Abstract
Background
Effective control of brain metastasis remains an urgent clinical need due a limited understanding of the mechanisms driving it. Although the gain of neuro-adaptive attributes in breast-to-brain metastases (BBMs) has been described, the mechanisms that govern this neural acclimation and the resulting brain metastasis competency are poorly understood. Herein, we define the role of neural-specific splicing factor Serine/Arginine Repetitive Matrix Protein 4 (SRRM4) in regulating microenvironmental adaptation and brain metastasis colonization in breast cancer cells.
Methods
Utilizing pure neuronal cultures and brain-naive and patient-derived BM tumor cells, along with in vivo tumor modeling, we surveyed the early induction of mediators of neural acclimation in tumor cells.
Results
When SRRM4 is overexpressed in systemic breast cancer cells, there is enhanced BBM leading to poorer overall survival in vivo. Concomitantly, SRRM4 knockdown expression does not provide any advantage in central nervous system metastasis. In addition, reducing SRRM4 expression in breast cancer cells slows down proliferation and increases resistance to chemotherapy. Conversely, when SRRM4/REST4 levels are elevated, tumor cell growth is maintained even in nutrient-deprived conditions. In neuronal coculture, decreasing SRRM4 expression in breast cancer cells impairs their ability to adapt to the brain microenvironment, while increasing SRRM4/RE-1 Silencing Transcription Factor (REST4) levels leads to greater expression of neurotransmitter and synaptic signaling mediators and a significant colonization advantage.
Conclusions
Collectively, our findings identify SRRM4 as a regulator of brain metastasis colonization, and a potential therapeutic target in breast cancer.
Keywords: breast-to-brain metastasis, cancer neuroscience, REST, REST4, SRRM4
Graphical Abstract
Graphical Abstract.
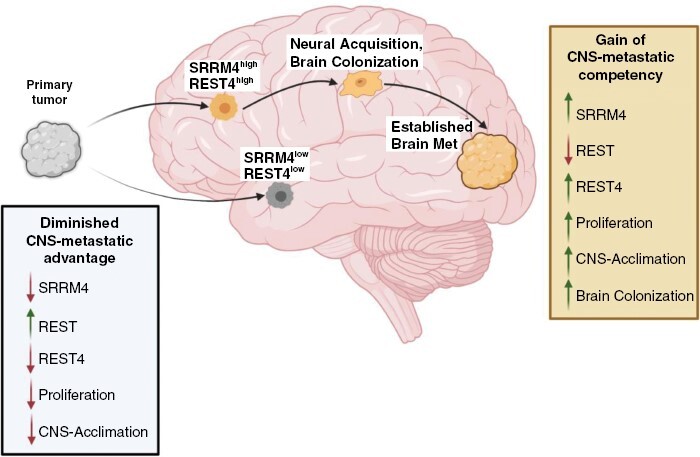
Key Points.
Serine/Arginine Repetitive Matrix Protein 4 (SRRM4) regulates central nervous system (CNS) metastatic potential in breast cancer.
Elevated SRRM4/RE-1 Silencing Transcription Factor (REST4) augments proliferation and CNS microenvironmental acclimation.
Reduced SRRM4/REST4 confers quiescence, chemoresistance in breast cancer cells.
Importance of the Study.
This study highlights the dual role of Serine/Arginine Repetitive Matrix Protein 4 (SRRM4) in breast cancer progression; where high SRRM4 levels in breast cancer promote brain metastasis through upregulation of RE-1 Silencing Transcription Factor leading to gain of neural characteristics in tumor cells, while low SRRM4 levels result in slower-growing tumor cells with diminished central nervous system metastatic advantage. The findings identify SRRM4 as a potential therapeutic target in advanced breast cancer, due to its role in promoting neurotransmitter and synaptic signaling mediators in epithelial tumors and regulating brain metastasis colonization.
Brain metastases (BMs) are the main cause of intracranial neoplasms in adults with invasive cancers.1 Although progress has been made in understanding the molecular factors that drive metastasis, the mechanisms responsible for microenvironmental adaptation and colonization, especially in brain metastasis, are not well understood.
Breast cancer (BC) is unique in its longer latency in forming BMs2,3 indicating that tumor cells, which successfully form detectable metastases within the central nervous system (CNS), are able to overcome dormancy, immune clearance, and cell death and can acclimate to the neural microenvironment.4 Moreover, adaptation to the CNS microenvironment appears to be requisite for successful breast-to-brain metastasis (BBM). Specifically, previous studies have reported acquisition of neuronal characteristics in BBMs,5,6 and enhanced expression of neurotransmitter (NT) receptors in aggressive breast cancers that prime them to successfully metastasize to the brain.7 The gain of CNS-specific attributes in BBMs indicates that neural regulatory pathways may be activated in these tumor cells. However, up to now, there are no studies investigating the contribution of neurodevelopmental programs in promoting BM competency in breast cancer.
RE-1 Silencing Transcription Factor (REST) is a regulator of neuronal development, homeostasis, and response to cellular stress, which represses target genes by binding to the Neuron Restrictive Silencer Element (RE-1 element).8 It inhibits the expression of neural genes in nonneuronal cells9 and regulates neuronal differentiation in CNS-progenitor cells.10,11 Owing to its dual functions, REST is an oncogene in primary brain tumors like medulloblastoma and glioblastoma,12,13 but a tumor suppressor in epithelial cancers, where loss of REST function promotes invasiveness in breast, lung, and colon cancer.14 However, the role of REST dysregulation remains uncharacterized in brain metastasis, particularly in breast cancer.
Serine/Arginine Repetitive Matrix Protein 4 (SRRM4), a neural-specific RNA splicing factor, regulates REST expression by inserting neural exon “N” into the REST transcript, resulting in alternative splice variants including truncated isoform REST4.15 REST4, although able to enter the nucleus like REST, shows weak binding to the RE-1 element, and is able to competitively reverse REST-mediated repression of target genes.16 SRRM4-mediated REST-to-REST4 splicing is crucial in the developing brain allowing progenitor cells to differentiate into functional neurons by enabling the expression of neuronal genes.15,17,18
In this study, we investigated the role of SRRM4-mediated REST dysregulation in driving CNS acclimation, dormancy, and BM colonization in breast cancer cells. We show that SRRM4 expression predicts overall patient survival in advanced breast cancer and is elevated in BBMs relative to brain-naive BC cells. This is accompanied by enhanced expression and nuclear localization of alternative splice form REST4, concurrent with reduced expression of full-length REST. SRRM4 overexpression in BC cells significantly accelerates BM incidence and poor survival in vivo, while SRRM4 knockdown reduces tumor burden and provides no CNS-metastatic advantage. Mechanistically, elevated SRRM4 in BC cells increases their proliferative potential through increased nuclear REST4 promoting CNS adaptation and colonization by inducing expression of neural-specific genes, many regulated by the SRRM4/REST axis. In contrast, SRRM4 depletion is consistent with slow-growing phenotype, increased chemoresistance, and suppressed neuronal pathway activation in BC cells. Collectively, our work shows that SRRM4-mediated REST-to-REST4 splicing augments microenvironmental acclimation and BM colonization in breast cancer.
Materials and Methods
Cell Culture
Low-passage patient-derived BM breast (BBM 3.1), lung (LuBM5), and melanoma (MBM2) cells were propagated in our lab from surgically resected BM tissue from consenting patients. On receipt from USC Neurosurgery, tumor tissue was mechanically dissociated, trypsinized at 37°C for 10 minutes, and triturated. The tissue was centrifuged (500g for 2 minutes) and resuspended in cell culture media (50% DMEM-F12, 50% Neurobasal-A, 5% FBS, 1% glutamine, 1% Antibacterial-Antimycotic, 05.X B-27). For non-BM cells the following commercially available cell lines were used: breast cancer (SKBR3, BT474, MDA-MB-231), lung cancer (A549), and melanoma (A2058). All cell lines were maintained in DMEM-F12 (Thermo, 12634028) supplemented with 10% FBS, 1% Glutamine (Thermo, 35050061) and 1% Antibacterial-Antimycotic agent (Sigma, 15240062), and were tested for mycoplasma using the DAPI test before use.
Primary Neuron Cultures
Neural cells were isolated from whole brain tissue of postnatal day 1–4 mice and in vitro cultured as previously described.19
Lentiviral Transduction
Lentivirus were constructed in 293T cells using SRRM4 overexpression ORF cDNA clone (Genecopoeia EX-Y3278-Lv224), scrambled and SRRM4 knockdown shRNA clone set (Genecopoeia HSH114130-LVRU6GP). SKBR3 BC cells were transduced to stably overexpress (SRRM4OE) or knockdown (SRRM4KD) SRRM4 or scrambled. These cells were further lentiviral transduced to express GFP-FF Luciferase to facilitate bioluminescent imaging of tumors in vivo.
Establishment of Nutrient-Deficient Media-Acclimated Breast Cancer Cells
SKBR3 (control, SRRM4OE and SRRM4KD) cells were gradually acclimated to nutrient-deficient (ND) medium over 1 week as in previous studies.6 Specifically, ND media composition was as follows: (DMEM with no glucose, no glutamine, Thermo A1413004) supplemented with 5% Fetal Bovine Serum (FBS), and 1% Antibacterial-Antimycotic agent (Sigma, 15240062).
Tumor–Neuron Cocultures
Tumor cells were resuspended in complete neuron culture medium and seeded onto established neuronal cultures (1:60 tumor cell to neuron ratio) to model tumor–neuron interaction. Forty-eight hours postseeding, tumor cells were collected from cocultures for qPCR, or cocultures were fixed with 4% formaldehyde for immunofluorescence studies. For in vitro competitive colonization model, the following combinations of tumor cells were cocultured with neurons: (1) SKBR3 control scrambled (GFP) and SRRM4OE (mCherry), (2) SRRM4OE (mCherry) and SRRM4KD (GFP), or (3) SKBR3 (mCherry) and SRRM4KD (GFP). Thousand cells of each type were plated in all cocultures at day 0. Cultures were fixed every 24 hours up to 120 hours.
Cell Viability Assay
SKBR3 cells (control, SRRM4 OE, SRRM4 KD) were treated with 20 µM Paclitaxel (Sigma, T7402-1MG), and 500 µM 5-Fluorouracil (Sigma, 343922), and cell viability was measured using the Live/Dead Viability/Cytotoxicity kit (Thermo, L3224) at 48 hours posttreatment. DMSO (Sigma, D8418-100 mL) was used as vehicle. Percent viability data were normalized to vehicle (DMSO)-treated SKBR3ctrl cells.
Animals
Eleven-week-old adult female NSG mice on BalbC background were purchased from Jackson Laboratories and used for in vivo intracardiac injection experiments to model brain metastasis. Animal procedures were performed under approved IACUC protocols and guidelines.
Intracardiac Injection of Tumor Cells and Animal Imaging
SKBR3 (control, SRRM4OE, and SRRM4KD) cells were injected intracardiac into mice to evaluate the role of SRRM4 in the development of BBMs. Each mouse was injected with 1 × 105 tumor cells in 100 µL of sterile 1X PBS, with a 25-gauge needle. Development of brain lesions and other distant metastases was monitored by optical imaging of injected animals. Luciferin (1 µL/gram bodyweight of animal) was injected into the tail vein of each mouse and bioluminescence was imaged (dorsal and ventral) 15 minutes after injection. Optical signal from BMs was quantified by measuring the same brain region of interest (ROI) for all experimental animals on all imaging days.
RNA Isolation and qPCR Analysis
Cells from various conditions were harvested by trypsinization for 3–5 minutes at 37 °C, followed by trypsin neutralization in FBS, and centrifugation at 1000 rpm for 1 minute. Resulting pellet was processed immediately or frozen for subsequent use in qPCR (performed in triplicate per sample) as previously described.20 All primers used for qPCR analysis were purchased from IDT (Supplementary Table 1).
Immunofluorescence
Human primary tumor tissue microarrays were obtained from Biomax US (Breast PM2a-ER, Lung LC241l, Melanoma ME242c). Brain-metastatic tumor tissues from patients were acquired via USC Neurosurgery. These tissues were formalin fixed, paraffin embedded, and processed into 10 µm-thick sections by the histology core at USC. Immunofluorescence protocol on tissue and cells was performed as previously described.20 Primary antibodies used were as follows: Rabbit Anti-REST (1:100, LSBio), Rabbit Anti-REST4 (1:300 IHC, 1:500 ICC, generous gift from Dr. Noel J. Buckley at Oxford University), Rabbit Anti-SRRM4 (1:500, Biorbyt), Rabbit Anti-RILP (1:500, Abcam), Goat Anti-SOX9 (1:500, Novus Biologicals), Rabbit Anti-Ki67 (1:500, Biocare Medical). Secondary antibodies used at 1:300 (Goat Anti-rabbit Cy3, Goat Anti-rabbit 647, Donkey Anti-Goat 647). Phalloidin (488 or 647 conjugated, Thermofisher) was used to stain cell membrane where necessary. Stained cells and tissue sections were mounted in Prolong Gold Antifade reagent with DAPI (Invitrogen) for imaging and long-term storage.
Microscopy and Imaging
Confocal imaging was performed using the Leica confocal SP8 LIAchroics CSU RGB HyD 405 FOV DMi8. Nuclear colocalization studies were performed by analyzing colocalization between protein of interest (REST, SRRM4, REST4) and DAPI, which stains cellular nuclei. Channel background and thresholds were set and normalized so that different groups could be compared to one another for statistical analysis. For quantification of SRRM4, mean intensity per cell was measured in ImageJ, by drawing a region of interest (ROI) around each cell. Light microscopy images (H&E) were obtained using Zeiss SN: 3834000524, Zen 2.3 imaging software version 2.3.69.1013.
Bioinformatics
Kaplan-Meier survival curve analyses for target genes were performed using UCSC Xena, an online exploration tool for public/private multiomic and clinical/phenotypic data. Publicly available TCGA data for breast cancer, lung cancer, and melanoma were analyzed using this open-source platform.21 Venn diagrams were constructed using a freely available web tool designed by the Bioinformatics and Evolutionary Genomics (BEG) department at Ghent University, Belgium.
Statistical Analysis
Statistics were performed using GraphPad Prism. t tests, 1-way ANOVA, an2-way ANOVA were used to calculate statistical significance as appropriate throughout the study. All data were shown as mean and SEM. Statistical significance and Hazard Ratio for survival data from in vivo experiments were calculated using Log-Rank Test.
Ethics Statement
The current investigation was carried out in adherence to the Declaration of Helsinki and in accordance with the regulations of the institutional ethics committee at the University of Southern California, as well as the local ethical committees of the relevant hospitals. All patients and/or their legal representatives provided written informed consent.
Results
Breast-to-Brain Metastases Show Reduced Expression and Nuclear Localization of REST
Previous studies have demonstrated the gain of neuronal cell characteristics in BBMs.5,7 Of these, several genes (BDNF, RELN, GABBR1, GRIN2B) are normally repressed by REST.22,23 We hypothesized that REST would be downregulated in BBMs, allowing for the expression of CNS-specific genes. First, we examined whether REST expression in tumors was associated with overall patient survival. Kaplan-Meier survival analysis from TCGA datasets shows REST was not prognostic in breast and lung cancer, or in melanoma (Supplementary Figure S1A–C). In vitro REST expression was reduced in BM cells compared to non-BM cells in melanoma, and in BM cells compared to 2 out of 3 non-BM cells in breast cancer (Supplementary Figure S1D; Figure 1A), but not in lung cancer cells (Supplementary Figure S1E). Patient tissues revealed reduced levels of REST protein in BM breast cancer and melanoma, compared to their non-BM counterparts (Figure 1B; Supplementary Figure S1F). This difference was not observed in lung cancer (Supplementary Figure S1G).
Figure 1.
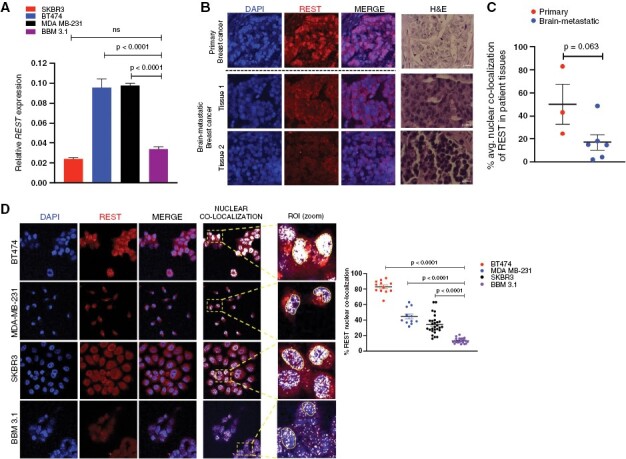
BBMs show reduced nuclear REST. (A) Relative REST mRNA expression in brain-metastatic (BM) breast BBM 3.1 cancer cells, relative to their primary (non-BM) counterparts (mean and SEM, n = 3 per sample) (B) Representative immunofluorescence (IF) images comparing REST protein expression in patient tissues from primary and BM breast cancer. H&E images for each representative tumor tissue included. (C) Quantification for nuclear REST localization in primary (non-BM, n = 3) versus brain metastatic (BM, n = 6) patient tissues. Graph shows comparison of average nuclear REST percentage in each patient in respective groups. (D) Representative IF images, and quantification of nuclear REST localization in non-BM (BT474, MDA MB-231, SKBR3) and patient-derived BM (BBM 3.1) breast cancer cells (magnification ×63; scale 20 µm). Nuclear REST ROI highlighted. Graph shows comparison of percent nuclear REST in individual cells (4 images per sample, 9 cells per image).
Moreover, REST acts as a transcriptional repressor in cellular nucleus; with loss of nuclear REST promoting the expression of its target genes.24 We thus evaluated nuclear REST in non-BM and BM tumor cells. Nuclear REST localization levels measured in patient tissues were heterogenous in primary breast cancer (n = 3), but uniformly diminished in BMs (n = 6) (Figure 1C), indicating the importance of reduced REST activity in CNS adaptation and colonization potential in breast cancer. Correspondingly, in vitro, nuclear REST was significantly reduced in BBM 3.1 cells derived from a breast tumor that had successfully colonized the brain, compared to 3 different nonbrain trophic breast cancer cell lines (Figure 1D). In contrast, nuclear REST remained unchanged between BM versus non-BM cells in lung cancer and melanoma (Supplementary Figure S1H and I). These results suggest that REST is dysregulated at posttranscriptional and nuclear-localization level in BBMs.
REST Dysregulation in BBMs Is Mediated by SRRM4, Resulting in Alternative Splice Product REST4
REST expression is regulated by 2 mediators: REST/NRSF-Interacting LIM Domain Protein (RILP),25 or SRRM4.26 RILP has been shown to bind to REST and facilitate its function as a transcription factor by importing it into the nucleus.27 Our results show that ratio of REST/RILP nuclear colocalization remained unchanged between BM versus non-BM cells in breast and lung cancer, and melanoma (Supplementary Figure S2A–C). This suggests that reduced nuclear import of REST in BBMs is not mediated by RILP.
Thus, we next investigated the involvement of SRRM4, which regulates the alternative splicing of REST into its truncated isoform REST4, which in turn reverses REST-mediated repression of target genes15,16,28 (Figure 2A). Loss of REST concomitant with REST4 upregulation has been associated with increased breast cancer invasiveness.29 However, the mechanisms regulating REST-to-REST4 dysregulation in BC and its role in promoting BBMs were heretofore unknown.
Figure 2.
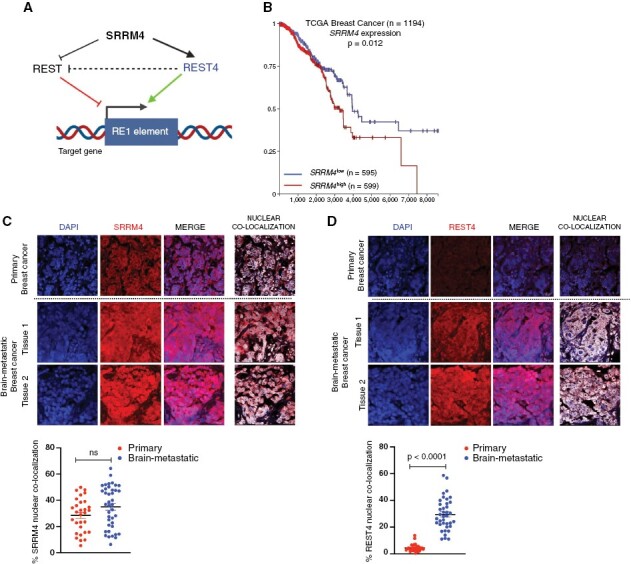
SRRM4 and REST4 expression in BBMs. (A) Schematic describing the SRRM4 mediated REST regulatory pathway. (B) Kaplan-Meier analysis for association between SRRM4 expression and overall survival in breast cancer. Representative immunofluorescence images comparing (C) SRRM4 and (D) REST4 protein expression and quantification of nuclear localization in patient tissues from primary (n = 3) and BM (n = 4) breast cancer. Graphs show comparison of percent nuclear localization of SRRM4 and REST4 in individual cells per image for all samples (mean and SEM).
We first evaluated whether SRRM4 expression in tumors was associated with overall survival (OS). Kaplan-Meier survival analysis from TCGA datasets showed that although not prognostic in lung cancer or melanoma (Supplementary Figure S3A and B), high SRRM4 expression was significantly associated with worse OS in patients with breast cancer (Figure 2B). Furthermore, the dichotomy between high and low expression of SRRM4 and OS in breast cancer patients becomes larger later in the course of the disease (>6 years of follow-up), suggesting a role for SRRM4 in advanced breast cancer, and potentially brain metastasis.
Given the association of SRRM4 with poor survival, and the reduced expression and nuclear localization of REST in BBMs as shown previously, we postulated that SRRM4 expression would be enhanced in BBMs, accompanied by increased expression of REST4. Results show that total SRRM4 and REST4 expression were enhanced in BBM tissues compared to non-BM breast cancer (Figure 2C and D). Quantification of nuclear SRRM4 did not show statistically significant increase in BBMs compared to primary breast cancer tissues (Figure 2C). However, nuclear REST4 was significantly increased in BBMs, indicating enhanced SRRM4 expression may mediate increased REST4 functionality in these tumors (Figure 2D). Expression and nuclear localization of SRRM4 remained unchanged, while REST4 reduced in BM and non-BM lung cancer (Supplementary Figure S3C and D). Moreover, nuclear SRRM4 and REST4 were significantly reduced in BM melanoma (Supplementary Figure S3E and F) patient tissues.
In vitro, while SRRM4 and REST4 expression remained unchanged between primary and BM in lung cancer and melanoma cells (Supplementary Figure S3G and H), there was significant increase in the expression of both mRNAs in BBM 3.1 cells compared to 3 different non-BM breast cancer cell types (Figure 3A and B). SRRM4 regulates splicing in the cellular nucleus, and nuclear colocalization analysis revealed no difference in nuclear SRRM4 levels in BM versus non-BM cells in lung cancer (Supplementary Figure S3I), while BM melanoma showed significant reduction of nuclear SRRM4 compared to its primary (Supplementary Figure 3J). However, nuclear SRRM4 was significantly enhanced in BBM 3.1 cells compared to non-BM SKBR3 breast cancer cells (Figure 3C).
Figure 3.
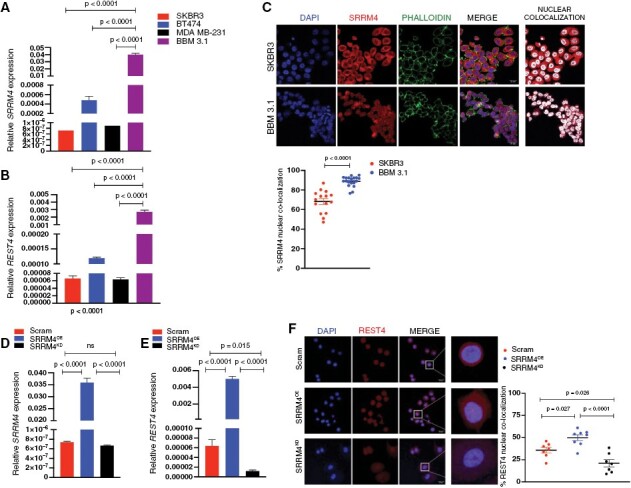
REST dysregulation in BBMs is mediated by SRRM4 and alternative splice isoform REST4. (A) Relative SRRM4 and (B) REST4 mRNA expression in BM BBM 3.1 breast cancer cells, relative to their non-BM counterparts (mean and SEM, n = 3 per sample). (C) Representative IF images, and quantification of nuclear SRRM4 localization in primary (non-BM; SKBR3) and patient-derived BM BBM 3.1 breast cancer cells. Graphs show comparison of percent nuclear localization of SRRM4 in individual cells (2 images per sample, 10 cells per image). Confirmation of successful transduction of SKBR3 cells by mRNA expression analysis of (D) SRRM4 and (E) REST4. mRNA overexpression or knockdown was confirmed by qPCR, with data represented in bar graphs as fold change in expression relative to SKBR3 scrambled controls (mean and SEM, n = 3 per sample). (F) Representative IF images of REST4 protein expression and nuclear localization in SKBR3 cells—scrambled (SKBR3ctrl), SRRM4 overexpression (SRRM4OE), and knockdown (SRRM4KD). Graphs show comparison of percent nuclear localization of REST4 in individual cells.
We next sought to determine a definitive role of SRRM4 in REST-to-REST4 dysregulation and regulating potential brain tropism in breast cancer. To elucidate whether increased SRRM4 promotes BM competency, we conducted our studies in nonbrain trophic SKBR3 cells, which showed high REST, and low endogenous SRRM4/REST4 expression. We first established stably transduced SKBR3 cells with SRRM4 overexpression (SRRM4OE) or knocked down using 3 different shRNA clones (SRRM4KD). SKBR3KD cells derived using shRNA clone B were used for knockdown studies, based on mitigated SRRM4 expression and nuclear localization in these cells (Supplementary Figure 3K). SRRM4 mRNA expression was significantly enhanced in SRRM4OE and reduced in SRRM4KD cells, compared to control (scrambled) SKBR3 cells (Figure 3D). Appropriately, REST4 analysis showed increase in SRRM4OE cells and diminished mRNA expression (Figure 3E) and nuclear localization in SRRM4KD cells (Figure 3F). Therefore, we conclude that SRRM4 mediates the dysregulation of REST to REST4 in breast cancer.
Next, we elucidated the contribution of SRRM4 to BBM colonization in vivo. All SRRM4OE xenografted mice developed BMs by 98 days post-intracardiac injection, compared to 2 mice from the SKBR3CTRL (control) group and only 1 from the SRRM4KD group (Figure 4A–C). Extracranial tumor load remained comparable between SKBR3CTRL and SRRM4OE xenografts, whereas SRRM4KD showed lower tumor load overall. Although 4 out of 5 mice in the control group developed BMs eventually, this was a delayed event compared to SRRM4OE group, while mice from SRRM4KD group remained BM-free significantly longer than SKBR3OE group (Figure 4D). Furthermore, SRRM4OE xenografted mice showed significantly worse OS compared to control (Figure 4E), suggesting that even in the presence of comparable extracranial tumor load, increased SRRM4 expression in tumor cells contributes to worse prognosis by triggering enhanced BM competency in breast cancer. SRRM4OE xenografted mice also showed worse OS compared to SRRM4KD group. Analysis of SKBR3CTRL xenografts showed areas of cells with high expression and nuclear localization of SRRM4 (Figure 4F) and REST4 (Figure 4G), and low REST (Figure 4H) in an otherwise heterogenous BM tumor population. In contrast, SRRM4OE BMs showed uniform and significantly higher nuclear localization of SRRM4 and REST4, concurrent with diminished REST expression. Overall, these results show increased SRRM4 expression enhances brain-metastatic potential in breast cancer cells and contributes to worse OS in vivo.
Figure 4.
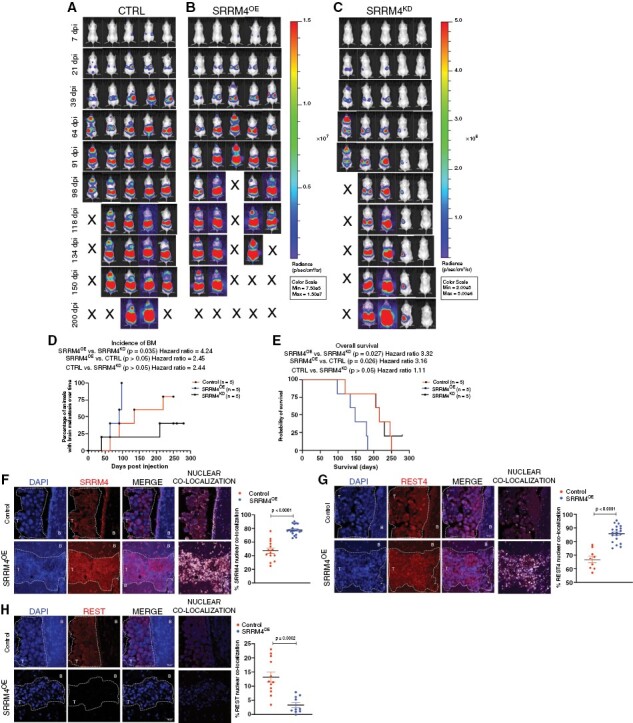
Enhanced SRRM4 expression augments breast-to-brain metastatic competency and contributes to worse overall survival in vivo: representative images of bioluminescent imaging (BLI) for brain metastasis and overall tumor load, post-intracardiac injection of SKBR3 (A) control scramble (CTRL), (B) SRRM4OE, and (C) SRRM4KD cells in vivo (n = 5 per group). BLI intensity scale was normalized in control and SRRM4OE groups (Min: 7.5e5; Max: 1.5e7). Scale for SRRM4KD group was reduced (Min: 2e5; Max: 5e6) since mice showed lower overall tumor load, and any incidence of BMs in these animals would go unnoticed if scale was as per groups A and B. (D) Graphical comparison of BM incidence in animals over time (n = 5 per group). Mice in SRRM4OE group showed accelerated incidence of BMs compared to SRRM4KD group (P = 0.035, HR = 4.24) and SKBR3ctrl group (P > 0.05, HR = 2.45). (E) Kaplan-Meier survival analysis in SKBR3ctrl, SRRM4OE, and SRRM4KD xenografted mice (n = 5 per group). Mice in SRRM4OE group (blue line) showed significantly worse overall survival compared to SKBR3ctrl cells (P = 0.026, HR = 3.16), and SRRM4KD cells (P = 0.027, HR = 3.32) (Statistics—Log-Rank test). Representative IF images for expression and comparison of nuclear (F) SRRM4, (G) REST4, and (H) REST in brain-metastatic lesions from SKBR3ctrl and SRRM4OE xenografted mice (magnification ×40; scale 30 µm). Graphs show comparison of percent nuclear SRRM4, REST4, and REST in individual cells for both groups. T = tumor, B = brain tissue.
SRRM4 Regulates Proliferative Capacity in Breast Cancer Cells
Clinically, breast cancer patients have longer latency to brain metastasis compared to more aggressive cancers like lung cancer, and BMs can occur following successful control of extracranial disease.30 This can be attributed partly to the presence of slow-growing tumor cells, that evade chemotherapy and cell death, and can eventually be reactivated for successful colonization and distant relapse.31,32 Therefore, we wanted to determine whether the reduced metastatic competency we observed in vivo in SRRM4-low (SRRM4KD) breast cancer cells was because of a conferring of a slow-proliferating phenotype in these cells. Initial cellular-doubling time and BrDU incorporation analysis showed enhanced proliferation in SRRM4OE cells, but slower growth in SRRM4KD cells, relative to control SKBR3 cells (Figure 5A; Supplementary Figure S4A). This was further confirmed by the presence of significantly enhanced Ki67 positivity in SRRM4OE cells compared to SRRM4KD and control SKBR3 cells (Supplementary Figure S4B). These results indicate that low SRRM4 expression confers slower proliferative potential in breast cancer cells.
Figure 5.
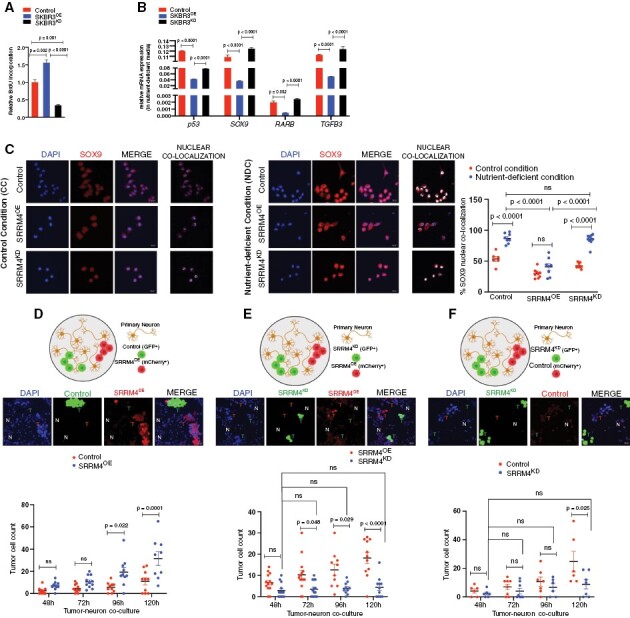
Enhanced SRRM4 expression confers more proliferative phenotype to breast cancer cells: (A) relative BrdU incorporation (luminescence) in SKBR3 control scramble, SRRM4OE, SRRM4KD cells (n = 2 per sample, 3 wells per experiment). (B) Relative mRNA expression of dormancy markers p53, SOX9, RARB, TGFB3 in SKBR3, SRRM4OE, SRRM4KD cells acclimated to nutrient-deficient media (mean and SEM, n = 3 per sample). (C) Representative IF images and comparison of nuclear colocalization of SOX9 in SKBR3 scramble SRRM4OE, SRRM4KD cells in control (CC) and nutrient-deficient (NDC) conditions, respectively. Data presented as percent nuclear colocalization per cell in both conditions. (D–F) In vitro competitive colonization model. Representative IF images and quantification of total cell numbers for each tumor cell type in 3 different combination cocultures with primary neurons: (D) SKBR3 (GFP)/SRRM4OE (mCherry), (E) SRRM4OE (mCherry)/SRRM4KD (GFP), (F) SKBR3 (mCherry)/SRRM4KD (GFP). Individual values in graph represent number of cells of each type (mean and SEM) per image quantified (>6 images/coculture sample/timepoint).
Next, SKBR3 were acclimated to ND media20 to recapitulate the microenvironment encountered by metastatic tumor cells. SKBR3 control and SRRM4KD cells showed enhanced mRNA expression of previously validated dormancy markers p53, SOX9, RARB, and TGFB333 compared to SRRM4OE cells (Figure 5B). Consistent with mRNA data, control and SRRM4KD cells showed enhanced SOX9 expression and nuclear localization compared to SRRM4OE cells in ND-conditions (Figure 5C), indicating propensity for metastatic dormancy in cells with low SRRM4/REST4 expression. SRRM4OE cells showed sustained nuclear REST4, while control and SRRM4KD cells displayed diminished REST4 expression with significant loss of nuclear REST4 in ND-conditions (Supplementary Figure S4C).
Next, we treated SKBR3 cells with 5-Fluorouracil and Paclitaxel which are standard chemotherapeutic drugs used for breast cancer patients.34 Slower-growing SRRM4KD cells were significantly more resistant to both chemotherapies compared to control, whereas SRRM4OE cells showed significant enhanced sensitivity to Paclitaxel compared to control and SRRM4KD (Supplementary Figure S4D and E). Taken together, these results suggest low SRRM4 expression confers slower growth, whereas enhanced SRRM4 expression leading to REST4 activity increases proliferative capacity and chemo-sensitization in breast cancer cells.
We next asked whether interaction with the neuronal niche along with enhanced SRRM4 in tumor cells provides a growth advantage and early colonization to breast cancer cells in the CNS-metastatic microenvironment. To determine this, we established a competitive in vitro colonization model where primary neurons were cocultured with combination of either: (1) SKBR3 control and SRRM4OE (Figure 5D), (2) SRRM4OE and SRRM4KD (Figure 5E), or (3) SKBR3 control and SRRM4KD (Figure 5F) cells and then evaluated for growth. Results show SRRM4OE cells have significant proliferative advantage over both control and SRRM4KD cells (Figure 5D and E). Additionally, control SKBR3 cells have significant growth advantage compared to SRRM4KD cells (Figure 5F). Furthermore, SRRM4KD remained quiescent throughout the various competitive models (Figure 5E and F). Taken together, increased SRRM4 promotes CNS acclimation in breast cancer cells resulting in proliferative advantage in the neuronal microenvironment.
SRRM4 Facilitates CNS Acclimation in BC Cells in the Neuronal Microenvironment
Since upregulation of SRRM4 provides a proclivity for breast cancer growth in the neural niche, we next asked whether this neuronal master regulator facilitates tumor-CNS acclimation through enhanced tumor–neuron interaction. We observed an increase in SRRM4 and REST4 expression in control SKBR3 cells cultured with neuronal conditioned media (Figure 6A), concomitant with a gain of nuclear SRRM4 in these cells when cocultured with neurons (Supplementary Figure 5A). Furthermore, SRRM4OE cells cocultured with neurons showed significantly enhanced nuclear SRRM4 and REST4 (Figure 6B; Supplementary Figure 5B and C). These results indicate that exposure to the neuronal microenvironment promotes increased SRRM4 and REST4 activity in breast cancer cells. Furthermore, SRRM4OE cells maintained their inherently high expression of SRRM4 and REST4, while SRRM4KD cells were unable to upregulate the expression of either gene (Figure 6C) when exposed to neuron-conditioned media.
Figure 6.
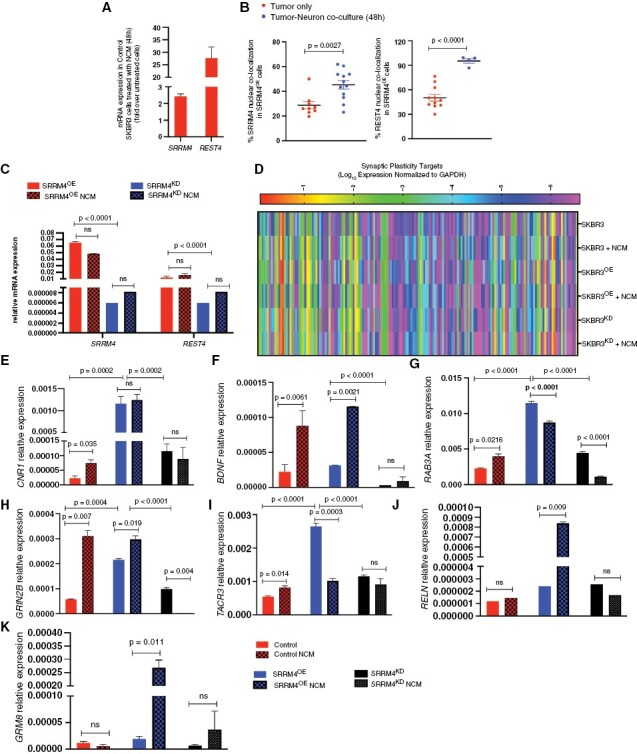
SRRM4 facilitates CNS acclimation in breast cancer cells in the neuronal microenvironment: (A) SRRM4 and REST4 expression in SKBR3ctrl cells in neuron-conditioned media (NCM). Data represented as fold change in expression relative to untreated SKBR3 cells. (B) Comparison of nuclear SRRM4 and REST4 in SRRM4OE cells in 2 conditions (tumor only, tumor–neuron coculture). Data represented in graph as percent nuclear colocalization per cell for both conditions (3 images per sample, 4 tumor cells per image). (C) Relative expression of SRRM4 and REST4 in SRRM4OE and SRRM4KD cells in 2 conditions (cells only, cells treated with NCM for 48 hours). Red bars (plain, striped) represent SRRM4OE cells; blue bars (plain, striped) represent SRRM4KD cells (3 wells/sample/condition). (D) Clustergram representing differential mRNA expression of CNS-specific (neurotransmitter receptors and synaptic plasticity mediators) in SKBR3 cells (control, SRRM4OE, and SRRM4KD) in 2 conditions (tumor cells only, tumor cells grown in neuron-conditioned media). qPCR target validation for relative expression of SRRM4-dependent target genes (E) CNR1, (F) BDNF, (G) RAB3A, (H) GRIN2B, (I) TACR3, (J) RELN, and (K) GRM8 in tumor cells in 2 conditions (tumor only, tumor cells treated with NCM, 3 wells/sample/condition).
Since SRRM4 regulates the expression of CNS-specific genes, we next determined which targets in tumors are regulated by SRRM4 augmentation in presence of neurons to promote brain metastasis competency. To investigate this, we interrogated the expression of 153 CNS-specific mediators of NT signaling and synaptic plasticity in SKBR3 cells (control, SRRM4OE, SRRM4KD) cultured in the neuronal microenvironment (Figure 6D). Of these 153 targets, 45 were determined to be directly SRRM4-dependent based on association overlap of tumor-inherent SRRM4-regulated genes and neural microenvironment-induced SRRM4-regulated genes (Supplementary Figure 5D). We then focused on targets in breast cancer cells that would be critical in allowing them to mimic neuron-like synaptic plasticity function. SKBR3ctrl cells showed an increase in the expression of presynaptic regulator cannabinoid receptor CNR135 on exposure to the neuronal microenvironment, while SRRM4KD cells did not respond (Figure 6E). Overall, SKBR3ctrl and SRRM4OE cells showed upregulation of BDNF, a neurotrophin involved in regulating long-term synaptic plasticity36 (Figure 6F). SKBR3ctrl cells also upregulated vesicular trafficking protein RAB3A involved in regulating BDNF-mediated plasticity37 (Figure 6G), GRIN2B, an NMDAR-type glutamate receptor38 (Figure 6H), and TACR3 a receptor for neurokinins39 (Figure 6I). Specifically, SRRM4OE cells showed inherently enhanced expression of these genes, and thus retained the expression of most genes within the neuronal microenvironment. Furthermore, only SKBR3OE cells showed upregulation of synaptic master regulator RELN40 and metabotropic glutamate receptor GRM8 (Figure 6J and K) with exposure to the neuronal microenvironment. In contrast, SRRM4KD were unable to respond to the neuronal exposure and did not show upregulation of any CNS acclimatory genes mentioned above.
Discussion
Brain metastases are a significant cause of mortality in individuals with advanced breast cancer.41 Therefore, elucidating the mechanisms driving initial BM competency is imperative in improving patient outcomes. Our study sought to determine the regulators of neural acclimation and CNS colonization in brain-seeking BC cells. We uncovered enhanced expression of neural-specific splicing regulator SRRM4 in BBMs, concomitant with reduced functionality of REST, and increased expression and nuclear localization of REST4. SRRM4-mediated REST to REST4 splicing is critical in the brain for normal neuronal development, maturation, and maintenance,15 indicating that breast cancer cells co-opt neurodevelopmental pathways to successfully adapt to the CNS-metastatic niche.
In neurogenesis, inducible REST deletion triggers exit of neural stem cells from quiescence and increases proliferative potential,42 while REST4 is critical for inducing and maintaining a differentiated phenotype.43 Additionally, in tumor progression, recent evidence shows SRRM4-induced accelerated proliferation and neuroendocrine trans-differentiation in castrate-resistant prostate cancer.44 Correspondingly, we found that while SRRM4low cells exhibit a quiescent phenotype with reduced nuclear REST4, enhanced SRRM4 expression confers proliferative advantage to BC cells in normal and nutrient-deprived conditions through maintenance of elevated nuclear REST4. Owing to tumor heterogeneity, a subpopulation of BC cells may increase SRRM4 expression before extravasating into the CNS, which then allows them to successfully traverse the brain-metastatic cascade. This is evidenced by our results showing superior CNS-metastatic competency in BC cells transduced to endogenously overexpress SRRM4 that show depleted nuclear REST expression within the CNS. Thus, successful BMs are eventually established by tumor cells that can overcome nutritional stress and dormancy as they disseminate through the circulation.31,45 Our findings substantiate these observations and show that although SRRM4low BC cells persist for extended periods of time in vivo, only cells that upregulate SRRM4 activity ultimately colonize the brain contributing to overt brain metastasis and worse OS.
Moreover, since SRRM4 regulates neuronal gene activation, we hypothesized that its increased expression would facilitate BC colonization of the neuronal niche. Our findings demonstrate enhanced SRRM4/REST4 expression and nuclear localization, alongside REST loss in BC cells exposed to the neuronal microenvironment. Moreover, BC cells with higher SRRM4/REST4 activity within this niche showed improved colonization through increased expression of CNS-specific mediators involved in NT and synaptic response. In contrast, cells unable to upregulate SRRM4 remained quiescent and could not adapt to the neural environment. This SRRM4 upregulation can be attributed to feedback loop, which releases REST-mediated repression of the SRRM4 promoter.46 Potential regulators of SRRM4 expression and activity, particularly in metastatic progression, remain unexplored.
Our xenograft model shows faster CNS metastasis development and lower survival in mice implanted with SRRM4OE cells. This indicates that these breast cancer cells can successfully thrive in a CNS microenvironment with glia/neurons/oligodendrocyte populations intact.
Patient tumor analyses from multiple studies have shown reduced immune infiltration and enhanced protumoral microglial polarization in breast cancer brain metastases (BCBMs) compared to primary tumors.47,48 These changes, mediated by CNS-infiltrating tumor cells aid in brain colonization and establishing clinical metastasis. Furthermore, BCBMs interact with astrocytes in pseudosynapses facilitating nutrient utilization and NT-mediated signaling using GRIN2B7. Correspondingly, we show that SRRM4OE breast cancer cells upregulate neuro-adaptive genes, including GRIN2B, CNR1, and TACR3. Thus, it is likely that SRRM4 which provides early CNS-metastatic advantage to breast cancer cells, does so by stimulating the expression of mediators that allow BCBMs to influence the glial and immune microenvironment. Although beyond the scope of the current study, it would be meaningful to conduct proteomic and genomic analysis to determine cytokine expression and signaling pathways that are altered in SRRM4OE cells. This would shed light on their potential effects on the CNS glial/immune microenvironment during tumor colonization.
Targeting SRRM4 in brain metastases would be of immediate benefit to the patient since it would eliminate the SRRM4high proliferative tumor population and limit immediate progression. For the SRRM4low quiescent tumor population, which would be resistant to conventional cell-cycle targeted therapy, alternate cell-cycle independent targeted strategies would have to be utilized. Several avenues that target chemo-resistant dormant tumor cells (DCCs) to prevent relapse are currently under consideration.49These include combination strategies using conventional and targeted therapies, maintaining quiescence, or reactivation of DCCs and subsequent sensitization to anti-proliferative chemotherapy.
We thus show a dichotomy of SRRM4 in breast cancer progression, where enhanced SRRM4 and REST4 expression results in gain of proliferative capacity and neural attributes leading to increased BM; while loss of SRRM4 results in slower-growing tumor cells with diminished CNS-metastatic advantage. The findings presented in our study expand on the emerging role of SRRM4 as a promoter of neural characteristics in epithelial tumors and identify it as a regulator of BBM colonization, and a potential therapeutic target in advanced breast cancer.
Supplementary Material
Acknowledgments
The authors would like to acknowledge patient advocates (Ms. Michele Rakoff, Ms. Michelle Atlan, Ms. Andrea Hutton, and Ms. Sharon Schlesinger) and all breast cancer patients/survivors/family members for the invaluable role they have played in our research over the years.
Contributor Information
Krutika Deshpande, Department of Neurological Surgery, Keck School of Medicine, University of Southern California, Los Angeles, California, USA; USC Brain Tumor Center, Keck School of Medicine, University of Southern California, Los Angeles, California, USA).
Vahan Martirosian, Department of Neurological Surgery, Keck School of Medicine, University of Southern California, Los Angeles, California, USA; USC Brain Tumor Center, Keck School of Medicine, University of Southern California, Los Angeles, California, USA.
Brooke N Nakamura, Division of Gastrointestinal and Liver Diseases, Department of Medicine, Keck School of Medicine, University of Southern California, Los Angeles, California, USA; USC Brain Tumor Center, Keck School of Medicine, University of Southern California, Los Angeles, California, USA.
Diganta Das, Department of Neurological Surgery, Keck School of Medicine, University of Southern California, Los Angeles, California, USA; USC Brain Tumor Center, Keck School of Medicine, University of Southern California, Los Angeles, California, USA.
Mukund Iyer, Department of Neurological Surgery, Keck School of Medicine, University of Southern California, Los Angeles, California, USA; USC Brain Tumor Center, Keck School of Medicine, University of Southern California, Los Angeles, California, USA.
Max Reed, Department of Neurological Surgery, Keck School of Medicine, University of Southern California, Los Angeles, California, USA.
Ling Shao, Division of Gastrointestinal and Liver Diseases, Department of Medicine, Keck School of Medicine, University of Southern California, Los Angeles, California, USA.
Daniella Bamshad, Department of Neurological Surgery, Keck School of Medicine, University of Southern California, Los Angeles, California, USA.
Noel J Buckley, Department of Psychiatry, University of Oxford, Oxford, UK.
Josh Neman, Department of Neurological Surgery, Keck School of Medicine, University of Southern California, Los Angeles, California, USA; USC Brain Tumor Center, Keck School of Medicine, University of Southern California, Los Angeles, California, USA.
Conflict of interest statement
None declared.
Funding
This work was supported by grants from the National Institutes of Health/National Cancer Institute R01CA223544-01A1 to J.N., Department of Defense BCRP BC141728 to J.N., and Susan G Komen Career Catalyst Grant CCR15332673 to J.N.
Author Contributions
Conceptualization: K.D., J.N.; methodology: K.D., J.N., B.N., L.S., N.B.; investigation: K.D., J.N., V.M., B.N., D.D., M.I., M.R.; funding acquisition: J.N.; project administration: K.D., J.N.; supervision: J.N.; writing—original draft: K.D., J.N.; writing—review and editing: K.D., B.N., D.B., N.B., J.N.
Data Availability
Data from the current study will be made available from the corresponding author upon reasonable request.
References
- 1. Boire A, Brastianos PK, Garzia L, Valiente M.. Brain metastasis. Nat Rev Cancer. 2020;20(1):4–11. [DOI] [PubMed] [Google Scholar]
- 2. Saunus JM, Momeny M, Simpson PT, Lakhani SR, Da Silva L.. Molecular aspects of breast cancer metastasis to the brain. Genet Res Int. 2011;2011(1):219189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Leone JP, Lee AV, Brufsky AM.. Prognostic factors and survival of patients with brain metastasis from breast cancer who underwent craniotomy. Cancer Med. 2015;4(7):989–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Singh M, Manoranjan B, Mahendram S, et al. Brain metastasis-initiating cells: survival of the fittest. Int J Mol Sci. 2014;15(5):9117–9133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Neman J, Termini J, Wilczynski S, et al. Human breast cancer metastases to the brain display GABAergic properties in the neural niche. Proc Natl Acad Sci U S A. 2014;111(3):984–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Deshpande K, Martirosian V, Nakamura BN, et al. Neuronal exposure induces neurotransmitter signaling and synaptic mediators in tumors early in brain metastasis. Neuro Oncol. 2021;24(6):914–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zeng Q, Michael IP, Zhang P, et al. Synaptic proximity enables NMDAR signalling to promote brain metastasis. Nature. 2019;573(7775):526–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Baldelli P, Meldolesi J.. The transcription repressor REST in adult neurons: physiology, pathology, and diseases. eNeuro. 2015;2(4):ENEURO.0010–ENEU15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ballas N, Battaglioli E, Atouf F, et al. Regulation of neuronal traits by a novel transcriptional complex. Neuron. 2001;31(3):353–365. [DOI] [PubMed] [Google Scholar]
- 10. Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G.. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121(4):645–657. [DOI] [PubMed] [Google Scholar]
- 11. Soldati C, Bithell A, Johnston C, et al. Repressor element 1 silencing transcription factor couples loss of pluripotency with neural induction and neural differentiation. Stem Cells. 2012;30(3):425–434. [DOI] [PubMed] [Google Scholar]
- 12. Wagoner MP, Roopra A.. A REST derived gene signature stratifies glioblastomas into chemotherapy resistant and responsive disease. BMC Genomics. 2012;13(1):686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lawinger P, Venugopal R, Guo ZS, et al. The neuronal repressor REST/NRSF is an essential regulator in medulloblastoma cells. Nat Med. 2000;6(7):826–831. [DOI] [PubMed] [Google Scholar]
- 14. Coulson JM. Transcriptional regulation: cancer, neurons and the REST. Curr Biol. 2005;15(17):R665–R668. [DOI] [PubMed] [Google Scholar]
- 15. Raj B, O’Hanlon D, Vessey JP, et al. Cross-regulation between an alternative splicing activator and a transcription repressor controls neurogenesis. Mol Cell. 2011;43(5):843–850. [DOI] [PubMed] [Google Scholar]
- 16. Shimojo M, Lee J-H, Hersh LB.. Role of zinc finger domains of the transcription factor neuron-restrictive silencer factor/repressor element-1 silencing transcription factor in DNA Binding and Nuclear Localization*. J Biol Chem. 2001;276(16):13121–13126. [DOI] [PubMed] [Google Scholar]
- 17. Vuong CK, Black DL, Zheng S.. The neurogenetics of alternative splicing. Nat Rev Neurosci. 2016;17(5):265–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee J-H, Shimojo M, Chai Y-G, Hersh LB.. Studies on the interaction of REST4 with the cholinergic repressor element-1/neuron restrictive silencer element. Mol Brain Res. 2000;80(1):88–98. [DOI] [PubMed] [Google Scholar]
- 19. Deshpande K, Saatian B, Martirosian V, et al. Isolation of neural stem cells from whole brain tissues of adult mice. Curr Protoc Stem Cell Biol. 2019;49(1):e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Deshpande K, Martirosian V, Nakamura BN, et al. Neuronal exposure induces neurotransmitter signaling and synaptic mediators in tumors early in brain metastasis. Neuro Oncol. 2022;24(6):914–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Goldman MJ, Craft B, Hastie M, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38(6):675–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hara D, Fukuchi M, Miyashita T, et al. Remote control of activity-dependent BDNF gene promoter-I transcription mediated by REST/NRSF. Biochem Biophys Res Commun. 2009;384(4):506–511. [DOI] [PubMed] [Google Scholar]
- 23. Otto SJ, McCorkle SR, Hover J, et al. A new binding motif for the transcriptional repressor REST uncovers large gene networks devoted to neuronal functions. J Neurosci. 2007;27(25):6729–6739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shimojo M. Characterization of the nuclear targeting signal of REST/NRSF. Neurosci Lett. 2006;398(3):161–166. [DOI] [PubMed] [Google Scholar]
- 25. Shimojo M, Hersh LB.. REST/NRSF-interacting LIM domain protein, a putative nuclear translocation receptor. Mol Cell Biol. 2003;23(24):9025–9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Quesnel-Vallières M, Irimia M, Cordes SP, Blencowe BJ.. Essential roles for the splicing regulator nSR100/SRRM4 during nervous system development. Genes Dev. 2015;29(7):746–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shimojo M, Hersh LB.. Characterization of the REST/NRSF-interacting LIM domain protein (RILP): localization and interaction with REST/NRSF. J Neurochem. 2006;96(4):1130–1138. [DOI] [PubMed] [Google Scholar]
- 28. Spencer EM, Chandler KE, Haddley K, et al. Regulation and role of REST and REST4 variants in modulation of gene expression in in vivo and in vitro in epilepsy models. Neurobiol Dis. 2006;24(1):41–52. [DOI] [PubMed] [Google Scholar]
- 29. Wagoner MP, Gunsalus KT, Schoenike B, et al. The transcription factor REST is lost in aggressive breast cancer. PLoS Genet. 2010;6(6):e1000979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kodack DP, Askoxylakis V, Ferraro GB, Fukumura D, Jain RK.. Emerging strategies for treating brain metastases from breast cancer. Cancer Cell. 2015;27(2):163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lorger M, Felding-Habermann B.. Capturing changes in the brain microenvironment during initial steps of breast cancer brain metastasis. Am J Pathol. 2010;176(6):2958–2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ghajar CM, Peinado H, Mori H, et al. The perivascular niche regulates breast tumour dormancy. Nat Cell Biol. 2013;15(7):807–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sosa MS, Parikh F, Maia AG, et al. NR2F1 controls tumour cell dormancy via SOX9- and RARβ-driven quiescence programmes. Nat Commun. 2015;6(1):6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nicholson BP, Paul DM, Hande KR, et al. Paclitaxel, 5-fluorouracil, and leucovorin (TFL) in the treatment of metastatic breast cancer. Clin Breast Cancer. 2000;1(2):136–143; discussion 144. [DOI] [PubMed] [Google Scholar]
- 35. Carey MR, Myoga MH, McDaniels KR, et al. Presynaptic CB1 receptors regulate synaptic plasticity at cerebellar parallel fiber synapses. J Neurophysiol. 2011;105(2):958–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lu B, Nagappan G, Lu Y.. BDNF and synaptic plasticity, cognitive function, and dysfunction. Handb Exp Pharmacol. 2014;220(1):223–250. [DOI] [PubMed] [Google Scholar]
- 37. Thakker-Varia S, Alder J, Crozier RA, Plummer MR, Black IB.. Rab3A is required for brain-derived neurotrophic factor-induced synaptic plasticity: transcriptional analysis at the population and single-cell levels. J Neurosci. 2001;21(17):6782–6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shin W, Kim K, Serraz B, et al. Early correction of synaptic long-term depression improves abnormal anxiety-like behavior in adult GluN2B-C456Y-mutant mice. PLoS Biol. 2020;18(4):e3000717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cui W-Q, Zhang W-W, Chen T, et al. Tacr3 in the lateral habenula differentially regulates orofacial allodynia and anxiety-like behaviors in a mouse model of trigeminal neuralgia. Acta Neuropathol Commun. 2020;8(1):44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Beffert U, Weeber EJ, Durudas A, et al. Modulation of synaptic plasticity and memory by reelin involves differential splicing of the lipoprotein receptor apoer2. Neuron. 2005;47(4):567–579. [DOI] [PubMed] [Google Scholar]
- 41. Leone JP, Leone BA.. Breast cancer brain metastases: the last frontier. Exp Hematol Oncol. 2015;4(1):33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gao Z, Ure K, Ding P, et al. The master negative regulator REST/NRSF controls adult neurogenesis by restraining the neurogenic program in quiescent stem cells. J Neurosci. 2011;31(26):9772–9786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ovando-Roche P, Yu JSL, Testori S, Ho C, Cui W.. TRF2-mediated stabilization of hREST4 is critical for the differentiation and maintenance of neural progenitors. Stem Cells. 2014;32(8):2111–2122. [DOI] [PubMed] [Google Scholar]
- 44. Lee AR, Gan Y, Tang Y, Dong X.. A novel mechanism of SRRM4 in promoting neuroendocrine prostate cancer development via a pluripotency gene network. EBioMedicine. 2018;35(1):167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Heyn C, Ronald JA, Ramadan SS, et al. In vivo MRI of cancer cell fate at the single-cell level in a mouse model of breast cancer metastasis to the brain. Magn Reson Med. 2006;56(5):1001–1010. [DOI] [PubMed] [Google Scholar]
- 46. ENCODE Project Consortium. A user’s guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol. 2011;9(4):e1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Noh MG, Kim SS, Kim YJ, et al. Evolution of the tumor microenvironment toward immune-suppressive seclusion during brain metastasis of breast cancer: implications for targeted therapy. Cancers (Basel). 2021;13(19):4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ogiya R, Niikura N, Kumaki N, et al. Comparison of immune microenvironments between primary tumors and brain metastases in patients with breast cancer. Oncotarget. 2017;8(61):103671–103681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Damen MPF, van Rheenen J, Scheele C.. Targeting dormant tumor cells to prevent cancer recurrence. FEBS J. 2021;288(21):6286–6303. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data from the current study will be made available from the corresponding author upon reasonable request.


