J. C., Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 23, 327–341 (1977). [Google Scholar]
- 99.Hopkins C. W., Le Grand S., Walker R. C., Roitberg A. E., Long-time-step molecular dynamics through hydrogen mass repartitioning. J. Chem. Theory Comput. 11, 1864–1874 (2015). [DOI] [PubMed] [Google Scholar]
- 100.D. A. Case, H. M. Aktulga, K. Belfon, I. Ben-Shalom, S. R. Brozell, D. S. Cerutti, T. E. Cheatham, V. W. D. Cruzeiro, T. A. Darden, R. E. Duke, Amber 2021 (University of California, San Francisco, 2021). [Google Scholar]
- 101.L. Schrödinger, Release 2021-1: Maestro, Schrödinger, LLC, New York, NY, 2023. [Google Scholar]
- 102.Sastry G. M., Adzhigirey M., Day T., Annabhimoju R., Sherman W., Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 27, 221–234 (2013). [DOI] [PubMed] [Google Scholar]
- 103.Salam N. K., Adzhigirey M., Sherman W., Pearlman D. A., Structure-based approach to the prediction of disulfide bonds in proteins. Protein Eng. Des. Sel. 27, 365–374 (2014). [DOI] [PubMed] [Google Scholar]
- 104.Beard H., Cholleti A., Pearlman D., Sherman W., Loving K. A., Applying physics-based scoring to calculate free energies of binding for single amino acid mutations in protein-protein complexes. PLOS ONE 8, e82849 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 106.Meng E. C., Goddard T. D., Pettersen E. F., Couch G. S., Pearson Z. J., Morris J. H., Ferrin T. E., UCSF ChimeraX: Tools for structure building and analysis. Protein Sci. 32, e4792(2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC. [Google Scholar]
- 108.Croll T. I., ISOLDE: A physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. D. Struct. Biol. D74, 519–530 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Afonine P. V., Poon B. K., Read R. J., Sobolev O. V., Terwilliger T. C., Urzhumtsev A., Adams P. D., Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D. Struct. Biol. 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., UCSF Chimera--A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S17
Tables S1 to S5