Abstract
FtsH-mediated proteolysis against membrane proteins is processive, and presumably involves dislocation of the substrate into the cytosol where the enzymatic domains of FtsH reside. To study how such a mode of proteolysis is initiated, we manipulated N-terminal cytosolic tails of three membrane proteins. YccA, a natural substrate of FtsH was found to require the N-terminal tail of 20 amino acid residues or longer to be degraded by FtsH in vivo. Three unrelated sequences of this segment conferred the FtsH sensitivity to YccA. An artificially constructed TM9-PhoA protein, derived from SecY, as well as the SecE protein, were sensitized to FtsH by addition of extra amino acid sequences to their N-terminal cytosolic tails. Thus, FtsH recognizes a cytosolic region of sufficient length (∼20 amino acids) to initiate the processive proteolysis against membrane proteins. Such a region is typically at the N-terminus and can be diverse in amino acid sequences.
INTRODUCTION
Degradation of abnormal membrane proteins will comprise a quality control mechanism to keep the integrity of biological membranes. FtsH in Escherichia coli, a membrane-bound enzyme having ATPase and metalloproteinase activities, is responsible for degradation of certain integral membrane proteins, including the SecY subunit of the protein translocase (Kihara et al., 1995), the a subunit of the H+-ATPase (Akiyama et al., 1996) and the product of the yccA open reading frame (Kihara et al., 1998). It also degrades some short-lived cytosolic proteins (see Ogura et al., 1999 and references therein).
YccA of unknown function has seven transmembrane segments with a cytosolic N-terminus and a periplasmic C-terminus. FtsH initiates proteolysis of this protein at the N-terminal cytosolic region, and continues the processive degradation into the periplasmic regions (Kihara et al., 1999). Thus, the periplasmic PhoA (alkaline phosphatase) reporter domain in the YccA-(P3)-PhoA-His6-Myc fusion protein was rapidly degraded by FtsH when it lacked a tight folding. No detectable degradation intermediates were produced under such conditions. However, the FtsH-dependent degradation stopped at a site near to the N-terminus of the PhoA moiety under conditions that allowed the disulfide bond-induced tight folding of PhoA. Similar results were also obtained for the SecY-(P5)-PhoA fusion protein. It was suggested that, instead of being degraded successively by FtsH in conjunction with other protease(s), both transmembrane and periplasmic domains of membrane protein substrates are degraded by the processive proteolysis by FtsH (Kihara et al., 1999). We propose that FtsH dislocates the extracytoplasmic domains of substrates using its ATPase activity, provided that they are not tightly folded.
Given this mechanism, it is important to know how FtsH initiates proteolysis against a membrane protein. This work was aimed at characterizing features of substrate proteins that are recognized by FtsH for the initiation of proteolysis. The results revealed the importance of the length, but not exact amino acid sequence, at the N-terminal tail.
RESULTS
Unrelated amino acid sequences at the N-terminal region of YccA confer FtsH sensitivity
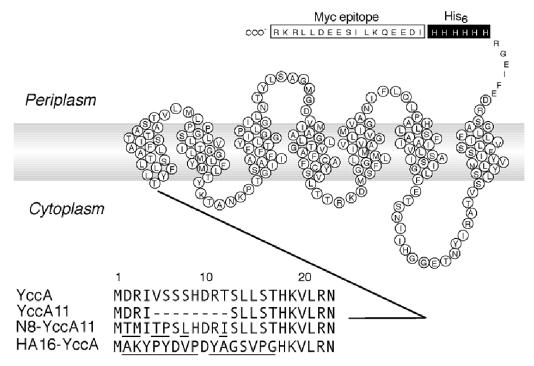
The N-terminal cytosolic tail of YccA (C1 domain) is important for the initiation of degradation (Kihara et al., 1999). We examined the stability of three different derivatives of YccA (Figure 1) by pulse–chase experiments. YccA11 and N8-YccA11 were described previously (Kihara et al., 1998, 1999), whereas HA16-YccA had the residues 2–17 replaced by a completely unrelated sequence of the same length derived from the HA epitope. All the YccA constructs in this study carried the His6-Myc tag sequence at the C-terminus, allowing their detection with anti-Myc.
Fig. 1. YccA derivatives used in this study. The YccA amino acid sequence, other than the N-terminal 23 residues followed by the C-terminally attached His6 and Myc sequences are shown schematically representing the topology model (Kihara et al., 1999). The amino acid sequences in the N-terminal region of YccA and its derivatives are shown in the lower part. Residues that are non-identical to the wild-type residues are underlined.
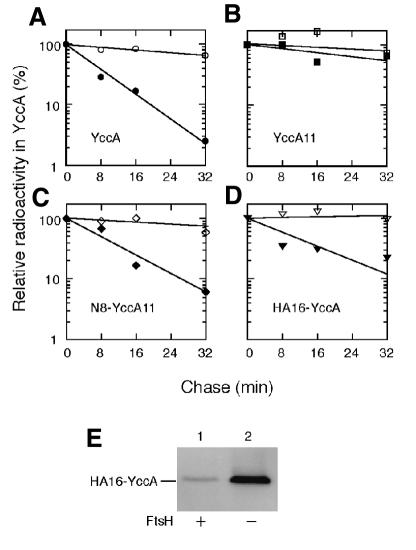
Whereas YccA11 was stable (Figure 2B), both YccA and N8-YccA11 were degraded with a half-life of ∼5–8 min in the ftsH+ cells (Figure 2A and C). In addition, HA16-YccA was degraded with a half-life of ∼10 min (Figure 2D). Degradation of HA16-YccA was FtsH dependent, since it was stabilized completely in the absence of FtsH (Figure 2D, open triangles). FtsH-dependent degradation of HA16-YccA was confirmed by examining its accumulation levels in the ftsH+ and ΔftsH cells (Figure 2E). Thus, at least three different sequences at the N-terminal region, those of wild-type YccA, N8-YccA11 and HA16-YccA, conferred FtsH sensitivity on this membrane protein.
Fig. 2. Stability of YccA derivatives with engineered N-terminal tails. (A–D) Pulse–chase examinations of YccA derivatives. Each of strains AR5087 (ftsH+; solid symbols) and AR5090 (ΔftsH; open symbols) was transformed with pKH330 (for YccA-His6-Myc; A), pKH331 (YccA11-His6-Myc; B), pKH356 (N8-YccA11-His6-Myc; C) or pCH55 (HA16-YccA-His6-Myc; D), grown at 37°C, and induced for lac transcription for 10 min. Cells were then pulse-labeled with [35S]methionine for 1 min followed by chase with unlabeled methionine for 1, 8, 16 and 32 min. Labeled proteins were immunoprecipitated with anti-Myc and separated by SDS–PAGE. Radioactivities were reported as values relative to that at the 1 min chase point. (E) Cellular accumulation of HA16-YccA. Strains AR5087 (ftsH+; lane 1) and AR5090 (ΔftsH; lane 2), each carrying pCH55 (HA16-YccA-His6-Myc), were grown in L-broth–glucose containing ampicillin at 37°C and induced for lac transcription for 2 h. Whole-cell proteins were subjected to SDS–PAGE and anti-Myc immunoblotting.
The length of the cytosolic tail is crucial for the FtsH susceptibility of YccA
To address the possibility that the length of the N-terminal cytosolic region, rather than the amino acid sequence, determines the FtsH susceptibility of YccA, we constructed a series of YccA mutants, in which the N-terminal tail was gradually shortened by removing the Asp2 residue and its C-terminal neighbors (Figure 3A). The abundance of each of the mutant proteins in the ftsH+ and ΔftsH cells was determined by immunoblotting (Figure 3B). Although some of the constructs were expressed poorly (e.g. Figure 3B, lane 8), due presumably to an altered messenger RNA structure, comparisons between the two host strains gave unequivocal information as to their FtsH susceptibility. YccAd1 (Figure 3B, lanes 5 and 6), YccAd2 (lanes 7 and 8) and YccAd3 (lanes 9 and 10) behaved like the wild-type YccA protein (lanes 3 and 4) in that they accumulated in the ΔftsH cells, but not significantly in the ftsH+ cells. In contrast, all the other mutant proteins, lacking four residues or more, accumulated to essentially identical levels in the two host strains (Figure 3B, lanes 11–18). Thus, FtsH sensitivity of YccA changed discontinuously as the N-terminal cytosolic tail was shortened. Assuming that Asn23 is the last cytosolic residue of this segment, FtsH requires a specific threshold length of 20 residues at the cytosolic N-terminal tail of YccA. Without structural information, this value should be regarded as an approximation. In addition, Met1 may be removed posttranslationally (especially for the d4, d5 and d14 constructs; Flinta et al., 1986), but this does not essentially affect our conclusion.
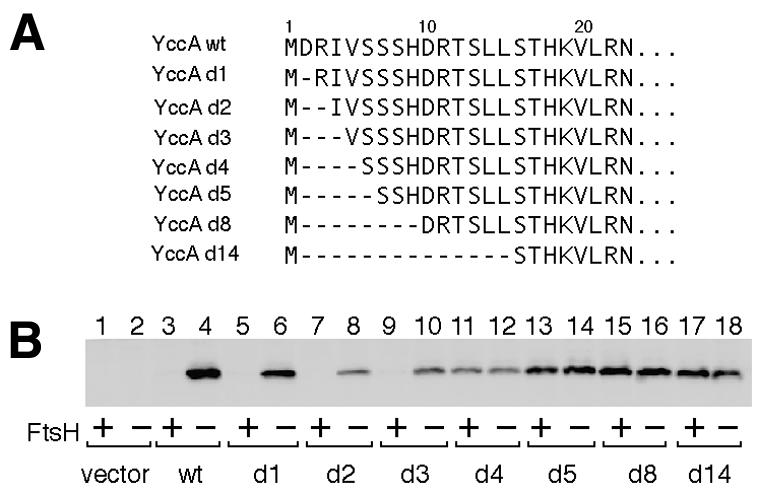
Fig. 3. Importance of the chain length at the N-terminal end of YccA for its degradation by FtsH. (A) Amino acid sequences of the N-terminal region of the YccA deletion mutants used. (B) Steady-state accumulation of YccA derivatives in the presence and absence of FtsH. Each of strains AR5087 (ftsH+; odd-numbered lanes) and AR5090 (ΔftsH; even-numbered lanes) was transformed either with pMW119H (vector; lanes 1 and 2), pCH10 (YccA-His6-Myc; lane 3 and 4), pCH20 (YccAd1-His6-Myc; lanes 5 and 6), pCH11 (YccAd2-His6-Myc; lanes 7 and 8), pCH21 (YccAd3-His6-Myc; lanes 9 and 10), pCH22 (YccAd4-His6-Myc; lanes 11 and 12), pCH12 (YccAd5-His6-Myc; lanes 13 and 14), pCH13 (YccAd8-His6-Myc; lanes 15 and 16) or pCH17 (YccAd14-His6-Myc; lanes 17 and 18). Plasmid-bearing cells were grown in L-broth–glucose supplemented with ampicillin at 37°C. Plasmid-encoded proteins were induced for 4.5 h. Anti-Myc-reacting proteins were visualized after SDS–PAGE of whole-cell proteins.
Sensitization of an artificial TM9-PhoA protein to FtsH by extension of its N-terminal tail
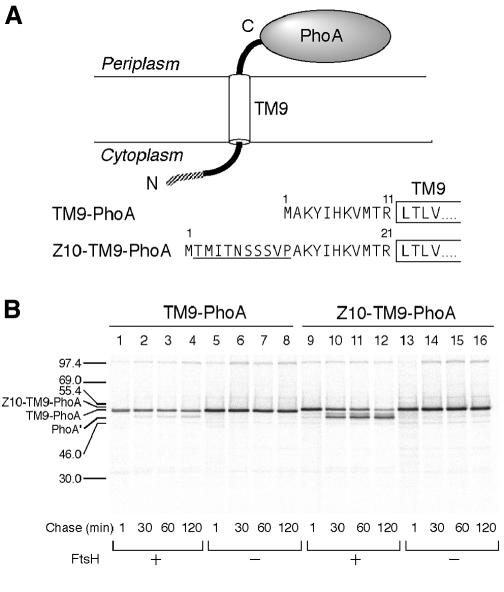
SecY has 10 transmembrane (TM1–TM10), six cytosolic (C1–C6) and five periplasmic (P1–P5) regions, with both of its termini facing the cytosol (Akiyama and Ito, 1987). The SecY-(P5)-PhoA fusion protein, composed of an N-terminal region of SecY up to TM9 followed by a part of P5 and PhoA, is degraded in an FtsH-dependent manner in vivo (Kihara et al., 1999). The degradation of the TM9-PhoA region requires its covalent attachment to the N-terminal region of SecY (Kihara et al., 1999). Thus, an independently expressed TM9-PhoA construct, having the predicted 11 amino acid residues at the N-terminal tail (Figure 4A), was insensitive to FtsH (Kihara et al., 1999; Figure 3B, lanes 1–8).
Fig. 4. Sensitization of TM9-PhoA to FtsH by addition of extra amino acids to its N-terminus. (A) Amino acid sequences of the predicted N-terminal tail of TM9-PhoA and Z10-TM9-PhoA. The extra sequence in the latter is underlined. (B) Stability of TM9-PhoA and Z10-TM9-PhoA. Each of strains AR5087 (ftsH+; lanes 1–4 and 9–12) and AR5090 (ΔftsH; lanes 4–8 and 13–16) was transformed either with pKH472 (TM9-PhoA; lanes 1–8) or pCH43 (Z10-TM9-PhoA; lanes 9–16). The plasmid-bearing cells were grown at 28°C, and induced for lac transcription for 9 min. Cells were pulse-labeled with [35S]methionine for 1.5 min followed by chase with unlabeled methionine for 1, 30, 60 and 120 min. Labeled proteins were immunoprecipitated with anti-PhoA, separated by SDS–PAGE, and visualized. PhoA′ indicates the degradation product.
We inserted a 10 residue sequence derived from LacZ into the N-terminal region of TM9-PhoA (Z10-TM9-PhoA in Figure 4A). Z10-TM9-PhoA was found to be converted into a lower molecular mass product in the ftsH+ cells (Figure 4B, lanes 9–12) but not in the ΔftsH cells (lanes 13–16). The product of this degradation was similar to the PhoA′ fragment we observed with the SecY-(P5)-PhoA protein in the dsbA+ cells; presumably, the tight folding of the PhoA domain interferes with the dislocation process (Kihara et al., 1999). Thus, TM9-PhoA can be converted to a substrate of FtsH when its N-terminal region is extended to a sufficient length (∼21 residues in this case).
Sensitization of SecE, a native membrane protein, to FtsH by extension of its N-terminal tail
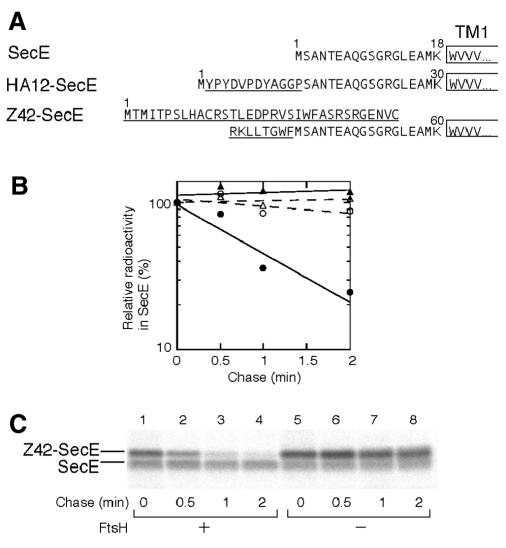
SecE, with three transmembrane segments, has the predicted 18 amino acid residues at the N-terminal cytosolic region (Schatz et al., 1989). We elongated this region by 12 residues (HA12-SecE in Figure 5A). HA12-SecE, overproduced in ftsH+ cells, was found to be degraded extremely rapidly, with a half life of <2 min (Figure 5B, solid circles). In contrast, SecE was stable within this timescale (Figure 5B, triangles). HA12-SecE was stable in the ΔftsH cells (Figure 5B, open circle), showing that FtsH was responsible for the rapid degradation.
Fig. 5. Sensitization of SecE to FtsH by addition of extra amino acids to its N-terminus. (A) Amino acid sequences at the N-terminal cytosolic regions of SecE, HA12-SecE and Z42-SecE. The extra sequences are underlined. (B) Stability of SecE and HA12-SecE. Each of strains AR5087 (ftsH+; solid symbols) and AR5090 (DftsH; open symbols) was transformed either with pCM134 (SecE; circles) or pHAsecE (HA12-SecE; triangles). Plasmid-bearing cells were grown at 37°C, and induced for 10 min for lac or ara transcription. Cells were then pulse-labeled with [35S]methionine for 0.5 min followed by chase with unlabeled methionine for 0, 0.5, 1 and 2 min. Labeled proteins were immunoprecipitated with anti-SecE and separated by SDS–PAGE. Radioactivities are reported as values relative to that at the 0 min chase point. (C) Stability of SecE and Z42-SecE. Cells of strains AR5087 (ftsH+; lanes 1–4) and AR5090 (ΔftsH; lanes 5–8), each carrying pKY271 (Z42-SecE/SecE), were grown, induced for lac transcription, and pulse-labeled for 0.5 min with [35S]methionine followed by chase for 0 (lanes 1 and 5), 0.5 (lanes 2 and 6), 1 (lanes 3 and 7) and 2 min (lanes 4 and 8). Labeled proteins were immunoprecipitated with anti-SecE.
We also used a secE clone in which secE had been fused in-frame to the upstream lacZ sequence on the vector, producing two products, authentic SecE and a variant SecE with an N-terminally attached 42 amino acid extension (Z42-SecE in Figure 5A). Z42-SecE was degraded rapidly (half life <1 min) in the presence of FtsH (Figure 5C, lanes 1–4) but not in its absence (lanes 5–8). The internally initiated SecE in the same set of cells was stable (Figure 5C), serving as an internal control. HA12-SecE and Z42-SecE had completely different sequences at their N-terminal regions. These results give additional examples for the N-tail-dependent but sequence-indifferent initiation of membrane protein degradation by FtsH.
DISCUSSION
The proteolytic and ATPase domains of FtsH are essentially cytosolic (Tomoyasu et al., 1993). The roles of the ATPase subfunctions in proteolysis are believed to be in the unfolding of a substrate polypeptide and its translocation to the protease active site present in the cavity of the assembly (De Mot et al., 1999). Like all the other known ATP-dependent proteases (De Mot et al., 1999), FtsH forms an oligomeric structure (Akiyama et al., 1995), probably ring-like (Shotland et al., 1997). The AAA ATPase of FtsH may function in substrate unfolding/presentation by dislocating the membrane-integrated substrate out of the membrane (Kihara et al., 1999). This mechanism will fit nicely a notion that extensive hydrolysis of peptide bonds as well as of ATP for thoroughly scavenging an abnormal membrane protein is best carried out in the aqueous cytosolic phase. Such a mechanism can be contrasted to the recently described proteolytic cleavage within the membrane integrated region, which leads to the liberation of a distinct product with a biological activity (Brown et al., 2000). The latter mode of proteolysis within the membranous environment could be energy independent and of low processivity, although this has not been shown experimentally.
We have shown that FtsH initiates proteolysis of membrane proteins only when they possess a cytosolic region of sufficient length (∼20 residues). While such an initiation region can definitely be at the N-terminal tail, this is probably not the exclusive rule. At least one substrate, subunit a of the H+-ATPase (Akiyama et al., 1996), has its N-terminus facing the periplasm (Long et al., 1998; Valiyaveetil and Fillingame, 1998; Wada et al., 1999). It was also reported that degradation of some cytosolic proteins by FtsH requires their C-terminal regions (Herman et al., 1998; Blaszczak et al., 1999). We observed that highly overproduced SecE was degraded slowly (half life ∼30 min) and FtsH contributed to this degradation (our unpublished results). Either the N-terminal tail of wild-type SecE (∼18 amino acids) or the internal cytoplasmic loop may be recognized inefficiently by FtsH. Overproduced SecG is also largely stable in the cell (our unpublished results). Among the translocase subunits, SecY is by far the best substrate of FtsH. Uncomplexed SecY may form an uncontrolled channel and must be eliminated (Kihara et al., 1995).
FtsH has an in vitro ability to bind denatured proteins (Akiyama et al., 1998), and such an ability may be related to the FtsH–substrate interaction, which leads to dislocation and proteolysis. A region of certain size may be required for a high-affinity FtsH binding, such that effective force for dislocation can be generated. In addition, the discontinuous length requirement suggests that an FtsH site acting to capture the substrate polypeptide is located at a certain distance from the membrane surface. This is analogous to the observation that, during co-translational protein translocation into the endoplasmic reticulum, a glycosylation site must be away from the membrane by at least 12–14 residues in order to be recognized by the glycosyltransferase (Nilsson and von Heijne, 1993). Our results raise an intriguing possibility that FtsH may act as a ruler that measures the N-terminal tail or other protruding region of a membrane protein for possible degradation. We assume that the hypothetical ruler site is located within the AAA domain of this protein. A substrate-binding site has been assigned to an N-terminal region of the AAA domain in Yme1, a yeast protein related to FtsH (Leonhard et al., 1999). To examine the ruler model, we are currently manipulating the distance between the AAA domain and the membrane by introducing deletions or insertions into a region following the membrane anchor; we will then ask whether the length requirement is altered accordingly.
The YccA11 mutant protein remains associated with the FtsH complex, thereby interfering with degradation of other membrane proteins (Kihara et al., 1998). We observed similar inhibitory effects for all the deletion derivatives of YccA that were FtsH insensitive, as well as for TM9-PhoA (our unpublished results), suggesting that they may still interact with the FtsH complex. Possibly, FtsH interacts not only with the N-terminal tail but also with the membrane-embedded regions of a substrate. The significance and the mechanism of the latter interaction are left for future studies.
Finally, it should be noted that folding/assembly states of the cytosolic protrusions as well as those of other parts of membrane proteins will be crucial to whether they are channeled to the FtsH pathway of degradation. Thus, every membrane protein is double-checked before it is eliminated by FtsH. The FtsH protease provides us with interesting lessons about how extensive degradation of membrane-embedded proteins can be achieved in the cytosolic environment that is optimized for hydrolytic reactions. The N-tail recognition is one strategy that this enzyme invented.
METHODS
Escherichia coli strains. Strains AR5087 (zad-220::Tn10 sfhC21) and AR5090 (ΔftsH zad-220::Tn10 sfhC21) were derivatives of JM103 (F′ lacIq lacZΔM15/Δlac-pro endA sbcB15 hsdR thi rpsL supE), and were provided by T. Karata and T. Ogura (personal communication). Growth of E. coli without FtsH was made possible by the sfhC21 suppressor mutation (Ogura et al., 1999).
Plasmids. Plasmids pKH330 (encoding YccA-His6-Myc), pKH331 (YccA11-His6-Myc), pKH356 (N8-YccA11-His6-Myc) and pKH472 (TM9-PhoA) were described previously (Kihara et al., 1998, 1999). They all carried the respective cloned genes under lac promoter control. pCH51 carried the sequence for the HA epitope, which was placed within the multi-cloning region of pTWV228, a pBR322-based lac promoter vector (Takara Shuzo); a synthetic duplex of 5′-CAGGAGGAAGAGCAAATGGCGAAGTATCCATATGATGTTCCAGATTATGCCGGCTCG with a 3′ overhanging sequence of GTAC-3′ was phosphorylated and cloned into the KpnI site of pTWV228. pCH10, a derivative of pSC101 encoding YccA-His6-Myc under lac promoter control, was constructed by treating pKH220 (Kihara et al., 1998) successively with AccI, T4 DNA polymerase and T4 DNA ligase (to prevent the in-frame continuation of the lacZ translation into yccA). pCH55, encoding HA16-YccA-His6-Myc, was constructed by amplifying a yccA-his6-myc region from pCH10 using primers 5′-AATTACCCGGGCATAAGGTGCTGCGTAATACC-3′ and 5′-GATGTGCTGCAAGGCGATTAA-3′, digesting the product with SmaI and HindIII, and cloning it into pCH51. The following plasmids were derivatives of pCH10 with a series of internal deletion mutations (Figure 2: pCH20, encoding YccAd1-His6-Myc; pCH11, YccAd2-His6-Myc; pCH21, YccAd3-His6-Myc; pCH22, YccAd4-His6-Myc; pCH12, YccAd5-His6-Myc; pCH13, YccAd8-His6-Myc; pCH17, YccAd14-His6-Myc). They were constructed using the Quick Change mutagenesis kit (Stratagene) and appropriate mutagenic primers.
pKY271 (Baba et al., 1994), carrying secE under the lac promoter control, proved to have secE fused in-frame to the vector-derived lacZ sequence such that it encoded both the Z42–SecE fusion protein (Figure 5) and the authentic SecE protein. pCM134, encoding only the authentic SecE, was constructed by replacing a 0.6 kb HindIII–XmaI fragment of pKY285 (Taura et al., 1993) by the corresponding fragment of pKY250 (Shimoike et al., 1992). For constructing pHAsecE (encoding HA12-SecE under the ara promoter), pHAsecEYG (Douville et al., 1995) was digested with SalI and HindIII to remove the secY-secG segment, blunted with T4 DNA polymerase, and ligated. pCH43 encoding Z10-TM9-PhoA was constructed as follows. The TM9-phoA fragment with added Z10 sequence (Figure 4) was amplified from pKH472 (Kihara et al., 1999) using primers 5′-AATGGTACCGGCGAAGTATATCCATAAAGTAATG-3′ and 5′-TTGTAAGCTTAAGCTTGGTAACCAGTAATGTTATT-3′, digested with KpnI and HindIII, and cloned back into pKH472.
Media. Media used were L (Davis et al., 1980) and M9 (Silhavy et al., 1984), with ampicillin (50 µg/ml) included for growing plasmid-bearing cells. For induction of lac or ara promoter-controlled genes, cells were first grown in the presence of glucose (0.4%) to an exponential phase and then either 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) plus 5 mM cyclic AMP (for induction of lac) or 0.4% arabinose (for induction of ara) was added.
Pulse–chase and immunoprecipitation. In vivo stability of the YccA, SecE and TM9-PhoA derivatives was examined by pulse–chase and immunoprecipitation experiments, essentially as described by Taura et al. (1993). SDS-solubilized whole-cell proteins were diluted with 50 mM Tris–HCl pH 8.1 containing 0.1 mM EDTA, 0.15 M NaCl and 2% Triton X-100 for immunoprecipitation. Antibodies used were anti-Myc (A-14) (Santa Cruz Biotechnology, Inc.), anti-PhoA (5 Prime 3 Prime, Inc.) and anti-SecE (Matsuo et al., 1998). [35S]methionine-labeled immunoprecipitates were separated by SDS–PAGE, and visualized and quantitated using a BAS1800 phosphoimager (Fuji Film).
Immunoblotting. Cellular contents of Myc-tagged proteins and SecE derivatives were assessed by separating a fixed amount of whole-cell proteins by SDS–PAGE and staining them immunologically using the respective antibodies, essentially as described previously (Shimoike et al., 1995). A Luminescence Image Analyzer (LAS-1000, Fuji Film) was used for visualization.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Kiyoko Mochizuki, Toshiki Yabe and Yusuke Shimizu for technical support, Akio Kihara for stimulating discussion, Teru Ogura for strains AR5087 and AR5090, and Bill Wickner for pHAsecEYG. This work was supported by grants from CREST, Japan Science and Technology Corporation (JST) and from the Ministry of Education, Science and Culture, Japan. E.-i.M. was supported by a JSPS Postdoctoral Fellowship for Young Scientists.
REFERENCES
- Akiyama Y. and Ito, K. (1987) Topology analysis of the SecY protein, an integral membrane protein involved in protein export in Escherichia coli. EMBO J., 6, 3465–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama Y., Yoshihisa, T. and Ito, K. (1995) FtsH, a membrane-bound ATPase, forms a complex in the cytoplasmic membrane of Escherichia coli. J. Biol. Chem., 270, 23485–23490. [DOI] [PubMed] [Google Scholar]
- Akiyama Y., Kihara, A. and Ito, K. (1996) Subunit a of proton ATPase F0 sector is a substrate of the FtsH protease in Escherichia coli. FEBS Lett., 399, 26–28. [DOI] [PubMed] [Google Scholar]
- Akiyama Y., Ehrmann, M., Kihara, A. and Ito, K. (1998) Polypeptide binding of Escherichia coli FtsH (HflB). Mol. Microbiol., 28, 803–812. [DOI] [PubMed] [Google Scholar]
- Baba T., Taura, T., Shimoike, T., Akiyama, Y., Yoshihisa, T. and Ito, K. (1994) A cytoplasmic domain is important for the formation of a SecY–SecE translocator complex. Proc. Natl Acad. Sci. USA, 91, 4539–4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaszczak A., Georgopoulos, C. and Liberek, K. (1999) On the mechanism of FtsH-dependent degradation of the σ32 transcriptional regulator of Escherichia coli and the role of the DnaK chaperone machine. Mol. Microbiol., 31, 157–166. [DOI] [PubMed] [Google Scholar]
- Brown M.S., Ye, J., Rawson, R.B. and Goldstein, J.L. (2000) Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell, 100, 391–398. [DOI] [PubMed] [Google Scholar]
- Davis R.W., Botstein, D. and Roth, J.R. (1980) Advanced Bacterial Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- De Mot R., Nagy, I., Walz, J. and Baumeister, W. (1999) Proteasomes and other self-compartmentalizing proteases in prokaryotes. Trends Microbiol., 7, 88–92. [DOI] [PubMed] [Google Scholar]
- Douville K., Price, A., Eichler, J., Economou, A. and Wickner, W. (1995) SecYEG and SecA are the stoichiometric components of preprotein translocase. J. Biol. Chem., 270, 20106–20111. [DOI] [PubMed] [Google Scholar]
- Flinta C., Persson, B., Jörnvall, H. and von Heijne, G. (1986) Sequence determinants of cytosolic N-terminal protein processing. Eur. J. Biochem., 154, 193–196. [DOI] [PubMed] [Google Scholar]
- Herman C., Thévenet, D., Bouloc, P., Walker, G.C. and D’Ari, R. (1998) Degradation of carboxy-terminal-tagged cytoplasmic proteins by the Escherichia coli protease HflB (FtsH). Genes Dev., 12, 1348–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A., Akiyama, Y. and Ito, K. (1995) FtsH is required for proteolytic elimination of uncomplexed forms of SecY, an essential protein translocase subunit. Proc. Natl Acad. Sci. USA, 92, 4532–4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A., Akiyama, Y. and Ito, K. (1998) Different pathways for protein degradation by the FtsH/HflKC membrane-embedded protease complex: an implication from the interference by a mutant form of a new substrate protein, YccA. J. Mol. Biol., 279, 175–188. [DOI] [PubMed] [Google Scholar]
- Kihara A., Akiyama, Y. and Ito, K. (1999) Dislocation of membrane proteins in FtsH-mediated proteolysis. EMBO J., 18, 2970–2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaus L., Stieger, A., Neupert, W. and Langer, T. (1999) Chaperone-like activity of the AAA domain of the yeast Yme1 AAA protease. Nature, 398, 348–351. [DOI] [PubMed] [Google Scholar]
- Long J.C., Wang, S. and Vik, S.B. (1998) Transmembrane topography of subunit a of the F1F0 ATP synthase as determined by labeling of unique cysteine residues. J. Biol. Chem., 273, 16235–16240. [DOI] [PubMed] [Google Scholar]
- Matsuo E., Mori, H., Shimoike, T. and Ito, K. (1998) Syd, a SecY-interacting protein, excludes SecA from the SecYE complex with an altered SecY24 subunit. J. Biol. Chem., 273, 18835–18840. [DOI] [PubMed] [Google Scholar]
- Nilsson I.M. and von Heijne, G. (1993) Determination of the distance between the oligosaccharyltransferase active site and the endoplasmic reticulum membrane. J. Biol. Chem., 268, 5798–5801. [PubMed] [Google Scholar]
- Ogura T. et al. (1999) Balanced biosynthesis of major membrane components through regulated degradation of the committed enzyme of lipid A biosynthesis by the AAA protease FtsH (HflB) in Escherichia coli. Mol. Microbiol., 31, 833–844. [DOI] [PubMed] [Google Scholar]
- Schatz P.J., Riggs, P.D., Jacq, A., Fath, M.J. and Beckwith, J. (1989) The secE gene encodes an integral membrane protein required for protein export in Escherichia coli. Genes Dev., 3, 1035–1044. [DOI] [PubMed] [Google Scholar]
- Shimoike T., Akiyama, Y., Baba, T., Taura, T. and Ito, K. (1992) SecY variants that inferfere with E. coli protein export in the presence of normal secY. Mol. Microbiol., 6, 1205–1210. [DOI] [PubMed] [Google Scholar]
- Shimoike T., Taura, T., Kihara, A., Yoshihisa, T., Akiyama, Y., Cannon, K. and Ito, K. (1995) Product of a new gene, syd, functionally interacts with SecY when overproduced in Escherichia coli. J. Biol. Chem., 270, 5519–5526. [DOI] [PubMed] [Google Scholar]
- Shotland Y. et al. (1997) Proteolysis of the phage λ CII regulatory protein by FtsH (HflB) of Escherichia coli. Mol. Microbiol., 24, 1303–1310. [DOI] [PubMed] [Google Scholar]
- Silhavy T.J., Berman, M.L. and Enquist, L.W. (1984) Experiments with Gene Fusions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Taura T., Baba, T., Akiyama, Y. and Ito, K. (1993) Determinants of the quantity of the stable SecY complex in the Escherichia coli cell. J. Bacteriol., 175, 7771–7775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoyasu T., Yamanaka, K., Murata, K., Suzuki, T., Bouloc, P., Kato, A., Niki, H., Hiraga, S. and Ogura, T. (1993) Topology and subcellular localization of FtsH protein in Escherichia coli. J. Bacteriol., 175, 1352–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valiyaveetil F.I. and Fillingame, R.H. (1998) Transmembrane topography of subunit a in the Escherichia coli F1F0 ATP synthase. J. Biol. Chem., 273, 16241–16247. [DOI] [PubMed] [Google Scholar]
- Wada T., Long, J.C., Zhang, D. and Vik, S.B. (1999) A novel labeling approach supports the five-transmembrane model of subunit a of the Escherichia coli ATP synthase. J. Biol. Chem., 274, 17353–17357. [DOI] [PubMed] [Google Scholar]