Abstract
Individuals affected by the autosomal recessive disorder Werner’s syndrome (WS) develop many of the symptoms characteristic of premature ageing. Primary fibroblasts cultured from WS patients exhibit karyotypic abnormalities and a reduced replicative life span. The WRN gene encodes a 3′–5′ DNA helicase, and is a member of the RecQ family, which also includes the product of the Bloom’s syndrome gene (BLM). In this work, we show that WRN promotes the ATP-dependent translocation of Holliday junctions, an activity that is also exhibited by BLM. In cells arrested in S-phase with hydroxyurea, WRN localizes to discrete nuclear foci that coincide with those formed by the single-stranded DNA binding protein replication protein A. These results are consistent with a model in which WRN prevents aberrant recombination events at sites of stalled replication forks by dissociating recombination intermediates.
INTRODUCTION
Patients with Werner’s syndrome (WS) exhibit pleiotropic properties characteristic of premature ageing, including the greying and loss of hair, cataract formation, osteoporosis, atherosclerosis, diabetes, hypogonadism and scleroderma (Epstein et al., 1966). Cultured WS cells display a limited capacity to proliferate and a prolonged S-phase (Martin et al., 1970). Moreover, WS cells exhibit evidence of genomic instability exemplified by chromosomal breaks, multiple large deletions, translocations and altered telomere dynamics (Fukuchi et al., 1989).
The gene mutated in WS, WRN, encodes a member of the RecQ family of DNA helicases. This family also includes the human RECQL, RECQ4, RECQ5 and BLM proteins, as well as the Saccharomyces cerevisiae Sgs1 protein (Chakraverty and Hickson, 1999). Mutations in the BLM gene lead to Bloom’s syndrome. Cells defective in BLM also exhibit genomic instability, the hallmark being an increased level of sister chromatid exchanges (German, 1993). In addition, BS patients are predisposed to all types of cancer.
Although the in vivo substrates of WRN remain unknown, in vitro studies indicate a role in the recognition and resolution of secondary structures in DNA. Of particular interest, WRN has been shown to interact with and dissociate guanine-rich tetraplex DNA structures (Fry and Loeb, 1999). Nevertheless, while tetraplex DNA can be readily formed in vitro, its existence in vivo has not been demonstrated, and it is difficult to reconcile this activity of the WRN protein with the gross chromosomal rearrangements observed in WS cells. Such rearrangements may be more indicative of aberrant recombination events caused by the lack of a functional WRN protein.
Recently, it has become clear that recombination plays an important role in the repair of stalled or broken replication forks, leading to the reinitiation of replication (reviewed in Haber, 1999; Rothstein et al., 2000). Defects in the processing of stalled replication forks could lead to aberrant recombination events culminating in genetic instability. Consistent with this proposal, S. cerevisiae sgs1 mutants exhibit a hyper-recombination phenotype, which can be partially suppressed by expression of WRN or BLM (Yamagata et al., 1998). WRN has been shown to co-purify with the DNA replication complex in ES cells and to interact directly with replication protein A (RPA), proliferating cell nuclear antigen (PCNA) and topoisomerase I (Brosh et al., 1999; Lebel et al., 2000).
To determine whether WRN might act upon replication forks that have been processed into structures resembling recombination intermediates, as predicted by current models, we analysed whether WRN protein could interact with and translocate Holliday junctions. By determining the subcellular distribution of WRN protein in HeLa cells arrested in S-phase, we also show that WRN is recruited to sites of blocked replication.
RESULTS AND DISCUSSION
Recombination intermediates (α-structures) were prepared by the actions of RecA protein with gapped circular and chimeric 3′-32P end-labelled linear duplex DNA (Figure 1A). In these experiments, the use of a [3′-32P]dideoxy end-label minimized complications due to the intrinsic exonuclease activity of WRN (Kamath-Loeb et al., 1998). Additionally, recombination intermediates formed between these substrates were stabilized by a region of sequence heterology at the distal end of the linear duplex that prevented the completion of RecA-mediated strand exchange (Eggleston et al., 1997). Proteins with branch migration activity, such as the Escherichia coli RuvAB complex, interact with recombination intermediates (Figure 1B, lane b) and catalyse translocation of the Holliday junction through the 2765 bp region of homology, leading to dissociation of the α-structures. The presence of 32P-labelled linear duplex product after analysis by agarose gel electrophoresis indicates completion of the branch migration reaction (Figure 1B, lane a).
Fig. 1. Dissociation of recombination intermediates by WRN. (A) Schematic diagram indicating recombination intermediates (α-structures) and the expected products of branch migration. A region of heterology (1670 bp) at the distal end of the linear duplex blocks strand exchange and is represented by a filled box. The positions of the 32P-labels are indicated with asterisks. (B) Activity of WRN protein on recombination intermediates. Purified recombination intermediates (1.5 µM, expressed as a concentration of nucleotide residues) were incubated with: lane a, 200 nM RuvA and 100 nM RuvB; lanes c–g, 0.7–10.5 nM WRN; lanes i–m, 5.2 nM WRN and RuvA as indicated. In the reactions indicated, purified recombination intermediates were pre-incubated for 3 min on ice with RuvA before addition of WRN. The positions of the recombination intermediates (α) and linear duplex DNA (L) are indicated.
Although WRN is a relatively poor helicase on DNA duplex substrates >50 bp (Brosh et al., 1999), we found that it dissociated the recombination intermediates by moving the junction through >2700 bp (Figure 1B, lanes c–g). In addition to making α-structures as shown, RecA also promotes the formation of DNA aggregates that cannot enter the gel, which are thought to represent secondary interactions between the recombination intermediates and linear or gapped duplex DNA (Müller et al., 1992). These complex DNA structures were also disrupted by WRN.
Given the weak processivity of WRN on duplex DNA substrates, it seemed unlikely that dissociation of the α-structures would be a consequence of non-specific helicase activity. However, to confirm that WRN initiated dissociation of the recombination intermediates at the junction, we determined whether RuvA protein could act as a competitor. RuvA is known to have a high affinity for Holliday junctions (Parsons et al., 1992) and was expected to block access to WRN. We found that a short pre-incubation of the α-structure with RuvA inhibited branch migration by WRN (Figure 1B, lanes i–m).
When reactions carried out in the presence of ATP were quantified by phosphoimaging, we found that ~50% of the α-structures were converted to products (32P-labelled linear duplex and gapped circular DNA) after 15 min (Figure 2A and C). Under these reaction conditions, the amount of dissociation due to spontaneous branch migration was negligible compared with that catalysed by WRN (data not shown). In reactions carried out in the absence of ATP (Figure 2C), or in the presence of a non-hydrolysable analogue of ATP, AMP-PNP (Figure 2B and C), we failed to observe WRN-mediated branch migration. These results show that ATP hydrolysis is required for WRN-catalysed dissociation of the α-structure.
Fig. 2. Dependence on ATP hydrolysis. Dissociation reactions were carried out as described in the legend to Figure 1 using 1.5 µM 32P-labelled recombination intermediates and 5 nM WRN, in (A) standard buffer or (B) standard buffer in which ATP was replaced with 2 mM AMP-PNP. At the times indicated, aliquots were withdrawn and analysed by gel electrophoresis. (C) Quantification of reactions carried out in standard buffer containing ATP or AMP-PNP (A and B), or in the absence of NTPs (primary data not shown), as indicated. The amount of 32P-labelled linear duplex branch migration product was quantified by phosphoimaging and expressed as a percentage of the total radiolabel.
The WRN protein is known to exhibit a higher affinity for single-stranded DNA (ssDNA) than for double-stranded DNA (dsDNA) (Orren et al., 1999). When 32P-labelled synthetic four-way DNA junctions, which resemble Holliday junctions, were incubated with WRN we observed complexes that exhibited retarded mobility during agarose gel electrophoresis (Figure 3A, lane b). The ability of unlabelled dsDNA and ssDNA to act as competitors to the formation of the junction–WRN complex is shown in Figure 3A and B, respectively. Duplex DNA was found to be a poor competitor compared with ssDNA. Indeed, ssDNA competed as effectively as unlabelled four-way junctions (Figure 3C). One interpretation of these data is that WRN is targeted to DNA containing secondary structure and that the actions of WRN on recombination intermediates are due at least in part to the recognition of the junction within the duplex DNA substrate. Consistent with this proposal, the S. cerevisiae homologue Sgs1 displays a strong affinity for branched DNA (Bennett et al., 1999).
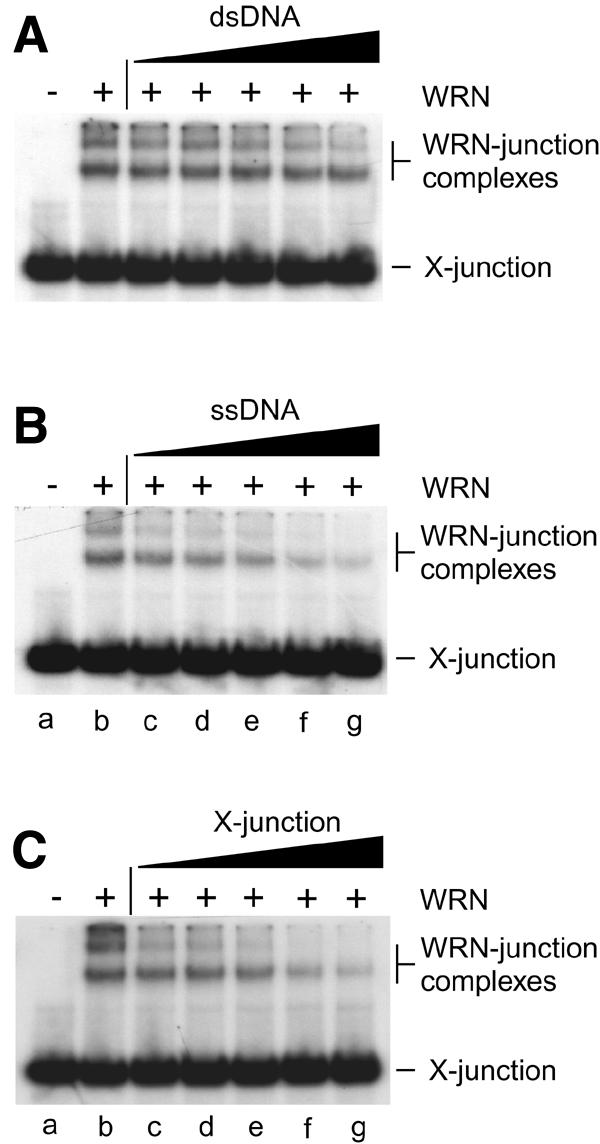
Fig. 3. Specificity of binding of WRN to synthetic Holliday junctions. Band-shift assays contained 7.3 nM WRN protein (indicated by +), 1 nM 32P-labelled synthetic Holliday junction (X-junction) and unlabelled DNA competitor as indicated. The competitors were (A) dsDNA, (B) ssDNA and (C) synthetic X-junction DNA, and were used at concentrations of 1.5 nM (lanes c), 3.1 nM (lanes d), 6.25 nM (lanes e), 12.5 nM (lanes f) and 25 nM (lanes g). DNA concentrations are expressed in moles of DNA substrate.
Since WRN lacks high specificity for the Holliday structure and is active upon a variety of substrates containing secondary structures (Brosh et al., 1999; Fry and Loeb, 1999), we do not suggest that the activities described in this paper indicate that WRN is a Holliday junction-specific branch migration protein like RuvAB. Instead, we suggest that WRN functions as a structure-specific helicase that has the ability to recognize and translocate branched structures. As such, the enzyme may be capable of dissociating a variety of structural distortions from duplex DNA.
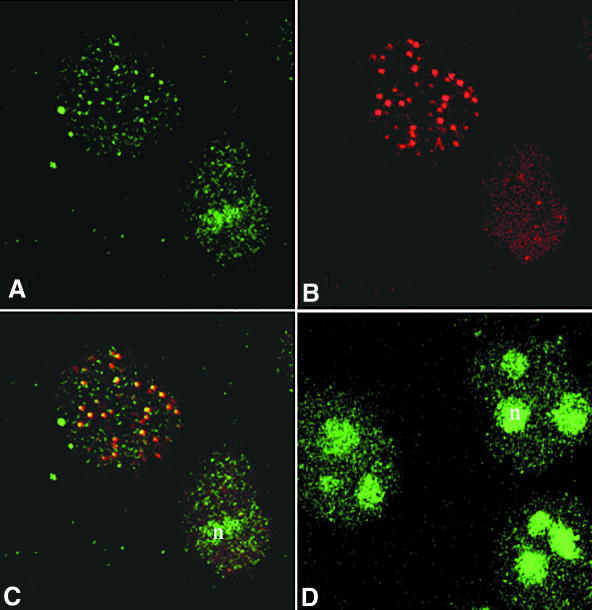
The human WRN protein has been shown to localize to the nucleoli in a variety of cell types, including HeLa, where anti-WRN antibodies give a diffuse staining of the nucleolar chromatin (Marciniak et al., 1998; Figure 4D). To determine whether WRN changes its localization when the cell cycle is blocked, we treated HeLa cells with hydroxyurea (HU), an agent known to deplete the cell of its dNTP supply, resulting in S-phase arrest with stalled replication forks. Cells in S-phase were identified by the distribution of the S-phase-specific PCNA protein (Jonsson and Hubscher, 1997), which was concentrated in distinct foci in 98% of the HU-treated HeLa nuclei (data not shown). In >50% of the treated cells, WRN was not detected in the nucleolus. Instead, the majority of cells exhibited a punctated pattern of nuclear staining characteristic of replication foci (Figure 4A). Staining with an anti-RPA antibody (Figure 4B) showed that the WRN foci coincided with RPA-formed foci (Figure 4C). This co-localization pattern indicates that WRN is recruited from the nucleoli and re-positioned at sites of stalled replication when a replication block is imposed.
Fig. 4. Co-localization of WRN and RPA proteins in cells arrested in S-phase with HU. (A–C) Cells were arrested in S-phase by HU treatment as described. Following lysis, fixation and permeabilization, cells were incubated with anti-WRN (A; green) or anti-RPA antibodies (B; red). In >50% of the cells, WRN localizes in nuclear foci that coincide with the RPA foci, as illustrated in the superimposed images (C). The yellow colour results from the overlapping of the red and green foci. In cells incubated for the same length of time in the absence of HU, WRN shows the characteristic diffuse nucleolar (n) localization (D).
Consistent with the immunofluorescence data, other lines of evidence also suggest that WRN may be associated with the DNA replication machinery: (i) WRN co-purifies with the multiprotein DNA replication complex (Lebel and Leder, 1998); (ii) purified WRN and RPA interact by co-immunoprecipitation, and the presence of RPA significantly increases the helicase activity of WRN on DNA duplex substrates (Brosh et al., 1999); (iii) WRN interacts with PCNA and topoisomerase I as part of a DNA replication complex (Lebel et al., 2000); and (iv) the Xenopus laevis FFA-1 protein (foci-forming activity-1), which is 66% similar to the WRN protein, has been shown to be essential for the formation of replication origin complexes and RPA aggregation (Yan et al., 1998).
In summary, we have shown that WRN can dissociate recombination intermediates and is redistributed upon replication arrest to distinct nuclear foci, where it co-localizes with RPA. These observations indicate a role for WRN in the processing of potentially recombinogenic broken replication forks. We suggest that WRN may be required for the prevention of illegitimate recombination events that could occur in regions of repeated sequences that would otherwise lead to chromosomal rearrangements. Given the recent finding that BLM can promote related branch migration reactions (Karow et al., 2000), we suggest that the human RecQ family members define a class of anti-recombinases that provide overlapping, although non-identical, functions critical for the maintenance of genomic stability during DNA metabolism.
METHODS
Proteins. Recombinant human WRN and E. coli RuvA, RuvB and RecA proteins were purified as described (Tsaneva et al., 1992; Eggleston et al., 1997; Orren et al., 1999). Terminal deoxynucleotidyl transferase, T4 polynucleotide kinase and restriction enzymes were obtained from commercial suppliers. Protein concentrations are indicated in moles of monomer.
DNA substrates. Recombination intermediates were generated by RecA-mediated strand exchange between gapped circular and 32P-labelled linear duplex DNA, deproteinized and purified as described (Eggleston et al., 1997). Gapped circular pDEA-7Z DNA was prepared as described (Eggleston et al., 1997). Linear duplex DNA was produced by Pst I digestion of pAKE-4M plasmid DNA (4674 bp) followed by 3′-32P end-labelling using terminal transferase and [α-32P]dideoxy ATP (Karow et al., 2000). The synthetic Holliday junction (X12) and linear duplex DNA were prepared by annealing 50mer oligonucleotides (Karow et al., 2000). The junction was 5′-32P-labelled in oligo 1. The ssDNA competitor was also 50 nucleotides long, as described (Karow et al., 2000).
Assay of Holliday junction translocation. WRN reactions contained 20 mM Tris–HCl pH 7.5, 1.25 mM MgCl2, 2 mM ATP, 0.1 mg/ml bovine serum albumin (BSA) and 1 mM dithiothreitol (DTT) (standard buffer). The RuvAB reaction was carried out in the same buffer except that the MgCl2 concentration was 15 mM. After 45 min at 37°C, DNA products were deproteinized and separated on 1% agarose gels in TAE buffer supplemented with 0.5 µg/ml ethidium bromide (Eggleston et al., 1997). 32P-labelled DNA was detected by autoradiography.
Band-shift assays. Proteins and DNA were incubated in 20 mM triethanolamine–HCl pH 7.5, 2 mM MgCl2, 1 mM ATPγS, 0.1 mg/ml BSA and 1 mM DTT. Incubation was for 30 min at room temperature. Protein–DNA complexes were fixed in the presence of 0.25% glutaraldehyde for 10 min at 37°C (Eggleston et al., 1997) and visualized by autoradiography after electrophoresis through a 5% native polyacrylamide gel.
Immunofluorescence studies. HeLa cells were grown on coverslips in MEM medium suplemented with 10% fetal bovine serum for 36 h, and then incubated for 15 h in the presence or absence of 2 mM HU (Sigma). Cells were washed three times in phosphate-buffered saline, hypotonically lysed in 0.5% NaCl pH 7.5, fixed in 4% paraformaldehyde for 30 min and permeabilized with 0.5% Triton X-100 for 10 min. Primary antibody incubation was for 2 h and secondary antibody for 1 h at room temperature. Primary antibodies were rabbit polyclonal anti-WRN (Novus) diluted 1:1000 and mouse monoclonal anti-RPA (a gift from M. Shivji and R. Wood) diluted 1:100. The antigens were visualized with the secondary antibodies Alexa488-conjugated goat anti-rabbit and Alexa546-conjugated goat anti-mouse (Molecular Probes), both used at a 1:1000 dilution. Images were obtained with a Zeiss Laser Scanning Microscope LSM 510 equipped with a photomultiplier.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by the Imperial Cancer Research Fund and the Human Frontiers Science Program. A.C. was supported by a European Science Exchange Fellowship from the Royal Society and the Swiss National Foundation, M.T. was supported by an EMBO Fellowship and J.K.K. is a Boehringer Ingelheim Fonds scholar.
REFERENCES
- Bennett R.J., Keck, J.L. and Wang, J.C. (1999) Binding specificity determines polarity of DNA unwinding by the Sgs1 protein of S. cerevisiae. J. Mol. Biol., 289, 235–248. [DOI] [PubMed] [Google Scholar]
- Brosh R.M., Orren, D.K., Nehlin, J.O., Ravn, P.H., Kenny, M.K., Machwe, A. and Bohr, V.A. (1999) Functional and physical interaction between WRN helicase and human replication protein A. J. Biol. Chem., 274, 18341–18350. [DOI] [PubMed] [Google Scholar]
- Chakraverty R.K. and Hickson, I.D. (1999) Defending genome integrity during DNA replication: a proposed role for RecQ family helicases. BioEssays, 21, 286–294. [DOI] [PubMed] [Google Scholar]
- Eggleston A.K., Mitchell, A.H. and West, S.C. (1997) In vitro reconstitution of the late steps of genetic recombination in E. coli. Cell, 89, 607–617. [DOI] [PubMed] [Google Scholar]
- Epstein C.J., Martin, G.M., Schultz, A.L. and Motulsky, A.G. (1966) Werner’s syndrome: a review of its symptomatology, natural history, pathologic features, genetics and relationship to the natural aging process. Medicine, 45, 177–221. [DOI] [PubMed] [Google Scholar]
- Fry M. and Loeb, L.A. (1999) Human Werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J. Biol. Chem., 274, 12797–12802. [DOI] [PubMed] [Google Scholar]
- Fukuchi K., Martin, G.M. and Monnat, R.J. (1989) Mutator phenotype of Werner syndrome is characterized by extensive deletions. Proc. Natl Acad. Sci. USA, 86, 5893–5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German J. (1993) Bloom syndrome: a mendelian prototype of somatic mutational disease. Medicine, 72, 393–406. [PubMed] [Google Scholar]
- Haber J.E. (1999) DNA recombination: the replication connection. Trends Biochem. Sci., 24, 271–275. [DOI] [PubMed] [Google Scholar]
- Jonsson Z.O. and Hubscher, U. (1997) Proliferating cell nuclear antigen: more than a clamp for DNA polymerases. BioEssays, 19, 967–975. [DOI] [PubMed] [Google Scholar]
- Kamath-Loeb A.S., Shen, J.C., Loeb, L.A. and Fry, M. (1998) Werner syndrome protein: II. Characterization of the integral 3′–5′ DNA exonuclease. J. Biol. Chem., 273, 34145–34150. [DOI] [PubMed] [Google Scholar]
- Karow J.K., Constantinou, A., Li, J.-L., West, S.C. and Hickson, I.D. (2000) The Bloom’s Syndrome gene product promotes branch migration of Holliday junctions. Proc. Natl Acad. Sci. USA, 97, 6504–6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel M. and Leder, P. (1998) A deletion within the murine Werner syndrome helicase induces sensitivity to inhibitors of topoisomerase and loss of cellular proliferative capacity. Proc. Natl Acad. Sci. USA, 95, 13097–13102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel M., Spillare, E.A., Harris, C.C. and Leder, P. (2000) The Werner syndrome gene product co-purifies with the DNA replication complex and interacts with PCNA and topoisomerase I. J. Biol. Chem., 274, 37795–37799. [DOI] [PubMed] [Google Scholar]
- Marciniak R.A., Lombard, D.B., Johnson, F.B. and Guarente, L. (1998) Nucleolar localization of the Werner syndrome protein in human cells. Proc. Natl Acad. Sci. USA, 95, 6887–6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G.M., Sprague, C.A. and Epstein, C.J. (1970) Replicative life-span of cultivated human cells. Effects of donor’s age, tissue, and genotype. Lab. Invest., 23, 86–92. [PubMed] [Google Scholar]
- Müller B., Burdett, I. and West, S.C. (1992) Unusual stability of recombination intermediates made by Escherichia coli RecA protein. EMBO J., 11, 2685–2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orren D.K., Brosh, R.M., Nehlin, J.O., Machwe, A., Gray, M.D. and Bohr, V.A. (1999) Enzymatic and DNA binding properties of purified WRN protein: high affinity binding to single-stranded DNA but not to DNA damage induced by 4NQO. Nucleic Acids Res., 27, 3557–3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons C.A., Tsaneva, I., Lloyd, R.G. and West, S.C. (1992) Interaction of Escherichia coli RuvA and RuvB proteins with synthetic Holliday junctions. Proc. Natl Acad. Sci. USA, 89, 5452–5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R., Michel, B. and Gangloff, S. (2000) Replication fork pausing and recombination or ‘gimme a break’. Genes Dev., 14, 1–10. [PubMed] [Google Scholar]
- Tsaneva I.R., Illing, G.T., Lloyd, R.G. and West, S.C. (1992) Purification and properties of the RuvA and RuvB proteins of Escherichia coli. Mol. Gen. Genet., 235, 1–10. [DOI] [PubMed] [Google Scholar]
- Yamagata K., Kato, J., Shimamoto, A., Goto, M., Furuichi, Y. and Ikeda, H. (1998) Bloom’s and Werner’s syndrome genes suppress hyper-recombination in yeast SGS1 mutant: implication for genomic instability in human diseases. Proc. Natl Acad. Sci. USA, 95, 8733–8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H., Chen, C.Y., Kobayashi, R. and Newport, J. (1998) Replication focus-forming activity 1 and the Werner syndrome gene product. Nature Genet., 19, 375–378. [DOI] [PubMed] [Google Scholar]