Abstract
Biochemical analysis has shown that mammalian Rad51 and Rad52 interact and synergize in DNA recombination reactions in vitro, but these proteins have not been shown to function together in response to DNA damage in vivo. By analysis of murine cells expressing murine Rad52 tagged with green fluorescent protein (GFP)–Rad52, we now show that DNA damage causes Rad51 and GFP–Rad52 to colocalize in distinct nuclear foci. Cells expressing GFP–Rad52 show both increased survival and an increased number of Rad51 foci, raising the possibility that Rad52 is limiting for repair. These observations provide evidence of coordinated function of Rad51 and Rad52 in vivo and support the hypothesis that Rad52 plays an important role in the DNA damage response in mammalian cells.
INTRODUCTION
DNA damage is repaired by nuclear proteins that are structurally and functionally conserved among eukaryotic cells. One of these proteins, Rad52, was first discovered in Saccharomyces cerevisiae (Adzuma et al., 1984). Saccharomyces cerevisiae rad52 mutants exhibit defects in repair and recombination, including homologous recombination in both meiotic and mitotic cells (Resnick, 1987; Petes et al., 1991), mating type switching (Petes et al., 1991) and recombination of the rDNA loci (Gottlieb and Esposito, 1989; Lin and Keil, 1991; Zou and Rothstein, 1997; Park et al., 1999). Structural homologs of Rad52 are present in higher eukaryotic organisms, including mouse and human (Muris et al., 1994; Shen et al., 1995). However, targeted deletion of murine Rad52 had no obvious physiological consequences or effect on DNA repair, and produced only a moderate decrease in homologous recombination (Rijkers et al., 1998). This raised the possibility that mammalian Rad52 was redundant, or that it did not play a critical role compared with its yeast homolog.
Rad51 is the eukaryotic homolog of the critical prokaryotic recombination protein RecA (reviewed by Kowalczykowski and Eggleston, 1994; Shinohara and Ogawa, 1995; Baumann and West, 1998). Rad51 and Rad52 have been shown to interact in vitro and in vivo (Shinohara et al., 1992; Milne and Weaver, 1993; Donovan et al., 1994; Shen et al., 1996; Sung, 1997) and to synergize in homologous DNA pairing reactions in vitro (Benson et al., 1998; New et al., 1998; Shinohara and Ogawa, 1998; Kurumizaka et al., 1999). Yeast and human Rad52 can bind to single-stranded and double-stranded DNA and promote DNA strand annealing (Mortensen et al., 1996; Sugiyama et al., 1998; Van Dyck et al., 1998, 1999), suggesting that, in vivo, Rad52 may bind at sites of double-strand breaks (DSBs) and recruit Rad51 to carry out homologous recombination (Van Dyck et al., 1999). Rad51 localizes to nuclear foci in response to DNA damage (Haaf et al., 1995; Bishop et al., 1998; Raderschall et al., 1999) and in cells carrying out developmentally regulated recombination, including immunoglobulin heavy chain switch recombination (Li et al., 1996; Li and Maizels, 1997) and meiosis (Ashley et al., 1995; Plug et al., 1996; Barlow et al., 1997). In both meiotic and mitotic mammalian cells, Rad51 colocalizes with the products of the tumor suppressor genes, BRCA1 and BRCA2 (Mizuta et al., 1997; Scully et al., 1997; Chen et al., 1998), and this has helped to establish the link between DNA repair and tumorigenesis, especially in the case of breast cancer (Sharan et al., 1997).
The fact that Rad52 and Rad51 synergize to function in vitro suggested that these proteins might colocalize in cells responding to DNA damage, but this has not been observed. Determining the localization of mammalian Rad52 is essential to understanding its function, particularly because genetic analysis has not provided clear information on the role of Rad52 in DNA repair. We recently showed that in mammalian cells treated with ionizing radiation (IR), Rad52 is induced to colocalize with Rad50 (Liu et al., 1999), one member of a complex that rapidly localizes to DNA breaks in response to DNA damage (Nelms et al., 1998; reviewed by Haber, 1998). We have now asked whether Rad51 and Rad52 function coordinately in vivo by studying the cellular localization of murine Rad51 and Rad52 in response to DNA damage. In this report, we show that treatment of murine cells with either IR or the alkylating agent, methylmethanesulfonate (MMS), induces Rad52 to form nuclear foci that colocalize with Rad51 foci. These observations support the hypothesis that mammalian Rad52 and Rad51 function together in recombinational repair of DSBs in vivo.
RESULTS
Induction of Rad51/GFP–Rad52 nuclear foci in response to IR
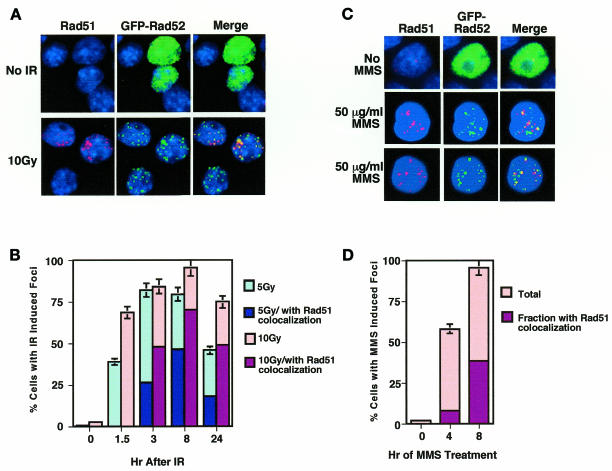
Cells expressing green fluorescent protein (GFP)–Rad52 were stained with anti-Rad51 antibodies before and after γ-irradiation. In untreated cells, few Rad51 foci could be visualized, and GFP–Rad52 was distributed throughout the nucleus. Irradiation caused Rad51 and GFP–Rad52 to relocalize to form distinct and bright nuclear foci (Figure 1A). Relocalization of GFP–Rad52 was rapid: GFP–Rad52 foci were evident within 1.5 h following irradiation in ~40% of cells irradiated with 5 Gy and 70% of cells irradiated with 10 Gy (Figure 1B). Rad51 foci appeared with slightly slower kinetics, but as soon as Rad51 staining was evident, Rad51 colocalized with GFP–Rad52; by 3 h post-irradiation, colocalization of Rad51 and GFP–Rad52 was evident in the majority of cells containing GFP–Rad52 foci (Figure 1A and B). In almost all cells containing both Rad51 and GFP–Rad52 foci (>90%), the foci colocalized. In some cells containing GFP–Rad52 foci, no Rad51 foci could be visualized, as is evident in one cell in Figure 1A. This probably reflects the fact that Rad50/GFP–Rad52 foci are also induced by irradiation (Liu et al., 1999), but both Rad50 and Rad51 foci are not observed in the same cell (Maser et al., 1997).
Fig. 1. Colocalization of Rad51/GFP–Rad52 in response to DNA damage. (A) Examples of DAPI (blue), Rad51 (red), GFP–Rad52 (green) and colocalized Rad51 and GFP–Rad52 (yellow in the merged image) fluorescent signals in unirradiated cells and cells at 8 h following irradiation with 10 Gy. Typically 20–40% of cells display a detectable GFP–Rad52 signal. (B) Percentage of cells expressing GFP–Rad52 that contained IR-induced nuclear foci at the times indicated after irradiation with 5 or 10 Gy, and the percentage of those foci that colocalized with Rad51 foci. (C) Examples of DAPI (blue), GFP–Rad52 (green), Rad51 (red) and colocalized Rad51/GFP–Rad52 (yellow in the merged image) fluorescent signals in cells treated with 50 µg/ml MMS. (D) Percentage of cells expressing GFP–Rad52 that contained MMS-induced nuclear foci at the times indicated after addition of 50 µg/ml MMS, and the percentage of those foci that colocalized with Rad51 foci.
The number of GFP–Rad52 foci per nucleus increased in the first few hours following irradiation, and by 8 h post-irradiation, most cells irradiated with 5 Gy contained ≥10 GFP–Rad52 foci per nucleus. Higher doses of IR induced greater numbers of foci: at 8 h, 25% of cells irradiated with 5 Gy contained >25 foci per nucleus, while 45% of cells irradiated with 10 Gy and 90% of cells irradiated with 75 Gy contained >25 foci per nucleus. Foci persisted from 3 to 8 h and diminished somewhat by 24 h. Essentially identical results were obtained by analyzing GFP–Rad52 transductants of the murine pre-B-cell line PD31 (shown) and the PE501 fibroblast line (not shown).
We have shown previously that in unirradiated cells, GFP–Rad52 is distributed diffusely throughout the nucleus during G1, but localized to the nucleolus in S phase and G0 (Liu et al., 1999). However, essentially no nucleolar localization of GFP–Rad52 was observed in irradiated cells (Figure 1). We have verified that nucleoli remain intact in these cells after irradiation, as assayed by staining with anti-nucleolin antibodies (unpublished observations). Our data therefore suggest that GFP–Rad52 may be released from the nucleolus upon irradiation.
Rad51/GFP–Rad52 colocalization induced by MMS
Treatment with the alkylating agent MMS also induced relocalization of GFP–Rad52 to nuclear foci (Figure 1C). Kinetic analysis showed that both induction of GFP–Rad52 foci and colocalization of Rad51 to these foci occurred more slowly following treatment with MMS than with IR. At 4 h post-treatment, relatively few GFP–Rad52 foci contained Rad51, but by 8 h colocalization of Rad51 and GFP–Rad52 foci was evident in ~40% of cells containing GFP–Rad52 foci (Figure 1D). As previously reported, MMS-induced Rad51 foci were more diffuse and stained less intensely than IR-induced Rad51 foci at early times (4 h) (Haaf et al., 1995) but not at later times (8 h) following MMS treatment. The delay in appearance of GFP–Rad52/Rad51 foci in response to MMS compared with IR may be due to the indirect action of MMS in generating DSBs.
Distinct conditions of induction of Rad51/GFP–Rad52 and Rad50/GFP–Rad52 foci
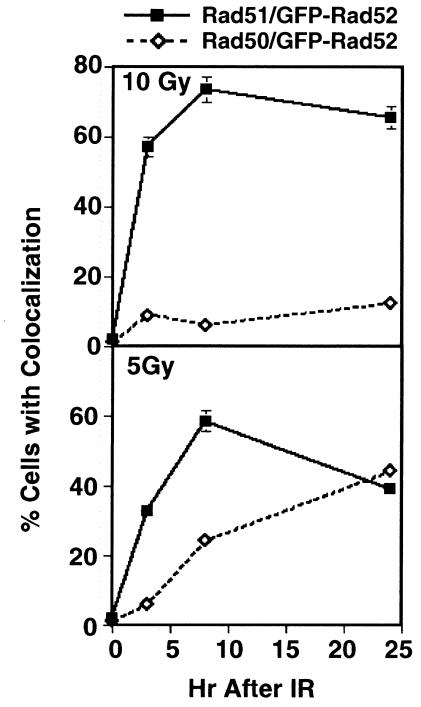
The Rad50/Mre11/Nbs1 complex has important functions in repair as well as in telomere maintenance (reviewed by Haber, 1998, 1999; Petrini, 1999). Petrini and collaborators have found that both Rad50 and Rad51 foci are not observed in the same cell (Maser et al., 1997), raising the question of whether the two classes of foci represent distinct DNA repair complexes. We have previously shown that IR (5 Gy) induces colocalization of GFP–Rad52 and Rad50 (Liu et al., 1999). Surprisingly, when we assayed induction of Rad50/GFP–Rad52 foci in the same conditions shown above to induce Rad51/GFP–Rad52 foci, we found that MMS did not induce Rad50 and GFP–Rad52 to colocalize (not shown). In addition, while the percentage of cells containing Rad51/GFP–Rad52 foci increased at the higher dose of IR (10 Gy), Rad50/GFP–Rad52 foci decreased at this radiation dose (Figure 2). We conclude that distinct conditions induce coordinated function of Rad52 with Rad50 or Rad51.
Fig. 2. Comparison of induction of Rad51/GFP–Rad52 and Rad50/GFP–Rad52 foci by IR. Percentage of cells containing colocalized Rad51/GFP–Rad52 nuclear foci or Rad50/GFP–Rad52 nuclear foci at the times indicated after irradiation with 5 or 10 Gy. Filled boxes, Rad51/GFP–Rad52; open diamonds, Rad50/GFP–Rad52 foci.
GFP–Rad52 expression enhances both cell survival and Rad51 foci formation
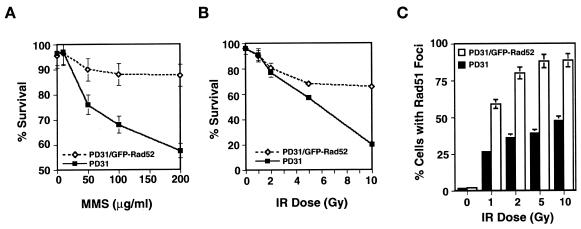
We tested the effect of GFP–Rad52 expression on cell survival by assaying viable cells following incubation with MMS or treatment with IR. GFP–Rad52 expression caused a moderate enhancement of cell survival following MMS treatment, an effect most evident at high doses of MMS (200 µg/ml; Figure 3A). A dose-dependent enhancement of survival was also evident in cells treated with IR: after irradiation with 10 Gy, cell survival was >3-fold higher in cells expressing GFP–Rad52 (Figure 3B). Increased survival was paralleled by an increase in the number of cells containing Rad51/GFP–Rad52 foci (Figure 3C). Expression of human Rad52 in monkey cell lines has similarly been shown to increase resistance to IR (Park, 1995). The increased resistance to DNA damage in cells expressing GFP–Rad52 may reflect increased efficiency of repair by pathways dependent upon Rad51. Alternatively, expression of GFP–Rad52 may alter efficiency of repair and Rad51 foci formation by some other mechanism, for example by enhancing the activity of another repair protein.
Fig. 3. Effect of GFP–Rad52 expression on cell survival and formation of Rad51 foci. (A) Comparison of survival of PD31 (filled boxes) and PD31 expressing GFP–Rad52 (open diamonds), assayed after 30 h of incubation with MMS at the concentration indicated. (B) Comparison of survival of PD31 (filled boxes) and PD31 expressing GFP–Rad52 (open diamonds), assayed 72 h after the dose of IR indicated. (C) Percentage of PD31 (filled bars) and PD31 expressing GFP–Rad52 (open bars) containing Rad51 foci 5 h after treatment with the dose of IR indicated.
DISCUSSION
The demonstration that Rad51 and GFP–Rad52 colocalize in response to DNA damage provides in vivo evidence to support the notion that these proteins function together in repair of DSBs. The potential for concerted function of mammalian Rad51 and Rad52 was suggested by biochemical analysis showing that these proteins interact physically and synergize in strand pairing reactions in vitro (Shinohara et al., 1992; Milne and Weaver, 1993; Donovan et al., 1994; Mortensen et al., 1996; Shen et al., 1996; Sung, 1997; Benson et al., 1998; New et al., 1998; Shinohara and Ogawa, 1998; Sugiyama et al., 1998; Van Dyck et al., 1998, 1999; Kurumizaka et al., 1999). In contrast to the biochemical analysis, genetic experiments had suggested that Rad52 does not play an important function in vertebrates: targeted deletion of murine Rad52 was reported to have no significant physiological consequences, and caused only a modest decrease (2-fold) in efficiency of homologous recombination (Rijkers et al., 1998); a similar lack of phenotype was produced by deletion of Rad52 in the chicken DT40 cell line (Yamaguchi-Iwai et al., 1998). Our results contrast with the genetic analysis, and argue for an important function of mammalian Rad52. Presuming that the genetic experiments with higher organisms provide an accurate picture of Rad52 function, then Rad52 may be dispensable because another gene product is redundant in function.
The fact that GFP–Rad52 colocalizes with both Rad51 and Rad50 is also consistent with a central function in DNA repair, especially because several lines of evidence show that Rad51 and Rad50/Mre11 function in independent pathways. In mammalian cells responding to IR, a cell appears to commit to one pathway or the other, as both classes of foci are not observed in a single cell (Maser et al., 1997; Nelms et al., 1998; Petrini, 1999). In addition, in yeast cells lacking the RNA component of telomerase, telomeres can be maintained by either Rad50- or Rad51-dependent pathways, but both pathways require the presence of functional Rad52 (Le et al., 1999). Our own results provide further support to the independence of the Rad50- and Rad51-dependent repair pathways, as we find that the Rad51/GFP–Rad52 foci are induced by treatment with MMS (Figure 2), while the Rad50/GFP–Rad52 foci are not, and that Rad50/GFP–Rad52 and Rad51/GFP–Rad52 foci are induced by different levels of irradiation and with different kinetics (Figure 3).
Eukaryotic cells can repair DNA DSBs using pathways dependent upon either homologous recombination or non-homologous end-joining (Liang et al., 1998; Takata et al., 1998). The fact that expression of GFP–Rad52 enhances formation of Rad51 foci suggests that the result of increased Rad52 expression might be to enhance the use of the homology-dependent repair pathway. Moreover, Rad52 may be limiting for repair, as expression of GFP–Rad52 enhances both formation of Rad51 foci and cell survival.
The nuclear foci that can be visualized upon induction of DNA damage contain multiple components. In addition to Rad52, proteins likely to regulate or be components of at least some classes of Rad51 foci are XRCC3 (Bishop et al., 1998), BRCA1 (Scully et al., 1997), RPA (Raderschall et al., 1999) and Rad54 (Tan et al., 1999). It is interesting that the dynamics of localization of GFP–Rad52 are in many respects similar to those of BRCA1: IR induces BRCA1 foci, and these foci colocalize predominantly with Rad51 and to a lesser extent with Rad50 (Zhong et al., 1999). Several recent reports have attempted to correlate mutations in Rad52 with early-onset breast cancer (Bell et al., 1999; Gonzalez et al., 1999) and prolymphocytic leukemia (Salomon-Nguyen et al., 1998). While no clear correlation has yet emerged, there is an intriguing loss of heterozygosity in the region of chromosome 12p within which Rad52 resides (Salomon-Nguyen et al., 1998; Gonzalez et al., 1999). These observations are consistent with the potential involvement of Rad52 in progression to malignancy, and point to the value of carrying out systematic screens for Rad52 mutation in malignant cells.
METHODS
Cell culture, staining and microscopy. In order to produce a clear fluorescent signal and avoid artifacts due to antibody cross-reactivity, we have studied Rad52 localization in cell lines stably expressing Rad52 fused at its N-terminus to GFP (Liu et al., 1999). Production of the LX–GFP–Rad52-N retrovirus, and generation and maintenance of stable transductants have been described (Liu et al., 1999). Fluorescence microscopy was carried out as described, except that Rad51 antiserum (a generous gift of Drs Charles Radding and Efim Golub, Yale) was used at 1:100 dilution in phosphate-buffered saline containing 1% bovine serum albumin. In all cases where secondary antibodies were used for staining, controls were carried out to show that the secondary antibody alone gave no signal. For kinetic analyses, at least 250 cells with green fluorescent signal were counted at each time point. A minimum of five foci per nucleus was required to score a cell as positive, and all cells containing five or more foci were included in the data. Cells were scored as positive for colocalization if >50% of the GFP–Rad52 foci stained with anti-Rad51 or anti-Rad50 antibodies.
Induction of DNA damage and assays of cell survival. Prior to IR or MMS treatment, cells were diluted to 5 × 105 cells/ml and incubated at 37°C for 1 h. For MMS treatment, the concentrations of MMS indicated were added to the culture, and cells were further incubated for the times indicated. For IR treatment, cells were exposed to a cesium (137Cs) irradiator at doses of 1, 2, 5 and 10 Gy. Irradiated cells were further diluted to 2 × 105 cells/ml and incubated as indicated. Viable cells were counted following staining with 0.4% Trypan Blue (Gibco-BRL). For each experimental point at least 250 cells were counted.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs E. Golub and C.M. Radding (Yale University School of Medicine) for anti-Rad51 antibodies, and Dr E. Lee (University of Texas Health Science Center) for the anti-Rad50 monoclonal antibody. Y.L. is an HHMI Predoctoral Fellow, and this research was supported by NIH R01 GM39799.
REFERENCES
- Adzuma K., Ogawa, T. and Ogawa, H. (1984) Primary structure of the RAD52 gene in Saccharomyces cerevisiae. Mol. Cell. Biol., 4, 2735–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley T., Plug, A.W., Xu, J., Solari, A.J., Reddy, G., Golub, E.I. and Ward, D.C. (1995) Dynamic changes in Rad51 distribution on chromatin during meiosis in male and female vertebrates. Chromosoma, 104, 19–28. [DOI] [PubMed] [Google Scholar]
- Barlow A.L., Benson, F.E., West, S.C. and Hulten, M.A. (1997) Distribution of the Rad51 recombinase in human and mouse spermatocytes. EMBO J., 16, 5207–5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P. and West, S.C. (1998) Role of the human RAD51 protein in homologous recombination and double-stranded-break repair. Trends Biochem. Sci., 23, 247–251. [DOI] [PubMed] [Google Scholar]
- Bell D.W., Wahrer, D.C.R., Kang, D.H., MacMahon, M.S., FitzGerald, M.G., Ishioka, C., Isselbacher, K.H., Krainer, M. and Haber, D. (1999) Common nonsense mutations in RAD52. Cancer Res., 59, 3883–3888. [PubMed] [Google Scholar]
- Benson F.E., Baumann, P. and West, S.C. (1998) Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature, 391, 401–404. [DOI] [PubMed] [Google Scholar]
- Bishop D.K., Ear, U., Bhattacharyya, A., Calderone, C., Beckett, M., Weichselbaum, R.R. and Shinohara, A. (1998) Xrcc3 is required for assembly of Rad51 complexes in vivo. J. Biol. Chem., 273, 21482–21488. [DOI] [PubMed] [Google Scholar]
- Chen J. et al. (1998) Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol. Cell, 2, 317–328. [DOI] [PubMed] [Google Scholar]
- Donovan J.W., Milne, G.T. and Weaver, D.T. (1994) Homotypic and heterotypic protein associations control Rad51 function in double-strand break repair. Genes Dev., 8, 2552–2562. [DOI] [PubMed] [Google Scholar]
- Gonzalez R., Silva, J.M., Dominguez, G., Garcia, J.M., Martinez, G., Vargas, J., Provencio, M., Espana, P. and Bonilla, F. (1999) Detection of loss of heterozygosity at RAD51, RAD52, RAD54 and BRCA1 and BRCA2 loci in breast cancer: pathological correlations. Br. J. Cancer, 81, 503–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb S. and Esposito, R.E. (1989) A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell, 56, 771–776. [DOI] [PubMed] [Google Scholar]
- Haaf T., Golub, E.I., Reddy, G., Radding, C.M. and Ward, D.C. (1995) Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc. Natl Acad. Sci. USA, 92, 2298–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber J.E. (1998) The many interfaces of Mre11. Cell, 95, 583–586. [DOI] [PubMed] [Google Scholar]
- Haber J.E. (1999) DNA repair. Gatekeepers of recombination. Nature, 398, 665–667. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski S.C. and Eggleston, A.K. (1994) Homologous pairing and DNA strand-exchange proteins. Annu. Rev. Biochem., 63, 991–1043. [DOI] [PubMed] [Google Scholar]
- Kurumizaka H., Aihara, H., Kagawa, W., Shibata, T. and Yokoyama, S. (1999) Human Rad52 amino acid residues required for Rad52 binding. J. Mol. Biol., 291, 537–548. [DOI] [PubMed] [Google Scholar]
- Le S., Moore, J.K., Haber, J.E. and Greider, C.W. (1999) RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics, 152, 143–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.-J. and Maizels, N. (1997) Nuclear Rad51 foci induced by DNA damage are distinct from Rad51 foci associated with B cell activation and recombination. Exp. Cell Res., 237, 93–100. [DOI] [PubMed] [Google Scholar]
- Li M.-J., Peakman, M.C., Golub, E.I., Reddy, G., Ward, D.C., Radding, C.M. and Maizels, N. (1996) Rad51 expression and localization in B cells carrying out class switch recombination. Proc. Natl Acad. Sci. USA, 93, 10222–10227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F., Han, M., Romanienko, P.J. and Jasin, M. (1998) Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc. Natl Acad. Sci. USA, 95, 5172–5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.H. and Keil, R.L. (1991) Mutations affecting RNA polymerase I-stimulated exchange and rDNA recombination in yeast. Genetics, 127, 31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Li, M.-J., Lee, E.Y. and Maizels, N. (1999) Localization and dynamic relocalization of mammalian Rad52 during the cell cycle and in response to DNA damage. Curr. Biol., 9, 975–978. [DOI] [PubMed] [Google Scholar]
- Maser R.S., Monsen, K.J., Nelms, B.E. and Petrini, J.H. (1997) hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Mol. Cell. Biol., 10, 6087–6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne G.T. and Weaver, D.T. (1993) Dominant negative alleles of RAD52 reveal a DNA repair/recombination complex including Rad51 and Rad52. Genes Dev., 7, 1755–1765. [DOI] [PubMed] [Google Scholar]
- Mizuta R., LaSalle, J.M., Cheng, H.L., Shinohara, A., Ogawa, H., Copeland, N., Jenkins, N.A., Lalande, M. and Alt, F.W. (1997) RAB22 and RAB163/mouse BRCA2: proteins that specifically interact with the RAD51 protein. Proc. Natl Acad. Sci. USA, 94, 6927–6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen U.H., Bendixen, C., Sunjevaric, I. and Rothstein, R. (1996) DNA strand annealing is promoted by the yeast Rad52 protein. Proc. Natl Acad. Sci. USA, 93, 10729–10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muris D.F. et al. (1994) Cloning of human and mouse genes homologous to RAD52, a yeast gene involved in DNA repair and recombination. Mutat. Res., 315, 295–305. [DOI] [PubMed] [Google Scholar]
- Nelms B.E., Maser, R.S., MacKay, J.F., Lagally, M.G. and Petrini, J.H. (1998) In situ visualization of DNA double-strand break repair in human fibroblasts. Science, 280, 590–592. [DOI] [PubMed] [Google Scholar]
- New J.H., Sugiyama, T., Zaitseva, E. and Kowalczykowski, S.C. (1998) Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature, 391, 407–410. [DOI] [PubMed] [Google Scholar]
- Park M.S. (1995) Expression of human RAD52 confers resistance to ionizing radiation in mammalian cells. J. Biol. Chem., 270, 15467–15470. [DOI] [PubMed] [Google Scholar]
- Park P.U., Defossez, P.A. and Guarente, L. (1999) Effects of mutations in DNA repair genes on formation of ribosomal DNA circles and life span in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 3848–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes T.D., Malone, R.E. and Symington, L.S. (1991) Genomic dynamics, protein synthesis and energetics. In Broach, J., Pringle, J. and Jones, E. (eds), The Molecular and Cellular Biology of the Yeast Saccharomyces. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 407–521.
- Petrini J.H. (1999) The mammalian Mre11–Rad50–nbs1 protein complex: integration of functions in the cellular DNA-damage response. Am. J. Hum. Genet., 64, 1264–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plug A.W., Xu, J., Reddy, G., Golub, E.I. and Ashley, T. (1996) Presynaptic association of Rad51 protein with selected sites in meiotic chromatin. Proc. Natl Acad. Sci. USA, 93, 5920–5924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raderschall E., Golub, E.I. and Haaf, T. (1999) Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage. Proc. Natl Acad. Sci. USA, 96, 1921–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick M.A. (1987) Meiosis. Academic Press, New York, NY.
- Rijkers T., Van Den Ouweland, J., Morolli, B., Rolink, A.G., Baarends, W.M., Van Sloun, P.P., Lohman, P.H. and Pastink, A. (1998) Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol. Cell. Biol., 18, 6423–6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon-Nguyen F., Brizard, F., Le Coniat, M., Radford, I., Berger, R. and Brizard, A. (1998) Abnormalities of the short arm of chromosome 12 in T cell prolymphocytic leukemia. Leukemia, 12, 972–975. [DOI] [PubMed] [Google Scholar]
- Scully R., Chen, J., Plug, A., Xiao, Y., Weaver, D., Feunteun, J., Ashley, T. and Livingston, D.M. (1997) Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell, 88, 265–275. [DOI] [PubMed] [Google Scholar]
- Sharan S.K. et al. (1997) Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature, 386, 804–810. [DOI] [PubMed] [Google Scholar]
- Shen Z., Denison, K., Lobb, R., Gatewood, J.M. and Chen, D.J. (1995) The human and mouse homologs of the yeast RAD52 gene: cDNA cloning, sequence analysis, assignment to human chromosome 12p12.2–p13, and mRNA expression in mouse tissues. Genomics, 25, 199–206. [DOI] [PubMed] [Google Scholar]
- Shen Z., Cloud, K.G., Chen, D.J. and Park, M.S. (1996) Specific interactions between the human RAD51 and RAD52 proteins. J. Biol. Chem., 271, 148–152. [DOI] [PubMed] [Google Scholar]
- Shinohara A. and Ogawa, T. (1995) Homologous recombination and the roles of double-strand breaks. Trends Biochem. Sci., 20, 387–391. [DOI] [PubMed] [Google Scholar]
- Shinohara A. and Ogawa, T. (1998) Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature, 391, 404–407. [DOI] [PubMed] [Google Scholar]
- Shinohara A., Ogawa, H. and Ogawa, T. (1992) Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell, 69, 457–470. [DOI] [PubMed] [Google Scholar]
- Sugiyama T., New, J.H. and Kowalczykowski, S.C. (1998) DNA annealing by RAD52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proc. Natl Acad. Sci. USA, 95, 6049–6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P. (1997) Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J. Biol. Chem., 272, 28194–28197. [DOI] [PubMed] [Google Scholar]
- Takata M., Sasaki, M.S., Sonoda, E., Morrison, C., Hashimoto, M., Utsumi, H., Yamaguchi-Iwai, Y., Shinohara, A. and Takeda, S. (1998) Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J., 17, 5497–5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan T.L., Essers, J., Citterio, E., Swagemakers, S.M., de Wit, J., Benson, F.E., Hoeijmakers, J.H. and Kanaar, R. (1999) Mouse Rad54 affects DNA conformation and DNA-damage-induced Rad51 foci formation. Curr. Biol., 9, 325–328. [DOI] [PubMed] [Google Scholar]
- Van Dyck E., Hajibagheri, N.M., Stasiak, A. and West, S.C. (1998) Visualisation of human Rad52 protein and its complexes with hRad51 and DNA. J. Mol. Biol., 284, 1027–1038. [DOI] [PubMed] [Google Scholar]
- Van Dyck E., Stasiak, A.Z., Stasiak, A. and West, S.C. (1999) Binding of double-strand breaks in DNA by human Rad52 protein. Nature, 398, 728–731. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Iwai Y., Sonoda, E., Buerstedde, J.M., Bezzubova, O., Morrison, C., Takata, M., Shinohara, A. and Takeda, S. (1998) Homologous recombination, but not DNA repair, is reduced in vertebrate cells deficient in RAD52. Mol. Cell. Biol., 18, 6430–6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Q., Chen, C.F., Li, S., Chen, Y., Wang, C.C., Xiao, J., Chen, P.L., Sharp, Z.D. and Lee, W.H. (1999) Association of BRCA1 with the hRad50–hMre11–p95 complex and the DNA damage response. Science, 285, 747–750. [DOI] [PubMed] [Google Scholar]
- Zou H. and Rothstein, R. (1997) Holliday junctions accumulate in replication mutants via a RecA homolog-independent mechanism. Cell, 90, 87–96. [DOI] [PubMed] [Google Scholar]