Abstract
Wnt molecules control numerous developmental processes by altering specific gene expression patterns, and deregulation of Wnt signaling can lead to cancer. Many Wnt factors employ β-catenin as a nuclear effector. Upon Wnt stimulation, β-catenin heterodimerizes with T-cell factor (TCF) DNA-binding proteins to form a transcriptional activator complex. As the activating subunit of this complex, β-catenin performs dual tasks: it alleviates repression of target gene promoters and subsequently it activates them. β-catenin orchestrates these effects by recruiting chromatin modifying cofactors and contacting components of the basal transcription machinery. Although β-catenin and TCFs are universal activators in Wnt signaling, their target genes display distinct temporal and spatial expression patterns. Apparently, post-translational modifications modulate the interactions between TCFs and β-catenin or DNA, and certain transcription factors can sequester β-catenin from TCFs while others synergize with β-catenin–TCF complexes in a promoter-specific manner. These mechanisms provide points of intersection with other signaling pathways, and contribute to the complexity and specificity of Wnt target gene regulation.
Introduction
β-catenin was originally identified as a component of cell–cell adhesion complexes, where it connects cadherins (Ca2+-dependent transmembrane proteins) to the cytoskeleton (Ozawa et al., 1989). However, much of the recent interest in β-catenin is based on the findings that it is an essential effector of Wnt signaling, and that the misregulation of its signaling activity contributes to the development of various forms of human cancer (Miller et al., 1999; Peifer and Polakis, 2000). Wnt factors constitute a large family of secreted proteins that control various developmental processes in a wide range of organisms (Cadigan and Nusse, 1997). A subset of Wnts and their receptors, members of the Frizzled protein family, initiate a chain of signaling events that culminates in the nuclear translocation of β-catenin and its heterodimerization with one of the four members of the T-cell factor (TCF) family of HMG-box proteins (reviewed by Cadigan and Nusse, 1997; Miller et al., 1999) (Figure 1). These transcription factor complexes control the activities of specific Wnt target genes, including developmental regulators and other genes involved in coordinating cell proliferation, cell–cell interactions, and cell–matrix interactions (Miller et al., 1999). Mutations altering the adenomatous polyposis coli (APC) tumor suppressor or β-catenin itself interfere with the degradative control of β-catenin and produce phenotypes equivalent to constitutive Wnt stimulation. Permanent activation of the c-myc or cyclin D1 genes by the β-catenin–TCF complex is likely to represent initial steps towards cancer (Peifer and Polakis, 2000). Clearly, both in Wnt signaling and oncogenesis, nuclear accumulation of β-catenin and its influence on gene expression are of key importance. However, β-catenin is ubiquitously expressed and, although TCFs display tissue-specific expression patterns, they all recognize the same DNA sequences. How then can Wnt target genes be differentially expressed in a temporally and spatially precisely controlled manner? And how does β-catenin function as a transcription factor? Recent studies from different organisms reveal various strategies to curb the nuclear activities of β-catenin–TCF complexes. In the following we first discuss how acetylation or phosphorylation of TCFs, limiting the available amount of β-catenin and pairing the β-catenin–TCF complex with other regulatory factors may generate tissue- and gene-specific Wnt responses, before we summarize what is known about the mechanisms of transactivation by β-catenin.
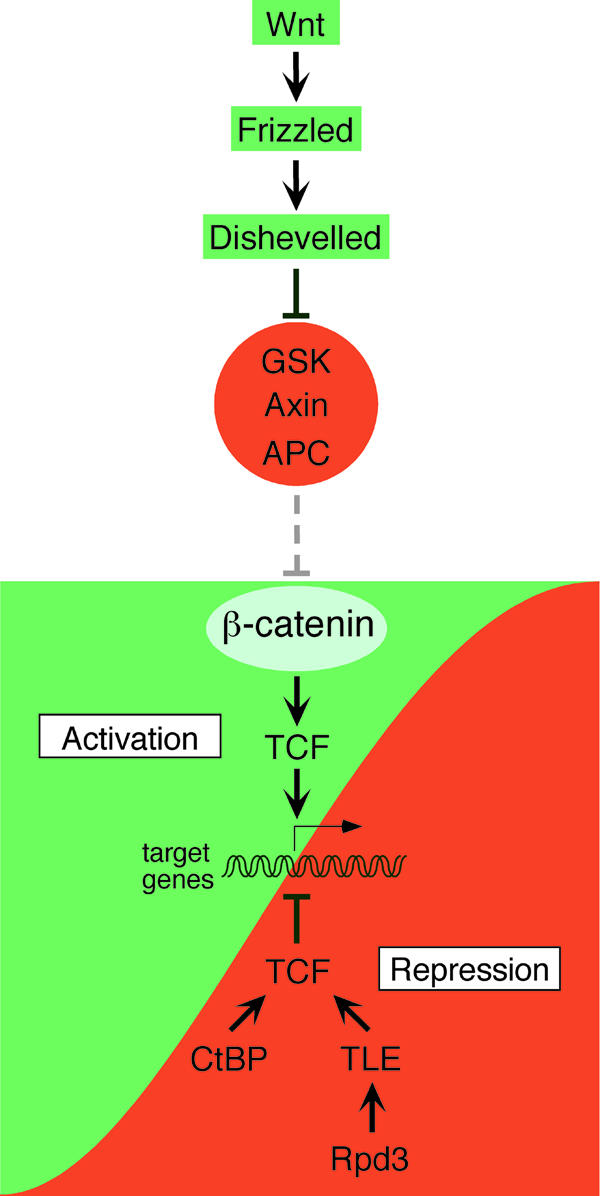
Fig. 1. Regulation of Wnt target gene expression. In the absence of Wnt signals, target gene promoters are kept in a repressed state by TCF factors and their corepressors CtBP, TLE and Rpd3 (red portion of the figure). Activation of Frizzled receptors and subsequently Dishevelled proteins relieves β-catenin from inhibition by a protein complex including glycogen synthase kinase-3β (GSK), Axin and APC. This allows β-catenin to form nuclear complexes with TCF proteins to counteract promoter repression and accomplish target gene activation (green portion of the figure).
Fine-tuning of β-catenin and TCF nuclear activities
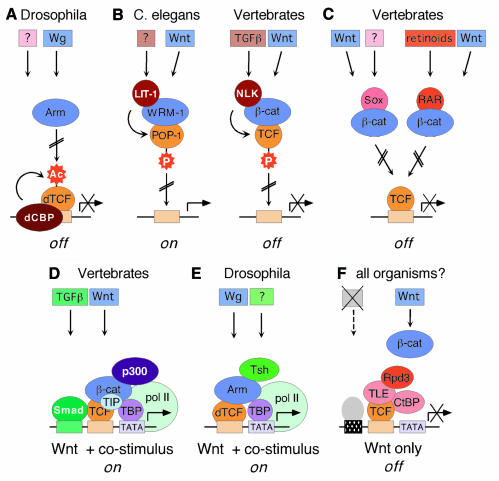
Acetylation of dTCF. In Drosophila, signaling by Wingless (Wg) and Armadillo (Arm) (fly orthologs of Wnt and β-catenin, respectively) is regulated by a relative of the cAMP-response-element binding protein (CREB)-binding protein (dCBP) (Figure 2A). Genetically, the dCBP acetylase behaves as a repressor of Wg signaling (Waltzer and Bienz, 1998). Consistent with this, dCBP acetylates the catenin-binding domain in dTCF, thereby weakening its interaction with Arm, and non-acetylatable forms of dTCF confer a Wg hyperactivation phenotype. In vertebrates, and perhaps in Drosophila as well, acetylase activity of CBP is regulated by changes in nuclear Ca2+-levels, MAP kinase-mediated phosphorylation, or interactions with inhibitors like Twist (Chawla et al., 1998; Ait-Si-Ali et al., 1999; Hamamori et al., 1999). Although the inhibitory role of dCBP so far appears to be specific for Drosophila, acetylation of dTCF may nonetheless represent one example of a combinatorial control mechanism for Arm–dTCF activity by multiple signaling events.
Fig. 2. Various mechanisms to restrict the nuclear activities of β-catenin–TCF complexes in different species (A–C) and to generate promoter-specific transcriptional responses (D–F). TIP49 (TIP), TBP and p300 may support β-catenin during promoter activation by facilitating changes in chromatin structure and by providing contacts with the basal transcription machinery (pol II). Question marks indicate that the particular signaling pathway leading to the activation or expression of a factor is not known. Whether Tsh can bind directly to specific promoter elements is unclear. Genes that do not receive an appropriate co-stimulatory signal remain transcriptionally silent (hatched box in F: inactive promoter element). See text for details.
Phosphorylation of TCFs. POP-1 is a relative of TCF that determines anterior/posterior cell-fate decisions in Caenorhabditis elegans. In posterior daughter cells POP-1 activity is downregulated by Wnt-signaling (Rocheleau et al., 1997; Thorpe et al., 1997). Mechanistically this is achieved by phosphorylation of POP-1 by the LIT-1 kinase and its activator MOM-4 (Meneghini et al., 1999; Rocheleau et al., 1999). LIT-1 and MOM-4 are related to the Nemo-like kinase (NLK) and the transforming-growth-factor (TGF)-β-activated kinase (Tak-1), components of a MAP kinase cascade in vertebrates (Ishitani et al., 1999). Phosphorylation of POP-1 by LIT-1 alters its subcellular distribution, and NLK-mediated phosphorylation of TCFs reduces their DNA-binding capacity (Ishitani et al., 1999; Meneghini et al., 1999; Rocheleau et al., 1999) (Figure 2B). Interestingly, LIT-1 interacts with WRM-1, a relative of β-catenin in C. elegans, as does NLK with β-catenin. Through these interactions, LIT-1 and NLK may be guided to their POP-1 and TCF targets. In C. elegans, alleviating promoter repression via cytoplasmic relocation of POP-1 seems to be the predominant mechanism for activating Wnt target genes. In vertebrates, NLK activity is more likely to provide a shut-off mechanism by removing β-catenin–TCF complexes from promoter regions. The activity of Tak1, the upstream regulator of NLK, can be triggered by members of the TGFβ family of growth factors and cytokines (Behrens, 2000). Hence, the NLK-mediated regulatory step may provide another mechanism for feeding additional signals into the Wnt pathway and thus generate distinct expression profiles of β-catenin–TCF target genes.
Sequestration of β-catenin. While dCBP and LIT-1/NLK regulate the β-catenin–TCF complex through their effects on the TCF component, there are also examples in which β-catenin itself is targeted. XSox17α/β and XSox3 are HMG-box-containing proteins, which like TCFs, participate in cell-fate decisions as context-dependent transcription factors and that require interactions with auxiliary factors to exert their functions (Pevny and Lovell-Badge, 1997). Although XSox17β normally does not play a role in the patterning of mesoderm, it was identified in a screen for factors affecting embryonic axis development in Xenopus (Zorn et al., 1999). Overexpression of XSox17β inhibits Wnt signaling, presumably because XSox17β competes (as do XSox17α and XSox3) with TCFs for interaction with β-catenin (Figure 2C). A similar mechanism appears to be employed by the retinoic acid receptor RAR, which binds to β-catenin in a ligand-dependent manner (Easwaran et al., 1999) (Figure 2C). Interestingly, the RAR may not only sequester β-catenin from TCFs, it may also utilize β-catenin for the activation of retinoic acid-responsive promoters. Although a physiological role for RAR- or Sox-mediated inhibition of Wnt signaling has not yet been shown, competitive binding to β-catenin may represent a prototypic mechanism whereby β-catenin–TCF activity can be temporally or spatially restricted by simultaneous inputs from other signaling cascades.
Gene-specific cooperation. The type of interference described above does not readily explain the differential activation of β-catenin–TCF regulated genes within a single cell. This appears to be achieved by other mechanisms. For example in Xenopus, Wnt stimulation of the twin promoter requires not only TCF-binding elements, but also sequences bound by Smad4, which is an essential mediator of signaling events triggered by members of the TGFβ superfamily (Nishita et al., 2000). Smad4 interacts with the HMG box of the TCF family member LEF-1, forming a Wnt-regulated β-catenin–LEF-1–Smad4 complex with a dual DNA recognition specificity (Figure 2D). This suggests a mechanism for selectively activating genes whose promoter regions contain binding sites for both LEF-1 and Smad4. Indeed, other Wnt target genes like siamois or nodal-related-3 were reported to be Smad4 independent (Nishita et al., 2000). Another example of a functional link between the Wnt/Wg and TGFβ signaling pathways is provided by the B enhancer of the Ultrabithorax (Ubx) gene. This regulatory region is composed of a Wg response element (WRE) that is targeted by dTCF, and TGFβ/Decapentaplegic response elements (DREs) recognized by a Drosophila CREB protein (Riese et al., 1997). Either one of these elements alone is insufficient to direct proper Ubx expression in the embryonic midgut, and, even more strikingly, a lacZ transgene driven by oligomerized WREs is not activated by Wg signaling at all (Riese et al., 1997). The zinc-finger protein Teashirt (Tsh), which binds to the C-terminus of Arm and modulates Wg signaling in a gene-specific manner, additionally exemplifies how distinct interaction partners can endow Arm or β-catenin and TCFs with a promoter-specific regulatory function (Gallet et al., 1999) (Figure 2E). Whether a recently described cooperation between β-catenin and CREB at the WISP-1 promoter fits into that scheme remains to be seen (Xu et al., 2000). But the picture emerging from all of these studies is that Wnt target genes appear to be generally activated in a combinatorial fashion. This concept would fit well with the frequently observed synergism between the Wnt and other signaling pathways in embryonic development (Nishita et al., 2000, and references therein), and would easily explain how β-catenin–TCF target genes can be differentially regulated within a given cell type by the presence or absence of the appropriate co-stimulatory signal or synergizing transcription factor (Figure 2D–F).
β-catenin as transcriptional activator
Once β-catenin has been delivered to a promoter, how does it activate transcription? In the absence of a Wnt signal, target genes are kept in a repressed state by TCFs and their associated corepressors, the vertebrate TLE proteins (‘transducin-like enhancer of split’, also known as ‘groucho-related genes’) or their Drosophila relative Groucho (Cavallo et al., 1998; Roose et al., 1998). TCF3 and TCF4 also bind another transcriptional repressor, CtBP (Brannon et al., 1999). Groucho additionally interacts with the histone deacetylase Rpd3 (Chen et al., 1999), suggesting that Wnt-regulated promoters are repressed through the formation of a chromatin structure that is non-permissive for transcription. To activate a target gene β-catenin might simply displace corepressors from promoter-bound TCFs. In vertebrates, however, antirepression does not suffice to activate Wnt-inducible genes since TCF mutants that no longer interact with TLE or CtBP are functionally neutral or behave as dominant negatives (McKendry et al., 1997; Vleminckx et al., 1999). Thus, activation of Wnt target genes requires genuine transcriptional stimulation. In support of this, β-catenin possesses multiple transactivating elements that also operate independently of TCFs, and, in all organisms tested, there is a strict correlation between the ability of β-catenin to function in Wnt signaling and its ability to transactivate (van de Wetering et al., 1997; Hsu et al., 1998; Hecht et al., 1999; Vleminckx et al., 1999).
Concordant with its role as transcriptional activator, β-catenin binds to the TATA box binding protein (TBP) at three different sites that map to some of its transactivating elements. However, certain C-terminal regions that are crucial for β-catenin’s function do not interact with TBP (Hecht et al., 1999). Direct binding to TBP appears not to be rate-limiting, and probably represents only a subordinate aspect of gene regulation by β-catenin. Perhaps this is because β-catenin can also interact with TBP through another factor TIP49 (also known as Pontin52) (Bauer et al., 1998; Wood et al., 2000). TIP49 is an evolutionarily conserved nuclear protein with sequence similarity to the bacterial DNA-dependent ATPase and helicase RuvB. It interacts with β-catenin and also binds to TBP in vitro. In addition, TIP49 copurifies with native RNA polymerase II, and hence it could bridge β-catenin and the basal transcription apparatus in at least two different ways. The apparent redundancy with which β-catenin can communicate with the basal transcription machinery could explain why there is at best a minor stimulatory influence of TIP49 on the transactivation capacity of β-catenin (Bauer et al., 1998). Alternatively, TIP49 may be required for promoter-specific activation by β-catenin, as has recently been proposed in the case of the c-Myc–TIP complex (Wood et al., 2000). So far, only a limited set of target genes has been used to study transactivation by β-catenin and it is quite possible that different experimental conditions will reveal a more critical role for the β-catenin–TIP interaction.
Aside from connecting to the basal transcription machinery via TBP or TIP49, β-catenin can stimulate transcription through at least one other mechanism. Like many other transcription factors that are regulated by extracellular signals, β-catenin cooperates with p300 and the closely related CBP, which, in contrast to the inhibitory role of dCBP in Drosophila, are required for the activation of certain promoters by β-catenin in vertebrates (Hecht et al., 2000; Takemaru and Moon, 2000). CBP and p300 are bimodal coactivators that may either link activator proteins to the basal transcription machinery or alter chromatin structure through their intrinsic or associated histone acetylase activities (Mannervik et al., 1999). The p300 or CBP acetylase complexes are likely to be used by β-catenin to destabilize repressive chromatin structures established by TLEs and Rpd3. Intriguingly, p300 and CBP are also coactivators of Smad proteins and of CREB (Mannervik et al., 1999), suggesting yet another way of integrating different signaling pathways at the level of individual promoters by synergistic recruitment of p300 or CBP by β-catenin and transcription factors mediating the response to TGFβ or other growth factors.
Summary and perspectives
A seemingly simple scheme of Wnt target gene activation by β-catenin has evolved quite rapidly into a rather complicated matter. The nuclear activities of β-catenin and TCFs are not only controlled by Wnts but are also coupled to other signaling pathways and depend on promoter-specific cooperation with various other transcription factors. DNA-binding and subcellular distribution of TCFs, as well as the interaction between β-catenin and TCFs, can be controlled by covalent modifications. Not all target genes are activated in the same fashion, and in different species similar players are used for different purposes as highlighted by the different emphasis on promoter derepression in C. elegans versus activation in vertebrates, and by the contrasting functions of p300/CBP in vertebrates versus dCBP in Drosophila. The candidate coactivators of β-catenin known so far—TBP, TIP49, p300 and CBP—are not sufficient to explain entirely how β-catenin stimulates transcription, and one can expect the identification of additional components of the basal transcription machinery or chromatin remodeling complexes that collaborate with β-catenin. The isolation and characterization of more β-catenin–TCF target genes is also likely to reveal new factors and signaling cascades that converge with the Wnt/β-catenin pathway in particular contexts. This will certainly pose new problems alongside other questions that remain. What exactly happens at a promoter region when it switches from a repressed to an activated state? Are resident TCF–corepressor complexes replaced by incoming β-catenin–TCF complexes? Can β-catenin and TLE or CtBP associate with TCFs simultaneously, or do they compete with each other? Since signals from TGFβ family members may have opposing effects on Wnt signaling (inhibition via NLK or activation as in the case of the Ubx DRE), what determines the ultimate outcome of these signaling events? Do β-catenin or TCFs constantly interact with the binding partners involved in these processes, or are their interactions dynamic, differing in a context-dependent manner? If dynamic, what mechanisms generate the selectivity? Understanding the nuclear activities of β-catenin will certainly remain a challenge for some time. On the other hand, the complexity that makes β-catenin so hard to understand is what also endows it with such versatility in Wnt signaling.
Acknowledgments
Acknowledgements
We thank R. Cassada for a critical reading of the manuscript and apologize to all colleagues whose work could not be mentioned due to the limitations of space.
References
- Ait-Si-Ali S., Carlisi, D., Ramirez, S., Upegui-Gonzalez, L.C., Duquet, A., Robin, P., Rudkin, B., Harel-Bellan, A. and Trouche, D. (1999) Phosphorylation by p44 MAP kinase/ERK1 stimulates CBP histone acetyl transferase activity in vitro. Biochem. Biophys. Res. Commun., 262, 157–162. [DOI] [PubMed] [Google Scholar]
- Bauer A., Huber, O. and Kemler, R. (1998) Pontin52, an interaction partner of β-catenin, binds to the TATA box binding protein. Proc. Natl Acad. Sci. USA, 95, 14787–14792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens J. (2000) Cross-regulation of the wnt signalling pathway: a role of MAP kinases. J. Cell Sci., 113, 911–919. [DOI] [PubMed] [Google Scholar]
- Brannon M., Brown, J.D., Bates, R., Kimelman, D. and Moon, R.T. (1999) XCtBP is a XTcf-3 co-repressor with roles throughout Xenopus development. Development, 126, 3159–3170. [DOI] [PubMed] [Google Scholar]
- Cadigan K.M. and Nusse, R. (1997) Wnt signaling: a common theme in animal development. Genes Dev., 11, 3286–3305. [DOI] [PubMed] [Google Scholar]
- Cavallo R.A., Cox, R.T., Moline, M.M., Roose, J., Polevoy, G.A., Clevers, H., Peifer, M. and Bejsovec, A. (1998) Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature, 395, 604–608. [DOI] [PubMed] [Google Scholar]
- Chawla S., Hardingham, G.E., Quinn, D.R. and Bading, H. (1998) CBP: a signal-regulated transcriptional coactivator controlled by nuclear calcium and CaM kinase IV. Science, 281, 1505–1509. [DOI] [PubMed] [Google Scholar]
- Chen G., Fernandez, J., Mische, S. and Courey, A.J. (1999) A functional interaction between the histone deacetylase rpd3 and the corepressor groucho in Drosophila development. Genes Dev., 13, 2218–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easwaran V., Pishvaian, M., Salimuddin and Byers, S. (1999) Cross-regulation of β-catenin-LEF/TCF and retinoid signaling pathways. Curr. Biol., 9, 1415–1418. [DOI] [PubMed] [Google Scholar]
- Gallet A., Angelats, C., Erkner, A., Charroux, B., Fasano, L. and Kerridge, S. (1999) The C-terminal domain of armadillo binds to hypophosphorylated teashirt to modulate wingless signalling in Drosophila. EMBO J., 18, 2208–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamori Y., Sartorelli, V., Ogryzko, V., Puri, P.L., Wu, H.Y., Wang, J.Y., Nakatani, Y. and Kedes, L. (1999) Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A. Cell, 96, 405–413. [DOI] [PubMed] [Google Scholar]
- Hecht A., Litterst, C.M., Huber, O. and Kemler, R. (1999) Functional characterization of multiple transactivating elements in β-catenin, some of which interact with the TATA-binding protein in vitro. J. Biol. Chem., 274, 18017–18025. [DOI] [PubMed] [Google Scholar]
- Hecht A., Vleminckx, K., Stemmler, M.P., van Roy, F. and Kemler, R. (2000) The p300/CBP acetyltransferases function as transcriptional coactivators of β-catenin in vertebrates. EMBO J., 19, 1839–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu S.C., Galceran, J. and Grosschedl, R. (1998) Modulation of transcriptional regulation by LEF-1 in response to wnt-1 signaling and association with β-catenin. Mol. Cell. Biol., 18, 4807–4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani T. et al. (1999) The TAK1–NLK–MAPK-related pathway antagonizes signalling between β-catenin and transcription factor TCF. Nature, 399, 798–802. [DOI] [PubMed] [Google Scholar]
- Mannervik M., Nibu, Y., Zhang, H. and Levine, M. (1999) Transcriptional coregulators in development. Science, 284, 606–609. [DOI] [PubMed] [Google Scholar]
- McKendry R., Hsu, S.C., Harland, R.M. and Grosschedl, R. (1997) LEF-1/TCF proteins mediate wnt-inducible transcription from the Xenopus nodal-related 3 promoter. Dev. Biol., 192, 420–431. [DOI] [PubMed] [Google Scholar]
- Meneghini M.D., Ishitani, T., Carter, J.C., Hisamoto, N., Ninomiya-Tsuji, J., Thorpe, C.J., Hamill, D.R., Matsumoto, K. and Bowerman, B. (1999) MAP kinase and Wnt pathways converge to downregulate an HMG-domain repressor in Caenorhabditis elegans. Nature, 399, 793–797. [DOI] [PubMed] [Google Scholar]
- Miller J.R., Hocking, A.M., Brown, J.D. and Moon, R.T. (1999) Mechanism and function of signal transduction by the Wnt/β-catenin and Wnt/Ca2+ pathways. Oncogene, 18, 7860–7872. [DOI] [PubMed] [Google Scholar]
- Nishita M., Hashimoto, M.K., Ogata, S., Laurent, M.N., Ueno, N., Shibuya, H. and Cho, K.W. (2000) Interaction between Wnt and TGF-β signalling pathways during formation of Spemann’s organizer. Nature, 403, 781–785. [DOI] [PubMed] [Google Scholar]
- Ozawa M., Baribault, H. and Kemler, R. (1989) The cytoplasmic domain of the cell adhesion molecule uvomorulin associates with three independent proteins structurally related in different species. EMBO J., 8, 1711–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer M. and Polakis, P. (2000) Wnt signaling in oncogenesis and embryogenesis—a look outside the nucleus. Science, 287, 1606–1609. [DOI] [PubMed] [Google Scholar]
- Pevny L.H. and Lovell-Badge, R. (1997) Sox genes find their feet. Curr. Opin. Genet. Dev., 7, 338–344. [DOI] [PubMed] [Google Scholar]
- Riese J., Yu, X., Munnerlyn, A., Eresh, S., Hsu, S.C., Grosschedl, R. and Bienz, M. (1997) LEF-1, a nuclear factor coordinating signaling inputs from wingless and decapentaplegic. Cell, 88, 777–787. [DOI] [PubMed] [Google Scholar]
- Rocheleau C.E., Downs, W.D., Lin, R., Wittmann, C., Bei, Y., Cha, Y.H., Ali, M., Priess, J.R. and Mello, C.C. (1997) Wnt signaling and an APC-related gene specify endoderm in early C. elegans embryos. Cell, 90, 707–716. [DOI] [PubMed] [Google Scholar]
- Rocheleau C.E., Yasuda, J., Shin, T.H., Lin, R., Sawa, H., Okano, H., Priess, J.R., Davis, R.J. and Mello, C.C. (1999) WRM-1 activates the LIT-1 protein kinase to transduce anterior/posterior polarity signals in C. elegans. Cell, 97, 717–726. [DOI] [PubMed] [Google Scholar]
- Roose J., Molenaar, M., Peterson, J., Hurenkamp, J., Brantjes, H., Moerer, P., van de Wetering, M., Destree, O. and Clevers, H. (1998) The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature, 395, 608–612. [DOI] [PubMed] [Google Scholar]
- Takemaru K.I. and Moon, R.T. (2000) The transcriptional coactivator CBP interacts with β-catenin to activate gene expression. J. Cell Biol., 149, 249–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe C.J., Schlesinger, A., Carter, J.C. and Bowerman, B. (1997) Wnt signaling polarizes an early C. elegans blastomere to distinguish endoderm from mesoderm. Cell, 90, 695–705. [DOI] [PubMed] [Google Scholar]
- van de Wetering M. et al. (1997) Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell, 88, 789–799. [DOI] [PubMed] [Google Scholar]
- Vleminckx K., Kemler, R. and Hecht, A. (1999) The C-terminal transactivation domain of β-catenin is necessary and sufficient for signaling by the LEF-1/β-catenin complex in Xenopus laevis. Mech. Dev., 81, 49–58. [DOI] [PubMed] [Google Scholar]
- Waltzer L. and Bienz, M. (1998) Drosophila CBP represses the transcription factor TCF to antagonize Wingless signalling. Nature, 395, 521–525. [DOI] [PubMed] [Google Scholar]
- Wood M.A., McMahon, S.B. and Cole, M.D. (2000) An ATPase/helicase complex is an essential cofactor for oncogenic transformation by c-Myc. Mol. Cell, 5, 321–330. [DOI] [PubMed] [Google Scholar]
- Xu L., Corcoran, R.B., Welsh, J.W., Pennica, D. and Levine, A.J. (2000) WISP-1 is a wnt-1- and β-catenin-responsive oncogene. Genes Dev., 14, 585–595. [PMC free article] [PubMed] [Google Scholar]
- Zorn A.M., Barish, G.D., Williams, B.O., Lavender, P., Klymkowsky, M.W. and Varmus, H.E. (1999) Regulation of Wnt signaling by Sox proteins: XSox17 α/β and XSox3 physically interact with β-catenin. Mol. Cell, 4, 487–498. [DOI] [PubMed] [Google Scholar]