Abstract
All eukaryotes so far studied, including animals, plants, yeasts and trypanosomes, have two pathways to target proteins to peroxisomes. These two pathways are specific for the two types of peroxisome targeting signal (PTS) present on peroxisomal matrix proteins. Remarkably, the complete genome sequence of Caenorhabditis elegans lacks the genes encoding proteins specific for the PTS2 targeting pathway. Here we show, by expression of green fluorescent protein (GFP) reporters for both pathways, that the PTS2 pathway is indeed absent in C. elegans. Lack of this pathway in man causes severe disease due to mislocalization of PTS2-containing proteins. This raises the question as to how C. elegans has accommodated the absence of the PTS2 pathway. We found by in silico analysis that C. elegans orthologues of PTS2-containing proteins have acquired a PTS1. We propose that switching of targeting signals has allowed the PTS2 pathway to be lost in the phylogenetic lineage leading to C. elegans.
INTRODUCTION
Proteins are targeted to peroxisomes via one of two different routes corresponding to the two types of peroxisome targeting signal (PTS). Most peroxisomal matrix proteins contain a PTS1, which consists of a somewhat degenerate C-terminal tripeptide. The original PTS1 consensus comprises (S/C/A)(K/R/H)(L) (Gould et al., 1989), but many tripeptides with a two-out-of-three fit with this consensus can target proteins to peroxisomes (Sommer et al., 1992; Motley et al., 1995; Elgersma et al., 1996; Mullen et al., 1997). The PTS2 is found in only a few peroxisomal proteins and is an N-terminally located, bipartite amino acid motif, the consensus of which comprises (R)(L/V/I)X5(H/Q)(L/A) (Osumi et al., 1992; Gietl, 1994; Glover et al., 1994; Tsukamoto et al., 1994; Kato et al., 1996; Flynn et al., 1998). Newly synthesized peroxisomal matrix proteins are recognized by cytosolic receptors specific for either PTS1 or PTS2. These receptors target the PTS-containing proteins to peroxisomes, where the two pathways converge and make use of a common translocation machinery (for reviews see Subramani, 1998; Hettema et al., 1999). Both protein targeting pathways have been identified in highly divergent eukaryotes, including trypanosomes, yeasts, plants and animals. The presence of a PTS2 pathway in such highly divergent eukaryotes implies that both PTS1 and PTS2 targeting pathways were present in the ancestral eukaryotic cell.
Three proteins, Pex7p (Marzioch et al., 1994; Zhang and Lazarow, 1995), Pex18p and Pex21p (Purdue et al., 1998), are required specifically for targeting PTS2 proteins to peroxisomes in Saccharomyces cerevisiae. Of these three proteins, the human PTS2 receptor protein Pex7p could be identified by similarity to the yeast receptor (Braverman et al., 1997; Motley et al., 1997; Purdue et al., 1997). Orthologues of Pex18p and Pex21p have not been found. Cells lacking Pex7p mistarget PTS2-containing proteins to the cytoplasm. In man, this mislocalization results in loss of activity of the PTS2-containing enzyme alkyldihydroxyacetonephosphate synthase (ADHAPS), causing the severe disorder rhizomelic chondrodysplasia punctata (RCDP) (Wanders et al., 1994; de Vet et al., 1998a). We were therefore surprised to find that orthologues of proteins required in the PTS2-targeting pathway are absent from the Caenorhabditis elegans genome databases.
Here we show, by expression of green fluorescent protein (GFP) reporter proteins, that the PTS2-specific targeting pathway is indeed absent in C. elegans. Furthermore, identification of C. elegans orthologues of PTS2-containing proteins shows that these proteins no longer have a PTS2, but have acquired a PTS1. We also found that ADHAPS, which has undergone targeting signal switching in C. elegans (de Vet et al., 1998b) and is required for normal development in man (Wanders et al., 1994; de Vet et al., 1998a), is also required for normal development in C. elegans. Because loss of the PTS2 targeting pathway (resulting in loss of ADHAPS activity) before rerouting of ADHAPS to the PTS1 pathway would have severe consequences for the survival of any individuals in which this occurred, we propose that the PTS2 targeting pathway must have been lost secondarily to targeting signal switching of at least ADHAPS.
RESULTS AND DISCUSSION
The PTS2 pathway is absent in C. elegans
We searched the C. elegans genome databases for any of the three proteins (Pex7p, Pex18p and Pex21p) required specifically for targeting PTS2-containing proteins to peroxisomes in S. cerevisiae. We did not find any orthologues for these peroxins, although we readily identified putative C. elegans orthologues of many other peroxisomal proteins including the PTS1 receptor. We were also able to identify putative Pex7p orthologues in divergent organisms such as Drosophila melanogaster (DDBJ/EMBL/GenBank accession No. AAF50379.1), Dictyostelium discoideum (slime mould, accession No. C257251.1) and Arabidopsis thaliana (accession No. AAD27848). The absence of components required specifically in the PTS2 targeting pathway was corroborated when we searched the C. elegans database for the PTS2-containing proteins themselves. 3-Ketoacyl-CoA thiolase contains a PTS2 in all organisms tested, but in C. elegans three candidate orthologues of this protein all contain a PTS1 (Table I). de Vet et al. (1998b) reported that ADHAPS, which contains a PTS2 in mammals (de Vet et al., 1997), contains a PTS1 in C. elegans. Phytanoyl-CoA hydroxylase also has a PTS2 in mammals and a PTS1 in C. elegans (Table I). Indeed, putative C. elegans orthologues of PTS2-containing proteins all contain a PTS1 (Table I). These observations suggested the complete absence of the PTS2 targeting pathway. We tested this in vivo by expression in C. elegans of GFP reporters for the two targeting pathways.
Table I. Peroxisomal proteins that have undergone targeting signal switches.
| Peroxisomal protein | Species | PTS1 consensus | PTS2 consensus | Accession No. | PTS1a of C. elegans orthologue |
|---|---|---|---|---|---|
| 3-Ketoacyl-CoA thiolase |
D. discoidium |
– |
+ |
AU060090 |
|
| |
yeasts |
– |
+b |
|
|
| |
plants |
– |
+b |
|
|
| |
mammals |
– |
+b |
|
|
| |
C. elegans |
+ |
– |
T02G5.8c |
-QKL |
| |
|
|
|
T02G5.7 |
-KKL |
| |
|
|
|
T02G5.4 |
-QKL |
| ADHAPS |
T. brucei |
+ |
– |
AAD19697 |
|
| |
D. discoidium |
+ |
– |
CAA09333 |
|
| |
zebra fish |
– |
+ |
AI667604 |
|
| |
zammals |
– |
+b |
|
|
| |
C. elegans |
+ |
– |
CAA05690.1 |
-CKL |
| Phytanoyl-CoA hydroxylase |
zammals |
– |
+b |
|
|
| |
C. elegans |
+ |
– |
yk151d7.3 |
-ANL |
| Peroxisomal malate dehydrogenase |
S. cerevisiae |
+b |
– |
|
|
| |
plants |
– |
+b |
|
|
| Peroxisomal citrate synthase |
S. cerevisiae |
+b |
– |
|
|
| |
plants |
– |
+b |
|
|
| Acyl-CoA oxidase |
Pichia pastoris |
+b |
– |
|
|
| |
plants |
– |
+b |
|
|
| |
mammals |
+b |
– |
|
|
| |
C. elegans |
+ |
– |
C48B4.1 |
-SKL |
| |
|
|
|
F58F9.7 |
-SKL |
| |
|
|
|
F59F4.1 |
-SKL |
| |
|
|
|
F08A8.3 |
-AKL |
| |
|
|
|
F08A8.2 |
-AKL |
| |
|
|
|
F08A8.1 |
-SKL |
| F08A8.4 | -AKL |
aThe C-termini of all the putative C. elegans orthologues have been shown to be functional in targeting a protein to peroxisomes in other organisms (Gould et al., 1989; Sommer et al., 1992; Motley et al., 1995; Elgersma et al., 1996; Mullen et al., 1997).
bThe functionality of these PTSs has been experimentally validated.
cThe protein encoded by this ORF is the most probable counterpart of the mammalian type I peroxisomal thiolase (Maebuchi et al., 1999).
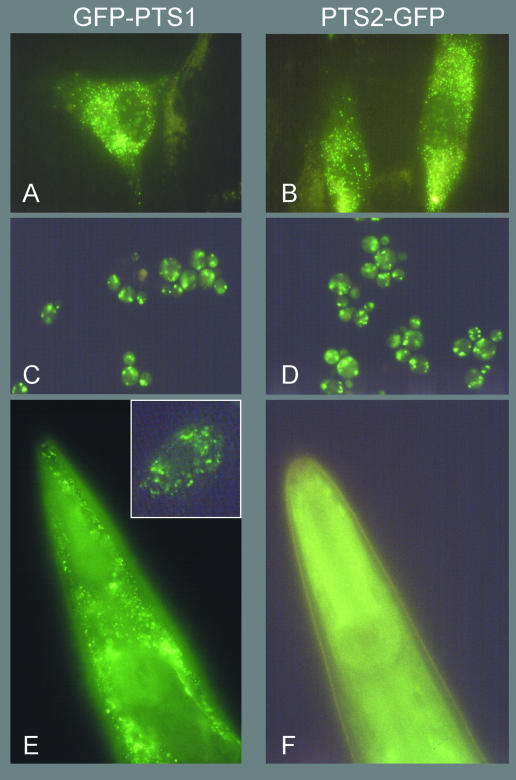
We used the presequence of rat 3-ketoacyl-CoA thiolase fused to GFP as a reporter protein for PTS2 import. Rat thiolase PTS2 has previously been shown to import a reporter protein into peroxisomes of plants (Flynn et al., 1998) and trypanosomes (Blattner et al., 1995). In C. elegans, however, a diffuse pattern of staining is seen after expression of PTS2–GFP (Figure 1F). The lack of import into peroxisomes indicates the absence of this targeting route in C. elegans. As a control, we expressed the reporter for the PTS1 pathway: GFP–PTS1 produces the punctate labelling pattern characteristic of peroxisomes (Figure 1E). We also expressed the two GFP reporters in such highly divergent cells as S. cerevisiae and human fibroblasts. Figure 1A–D shows the punctate pattern of staining indicating import into peroxisomes. We conclude that the PTS2 targeting pathway is absent in C. elegans.
Fig. 1. PTS2–GFP is not imported into peroxisomes in C. elegans. GFP–PTS1 (left panels) and PTS2–GFP (right panels) are imported into peroxisomes in human fibroblasts (A and B) and S. cerevisiae (C and D). Expression of GFP–PTS1 in C. elegans (E) shows a punctate labelling pattern indicative of peroxisome labelling. Inset: a single C. elegans cell in a backgound of GFP-negative cells showing punctate fluorescence. Single fluorescent cells can be observed with a low frequency in C. elegans carrying this GFP–PTS1 reporter transgene due to loss of the extrachromosomal plasmid array carrying the transgene. Expression of PTS2–GFP in C. elegans (F). A diffuse pattern of labelling after expression of PTS2–GFP is also seen in yeast (not shown) and human cells (Motley et al., 1997) lacking Pex7p.
ADHAPS is required for normal growth/development of C. elegans
In humans, three PTS2-containing proteins are currently known (Table I). In humans, loss of the PTS2 receptor, Pex7p, causes multiple enzyme deficiencies due to missorting of (at least) three (PTS2-containing) proteins, and results in the severe developmental disorder RCDP (Braverman et al., 1997; Motley et al., 1997; Purdue et al., 1997). It is the mislocalization and resulting loss of activity (Biermann et al., 1999) of one of these PTS2-containing enzymes, ADHAPS, that gives rise to the disease; in some RCDP patients, the only deficient activity is that of ADHAPS (Wanders et al., 1994) as a result of mutations in the gene encoding this enzyme (de Vet et al., 1998a).
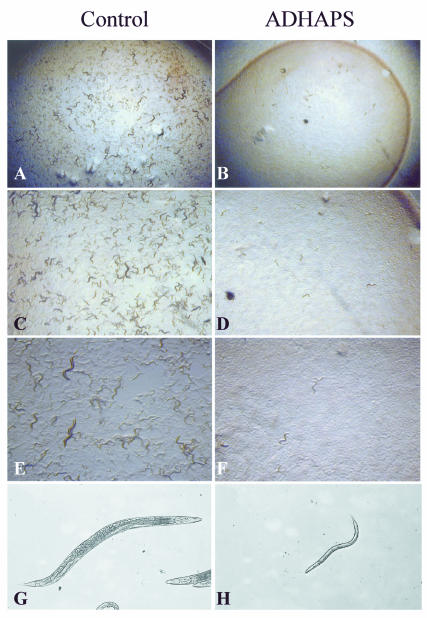
We wanted to determine whether ADHAPS is important for normal development of C. elegans. We inhibited the function of ADHAPS in C. elegans using RNA interference (Fire et al., 1998), a technique whereby double-stranded (ds)RNA of a target gene is injected into the gonads of adult hermaphrodites, resulting in strongly reduced activity of the target gene in the offspring of the injected animals. As shown in Figure 2, the growth of the offspring of ADHAPS-dsRNA-injected worms was strongly retarded compared with the growth of offspring of worms injected with a control dsRNA. Eight days after the eggs of the injected worms had hatched, the control offspring had completely cleared the bacterial lawn (on which they feed), and many eggs, larvae and adults were visible, indicating that the offspring of the injected worms had completed at least one life cycle. In contrast, the offspring of the ADHAPS-dsRNA-injected worms were still larval size, none of these worms had reached adulthood and no eggs were visible. The bacterial lawn was still intact on these plates. We conclude that ADHAPS is required for normal growth/development of C. elegans.
Fig. 2. ADHAPS is required for normal growth/development of C. elegans. Offspring of dsRNA-injected worms are shown 8 days after hatching, at increasing magnification. In the control panels, the animals had cleared the bacterial lawn (A), and there were large numbers of eggs, larvae and adults (A, C and E), indicating that the offspring of control-dsRNA-injected worms had completed at least one life cycle. The offspring of ADHAPS-dsRNA-injected worms (right-hand panels) were still larval sized at this time point, and no adult worms were visible (B, D and F). Also, the bacterial lawn was still intact (B). The highest magnification (G and H) shows offspring of (G) a control-dsRNA-injected worm (~1.5 mm in length) and (H) an ADHAPS-dsRNA-injected worm of the same age.
How did C. elegans lose the PTS2 route for protein targeting?
We propose that targeting signal switching from PTS2 to PTS1 must have occurred before loss of the PTS2-specific targeting pathway, as proper localization of peroxisomal enzymes is required for their biological activity, and loss of their function is a strongly disadvantageous event, as we have shown for C. elegans ADHAPS. Indeed, we think that proteins might acquire a PTS1 easily as this signal is highly degenerate and consists only of a C-terminal tripeptide. The acquisition of a PTS1 by a PTS2 protein could subsequently allow loss of the PTS2 without affecting its subcellular localization. Indeed, variation between possession of a PTS1 or a PTS2 does occur in peroxisomal matrix proteins in rare instances (see Table I). Targeting signals are usually found in N- or C-terminal extensions that crystallographic studies show protrude from the protein. Mutations in these extensions would not compromise the enzymatic activity of the protein, and may occasionally create a functional targeting signal. A striking example is alanine:glyoxalate aminotransferase I, which has changed its subcellular location between peroxisomes, mitochondria and cytoplasm several times during the evolution of mammals (Danpure, 1997). By analogy, C. elegans may have switched its PTS2 proteins to PTS1 proteins.
It remains a possibility that in the specific case of ADHAPS, the ancestral form had a PTS1 and acquired a PTS2 in the lineage leading to the chordates, as we found ADHAPS to have a PTS1 in all organisms except zebra fish and mammals, in which it has a PTS2 (Table I). However, we favour the reverse scenario for the reasons described above.
In summary, we propose that the surprisingly high degeneracy of PTS1 (Gould et al., 1989; Sommer et al., 1992; Motley et al., 1995; Elgersma et al., 1996; Mullen et al., 1997) allows switching from PTS2 to PTS1. Eventually, this could have led to switching of all PTS2-containing proteins to PTS1-containing proteins, and a subsequent loss of the PTS2 import machinery. With more eukaryotic genome sequences becoming available, it will be interesting to observe whether other organisms lacking a PTS2 targeting pathway will be identified. Indeed, it is intriguing that although D. melanogaster has a candidate PTS2 receptor, D. melanogaster ADHAPS and three D. melanogaster thiolase orthologues all contain a PTS1 rather than a PTS2. A very striking result of the recent molecular based reorganization of the metazoan phylogenetic tree has placed nematodes as a sister group of arthropods (for a perspective see Adoutte et al., 2000). Perhaps the targeting signal switching occurred in the common lineage leading to C. elegans and D. melanogaster, and D. melanogaster has yet to lose all remnants of the PTS2 targeting pathway, whereas this has already occurred in the rapidly evolving C. elegans.
METHODS
Plasmids. The GFP–PTS1 reporter for expression in C. elegans was constructed from plasmid pPD117.01 (a gift from A. Fire), in which the gene encoding GFP contains artificial introns for optimal expression in C. elegans. A linker encoding amino acids PLHSKLCOOH was ligated onto the 3′-end of GFP to create the C-terminal PTS1, and the mec7 promoter was replaced with a 1.6 kb genomic PCR fragment containing the SUR5 (house-keeping) promoter (obtained using forward primer CCGTCTAGACGTTAGGGTGGAATTGAACCC and reverse primer GGAATCGATTCTGAAAACAAAATGTAAAGTTC) into the XbaI ClaI sites for expression in many cell types.
The PTS2–GFP expression constructs were derived from the PTS2 of rat 3-ketoacyl-CoA thiolase, a plasmid kindly provided by S. Gould. The C. elegans PTS2–GFP reporter (pAM8) was constructed by replacing the 5′ region (ClaI–NcoI) of GFP in pPD117.01 (a gift from A. Fire) with a PTS2–GFP PCR fragment containing the appropriate sites. The mec7 promoter was then replaced with the house-keeping SUR5 promoter as described for the GFP–PTS1 expression construct. The yeast PTS2–GFP expression construct was constructed from the C. elegans GFP reporter construct pAM8: a ClaI–NcoI fragment containing the PTS2–GFP 5′-region was fused with an NcoI–HindIII fragment containing the remainder of the GFP open reading frame (ORF) (lacking C. elegans introns) into the AccI–HindIII sites of the yeast episomal plasmid Yeplac181 containing the catalase A promoter region. In this way, all three PTS2–GFP fusion proteins are identical.
Expression analysis. The C. elegans GFP reporter plasmids (pAM7 and pAM8) were each co-injected with the dominant marker rol-6 (pRF4) into the gonads of young hermaphrodites, and first generation transformants were examined for localization of GFP. The GFP expression constructs were introduced into yeast and human fibroblasts (Motley et al., 1997) as described previously. Fluorescent micrographs were recorded on a Zeiss microscope using standard filter sets.
RNA interference experiments. RNA was produced from a PCR template carrying the T7 and T3 promoter sequences at the 5′ and 3′ ends, respectively, using the Ambion MEGAscript T7 and T3 kits following the manufacturer’s instructions. The control template was amplified from C. elegans N2 genomic DNA (forward primer TAATACGACTCACTATAGGctttcgcgtcaagg-ctatc, reverse primer AATTAACCCTCACTAAAGGctcttcggcttcaccagaac, where the T7 and T3 promoter sequences are shown in upper case) and the ADHAPS template was amplified from a plasmid encoding the full-length cDNA sequence of ADHAPS (de Vet et al., 1998b) kindly provided by E. de Vet. The control template contains a segment of the coding region of the gene daf-12, which is required for dauer formation. After RNA production using the Megascript kit, the sense and antisense strands were pooled and purified using Qiagen RNEasy columns and annealed in 6 injection buffer for 30 min at 37°C as described previously (Fire et al., 1998). Formation of dsRNA was confirmed by testing the mobility of single-stranded relative to dsRNA on a standard (non-denaturing) agarose gel.
Young adult N2 hermaphrodites were injected into the gonads as described (Fire et al., 1998). After recovery, injected animals were transferred to fresh culture plates at 16 h intervals. This results in a series of approximately synchronous cohorts in which it is straightforward to identify phenotypic differences. After the first 16 h interval, all the progeny of the ADHAPS-dsRNA-injected worms showed the retarded growth phenotype.
Database analyses. Candidate Pex7p orthologues were identified by TBLASTN and PSI BLAST searches in the NCBI genome databases (non-redundant, other ESTS, D. melanogaster) using various search parameters. Candidate orthologues were confirmed by reciprocal TBLASTN searches in the same databases. A similar approach of reciprocal BLAST searching was used to identify orthologues of PTS2-containing proteins in the NCBI and C. elegans genome databases.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to The Wellcome Trust C. elegans course instructors, Patty Kuwabara in particular, for excellent instruction, to Edwin de Vet, Andy Fire and Steven Gould for plasmids, and to the Netherlands Organisation for Scientific Research for financial support, in the form of a fellowship to A.M.M.
REFERENCES
- Adoutte A., Balavoine, G., Lartillot, N., Lespinet, O., Prud’homme, B. and de Rosa, R. (2000) The new animal phylogeny: reliability and implications. Proc. Natl Acad. Sci. USA, 97, 4453–4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biermann J., Gootjes, J., Wanders, R.J. and van den Bosch, H. (1999) Stability of alkyl-dihydroxyacetonephosphate synthase in human control and peroxisomal disorder fibroblasts. IUBMB Life, 48, 635–639. [DOI] [PubMed] [Google Scholar]
- Blattner J., Dorsam, H. and Clayton, C.E. (1995) Function of the N-terminal import signal in trypanosome microbodies. FEBS Lett., 360, 310–314. [DOI] [PubMed] [Google Scholar]
- Braverman N., Steel, G., Obie, C., Moser, A., Moser, H., Gould, S.J. and Valle, D. (1997) Human PEX7 encodes the peroxisomal PTS2 receptor and is responsible for rhizomelic chondrodysplasia punctata. Nature Genet., 15, 369–376. [DOI] [PubMed] [Google Scholar]
- Danpure C.J. (1997) Variable peroxisomal and mitochondrial targeting of alanine glyoxylate aminotransferase in mammalian evolution and disease. BioEssays, 19, 317–326. [DOI] [PubMed] [Google Scholar]
- de Vet E.C.J.M., van den Broek, B.T.E. and van den Bosch, H. (1997) Nucleotide sequence of human alkyldihydroxyacetonephosphate synthase cDNA reveals the presence of a peroxisomal targeting signal 2. Biochim. Biophys. Acta, 1346, 25–29. [DOI] [PubMed] [Google Scholar]
- de Vet E.C., Ijlst, L., Oostheim, W., Wanders, R.J. and van den Bosch, H. (1998a) Alkyl-dihydroxyacetonephosphate synthase. Fate in peroxisome biogenesis disorders and identification of the point mutation underlying a single enzyme deficiency. J. Biol. Chem., 273, 10296–10301. [DOI] [PubMed] [Google Scholar]
- de Vet E.C., Prinsen, H.C. and van den Bosch, H. (1998b) Nucleotide sequence of a cDNA clone encoding a C. elegans homolog of mammalian alkyl-dihydroxyacetone phosphate synthase: evolutionary switching of peroxisome targeting signals. Biochem. Biophys. Res. Commun., 14, 277–281. [DOI] [PubMed] [Google Scholar]
- Elgersma Y., Vos, A., van den Berg, M., van Roermund, C.W.T., van der Sluijs, P., Distel, B. and Tabak, H.F. (1996) Analysis of the carboxy-terminal peroxisomal targetting signal 1 in a homologous context in S. cerevisiae. J. Biol. Chem., 271, 26375–26382. [DOI] [PubMed] [Google Scholar]
- Fire A., SiQun, X., Montgomery, M.K., Kostas, S.A., Driver, S.E. and Mello, C.C. (1998) Potent and specific genetic interference by double-stranded RNA in C. elegans. Nature, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- Flynn C.R., Mullen, R.T. and Trelease, R.N. (1998) Mutational analyses of a type 2 peroxisomal targeting signal that is capable of directing oligomeric protein import into tobacco BY-2 glyoxysomes. Plant J., 16, 709–720. [DOI] [PubMed] [Google Scholar]
- Gietl C.F., Faber, K.N., van der Klei, I.J. and Veenhuis, M. (1994) Mutational analysis of the N-terminal topogenic signal of watermelon glyoxysomal malate dehydrogenase using the heterologous host Hansenula polymorpha. Proc. Natl Acad. Sci. USA, 91, 3151–3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover J.R., Andrews, D.W., Subramani, S. and Rachubinski, R.A. (1994) Mutagenesis of the amino targeting signal of Saccharomyces cerevisiae 3-ketoacyl-CoA thiolase reveals conserved amino acids required for import into peroxisomes in vivo. J. Biol. Chem., 269, 7558–7563. [PubMed] [Google Scholar]
- Gould S.J., Keller, G.-A., Hosken, N., Wilkinson, J. and Subramani, S. (1989) A conserved tripeptide sorts proteins to peroxisomes. J. Cell Biol., 108, 1657–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema E.H., Distel, B. and Tabak, H.F. (1999) Import of proteins into peroxisomes. Biochim. Biophys. Acta, 1451, 17–34. [DOI] [PubMed] [Google Scholar]
- Kato A., Hayashi, M., Kondo, M. and Nishimura, M. (1996) Targeting and processing of a chimeric protein with the N-terminal presequence of the precursor to glyoxysomal citrate synthase. Plant Cell, 8, 1601–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maebuchi M., Togo, S.H., Yokota, S., Ghenea, S., Bun-ya, M., Kamiryo, T. and Kawahara, A. (1999) Type-II 3-oxoacyl-CoA thiolase of the nematode Caenorhabditis elegans is located in peroxisomes, highly expressed during larval stages and induced by clofibrate. Eur. J. Biochem., 264, 509–515. [DOI] [PubMed] [Google Scholar]
- Marzioch M., Erdmann, R., Veenhuis, M. and Kunau, W.-H. (1994) PAS7 encodes a novel yeast member of the WD-40 protein family essential for import of 3-oxoacyl-CoA thiolase, a PTS2-containing protein, into peroxisomes. EMBO J., 13, 4908–4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley A., Lumb, M.J., Patel, P.B., Jennings, P.R., De Zoysa, P.A., Wanders, R.J.A., Tabak, H.F. and Danpure, C.J. (1995) Identification of the peroxisomal targeting sequence of mammalian alanine:glyoxylate aminotransferase 1. Increased degeneracy and context specificity of the mammalian PTS1 motif and implications for the peroxisome-to-mitochondrion mistargeting of AGT in primary hyperoxaluria type 1. J. Cell Biol., 131, 95–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley A.M. et al. (1997) Rhizomelic chondrodysplasia punctata is a peroxisomal protein targeting disease caused by a non-functional PTS-2 receptor. Nature Genet., 15, 377–380. [DOI] [PubMed] [Google Scholar]
- Mullen R.T., Lee, M.S., Flynn, C.R. and Trelease, R.N. (1997) Diverse amino acid residues function within the type 1 peroxisomal targeting signal. Plant Physiol., 115, 881–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osumi T., Tsukamoto, T. and Hata, S. (1992) Signal peptide for peroxisomal targeting: replacement of an essential histidine residue by certain amino acids converts the amino-terminal presequence of peroxisomal 3-ketoacyl-CoA thiolase to a mitochondrial signal peptide. Biochem. Biophys. Res. Commun., 186, 811–818. [DOI] [PubMed] [Google Scholar]
- Purdue P.E., Zhang, J.W., Skoneczny, M. and Lazarow, P.B. (1997) Rhizomelic chondrodysplasia punctata is caused by deficiency of human PEX7, a homologue of the yeast PTS2 receptor. Nature Genet., 15, 381–384. [DOI] [PubMed] [Google Scholar]
- Purdue P.E., Yang, X. and Lazarow, P.B. (1998) Pex18p and Pex21p, a novel pair of related peroxins essential for peroxisomal targeting by the PTS2 pathway. J. Cell Biol., 143, 1859–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer J.M., Cheng, Q.L., Keller, G.A. and Wang, C.C. (1992) In vivo import of firefly luciferase into the glycosomes of T. brucei and mutational analysis of the C-terminal targeting signal. Mol. Biol. Cell, 3, 749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramani S. (1998) Components involved in peroxisome import, biogenesis, proliferation, turnover and movement. Physiol. Rev., 78, 171–188. [DOI] [PubMed] [Google Scholar]
- Tsukamoto T., Hata, S., Yokota, S., Miura, S., Fujiki, Y., Hijikata, M., Miyazawa, S., Hashimoto, T. and Osumi, T. (1994) Characterisation of the signal peptide at the amino terminus of the rat peroxisomal 3-ketoacyl-CoA thiolase precursor. J. Biol. Chem., 269, 6001–6010. [PubMed] [Google Scholar]
- Wanders R.J.A., Dekker, C., Horvath, V.A.P., Schutgens, R.B.H. and Tager, J.M. (1994) Human alkyldihydroxyacetonephosphate synthase deficiency: A new peroxisomal disorder. J. Inher. Metab. Dis., 17, 315–318. [DOI] [PubMed] [Google Scholar]
- Zhang J.W. and Lazarow, P.B. (1995) PEB1 (PAS7) in S. cerevisiae encodes a hydrophilic, intra-peroxisomal protein that is a member of the WD repeat family and is essential for the import of thiolase into peroxisomes. J. Cell Biol., 129, 65–80. [DOI] [PMC free article] [PubMed] [Google Scholar]