Abstract
An insertion sequence, IS1562, was identified in a Streptococcus pyogenes strain of the clinically important M1 serotype. IS1562 is located in the mga regulon between the genes coding for the M protein and the C5a peptidase, both important virulence factors. The same or similar insertion sequences were found in most S. pyogenes strains, but the chromosomal location differed among isolates.
Chromosomal rearrangements and horizontal gene transfer promote microbial evolution and can be facilitated by insertion sequences (IS). These mobile genetic elements, by definition, contain genes related only to insertion functions (4). Despite this definition, the phenotype of the recipient bacterium can be changed if the IS is inserted into a structural gene or if the insertion in front of a gene affects the expression of a downstream gene(s) (11). IS can also mediate deletions, duplications, and inversions and cointegrate formation contributing to changes in the bacterial genome. Virulence genes in pathogenic bacteria are often located together on the chromosome in so-called pathogenicity islands (12). In gram-negative bacteria these islands can contain IS which promote rearrangements and the horizontal transfer of virulence genes (12). Chromosomal regions in gram-positive bacteria also exhibit similarities to pathogenicity islands, but so far they have not been reported to contain IS (12).
Streptococcus pyogenes is an important human pathogen. Among significant virulence factors are the M or M-like proteins (10, 26) and the C5a peptidase (17, 21, 33), encoded by one to three emm-like genes and scpA, respectively. These genes are colocated on the chromosome and are referred to as the mga regulon. Their expression is controlled by the positive transcriptional regulator Mga encoded by mga located upstream from emm (5, 20, 31) (Fig. 1B). In some strains of S. pyogenes the emm-like genes and scpA are separated by a 3,300-bp DNA segment. In the strain of the M1 serotype studied here, this segment contains the gene coding for protein SIC, sic (1). In this study, sequencing of the remaining part of this segment led to the identification of an IS (IS1562) in a chromosomal locus important for the virulence of S. pyogenes.
FIG. 1.
The nucleotide sequence containing IS1562, deduced amino acid sequence of the ORF protein, and the relation to other genes in the mga regulon. (A) Nucleotide sequence and deduced amino acid sequence of the ORF from S. pyogenes AP1. The sequence of the PCR product generated with primers derived from scpA (A16) and sic (ES1) (1, 7) as shown. The deduced amino acid sequence predicts a protein with a molecular mass of 46,382 Da. Boxes mark cysteine residues arranged in a pattern resembling a zinc finger motif or an iron sulfur cluster (8, 9). Possible terminal inverted repeats are marked IR1. A star marks the end of the homology to the promoter region of scpA in other strains. A sequence which matches the consensus sequence to which Mga binds (20) is shown. (B) Schematic representation of the mga regulon of S. pyogenes AP1. The protein encoded by mga is a transacting positive regulator of transcription of the two emm-like genes, (emm1 and sph), sic, and scpA. The ORF is located on the opposite strand compared to the other genes in the mga regulon. Apart from the AP1 strain and other strains of the M1 serotype, the ORF sequence is also found immediately upstream of scpA in strains of the M12 and M55 serotypes. Primers used to screen for the ORF sequence and to identify the flanking genes are depicted (not in scale) in the positions at which they hybridize on the AP1 chromosome.
Bacterial strains, plasmids, and growth conditions.
Sequencing of the segment described above was done with S. pyogenes AP1 (strain 40/58 of the M1 serotype from the Institute of Hygiene and Epidemiology, Prague, Czech Republic). Forty-eight additional strains of different M serotypes were also obtained from this institute. A collection of strains of the M1 serotype isolated in Sweden in 1988 to 1989 from patients with septicemia were kindly provided by Stig Holm, Umeå University, Umeå, Sweden (16). For direct cloning of PCR products the pGEM-T vector (Promega, Madison, Wis.) was used. The pGEM-T vector was transformed into competent Escherichia coli JM109. Cells were grown in Luria-Bertani broth (27), supplemented with 100 μg of ampicillin (Sigma, St. Louis, Mo.) per ml when cells contained plasmids. S. pyogenes strains were grown in Todd-Hewitt broth (Difco, Detroit, Mich.) in 5% CO2 at 37°C.
DNA methods and homology searches.
Streptococcal DNA was isolated by the method described by Pitcher et al. (22), which was modified by adding a digestion with 1,000 U of mutanolysin (Sigma) per ml and 100 mg of lysozyme (Sigma) per ml in 0.1 M potassium phosphate buffer (pH 6.2) to replace the initial incubation step. PCR was performed with Taq DNA polymerase (Gibco-BRL, Gaithersburg, Md.). Standard ligation, transformation, and plasmid isolation procedures were used (27). Ligase and restriction enzymes were obtained from Gibco-BRL. Double-stranded DNA sequencing was done by the dideoxy nucleotide chain termination method (28) with the T7 sequencing kit (Pharmacia Biotech, Uppsala, Sweden) with the use of synthetic oligonucleotides that hybridize to the obtained sequence. Southern blotting was performed according to the methods of Sambrook et al. (27), with Hybond-N membranes (Amersham, Amersham, United Kingdom). The Megaprime DNA labeling system (Amersham) was used to generate probes from PCR products. The oligonucleotides used in this study (Fig. 1B) to generate different PCR products were named as follows (with the source indicated in parentheses): A16 (7), nucleotides (nt) 896 to 880; ES1 (1), nt 871 to 890; 1474 (7), nt 1114 to 1137; 1476 (7), nt 3960 to 3937; 3244 (Fig. 1), nt 601 to 624; 3266 (Fig. 1), nt 1265 to 1290; 3267 (Fig. 1), nt 1604 to 1575; and 4419 (15), 5′-TTA CCA/G TCA ACA GGT/C GAA A/GCA GCT/A AAC CCA TTG-3′, a partly degenerate oligonucleotide hybridizing with the consensus sequences of SF1 and SF3 coding for the LPXTGE motif (30). Homology searches were performed at the National Center for Biotechnology Information by using the BLAST network service, the Wisconsin package (version 8; Genetics Computer Group, Madison, Wis.), and the available sequences from the Streptococcal Genome Sequencing Project, Department of Chemistry and Biochemistry, The University of Oklahoma, Norman, Okla.
Characterization of the sequence between the sic and scpA genes.
Chromosomal DNA from the AP1 strain was used as the target in PCR with primers derived from the 3′ end of sic (ES1) and the strand complementary to the 5′ end of scpA (A16) (Fig. 1). PCR with these primers generated a product with a length of 2,300 bp. The sequence revealed an open reading frame (ORF) of 1,203 bp oriented in the direction opposite that of the other genes of the mga regulon (Fig. 1B). The ORF is flanked by three sets of inverted repeats. nt 379 to 404 are complementary to nt 1861 to 1836 (IR1 in Fig. 1A) in 21 of 26 nt. Two other possible inverted repeats are the sequences at nt 411 to 429 and 2217 to 2199 (15 of 19 nt hybridize) and sequences at nt 385 to 394 and 1830 to 1821, which match perfectly.
The nucleotide sequence obtained had the highest homology to GenBank entry J05229, coding for the C5a peptidase from an S. pyogenes strain of the M12 serotype. The sequence in Fig. 1 is 98% homologous to nt 1 to 887 in J05229. nt 1 to 380 show extensive homology to the region upstream from scpA in the S. pyogenes strains in which the gene sequence is known. When nt 1 to 380 were excluded, no homologies to any sequence in the database, except the previously mentioned scpA and sic sequences, were found (1).
The translation of the ORF predicts a protein with 401 amino acids and a molecular mass of 46,382 Da. Five of the eight cysteines in the protein are located in the COOH-terminal region, and the distribution of these cysteines (C-X2-C-X25-C-X2-C-X5-C) resembles DNA-binding zinc finger motifs (9) or iron sulfur clusters (8). Significant homologies with the protein were found only among peptides encoded by other IS or a sequence of unknown function (Z18920) (Table 1). The homologies to Z18920 (Yersinia enterocolitica) and U35635 (Staphylococcus haemolyticus) were found in different reading frames of these sequences. However, IS use programmed translational frameshift as a method to control expression and can consequently translate a protein from different reading frames (6). Only the Y. enterocolitica and the S. haemolyticus sequences showed homology regions distributed over the entire ORF protein sequence, including the putative zinc finger motif. Finally, the sequence determined in this study was compared to the sequences from the Streptococcal Genome Sequencing Project determined with a strain of the M1 serotype, and an almost identical sequence (different in 6 of 2,307 bases) was found. The region of homology is located upstream from scpA in the sequencing project. Within the ORF protein sequence, 400 of 401 amino acid residues were identical. No other significant homologies were identified.
TABLE 1.
Peptide sequences homologous to the ORF protein identified with the blastp and tfasta programs
| Organism | GenBank accession no. | Reference | Amino acid positions in the CDSa | Amino acid positions with homology to the ORF protein | % Identity (% similarity) |
|---|---|---|---|---|---|
| Y. enterocolitica | Z18920 | 35 | No CDS | 1–401 | 37 (78) |
| S. haemolyticus | U35635 | 3 | |||
| ORF 1 | 112–152 | 122–162 | 46 (58) | ||
| ORF 2 | 51–71 | 267–287 | 47 (71) | ||
| ORF 2 | 93–119 | 302–328 | 33 (53) | ||
| ORF 2 | 145–154 | 348–357 | 50 (70) | ||
| S. aureus | L14017 | 2 | 51–71 | 267–287 | 47 (71) |
| 93–119 | 302–328 | 33 (53) | |||
| 145–154 | 348–357 | 50 (70) | |||
| N. meningitidis | Z49092 | 14 | 12–25 | 140–153 | 42 (78) |
| 57–76 | 234–253 | 45 (60) | |||
| 89–116 | 268–295 | 32 (53) | |||
| B. fragilis | X72301 | 24 | 62–83 | 54–75 | 45 (72) |
| 96–118 | 89–111 | 43 (56) | |||
| 223–255 | 240–272 | 33 (57) | |||
| B. vulgatus | X71444 | 13 | 62–83 | 54–75 | 45 (72) |
| 96–118 | 89–111 | 43 (56) | |||
| 223–255 | 240–272 | 33 (57) | |||
| B. fragilis | X76949 | 32 | 59–83 | 51–75 | 40 (72) |
| 96–118 | 89–111 | 43 (56) |
CDS, coding region defined by the authors of the databank entry.
In search of the ORF sequence in other strains of S. pyogenes.
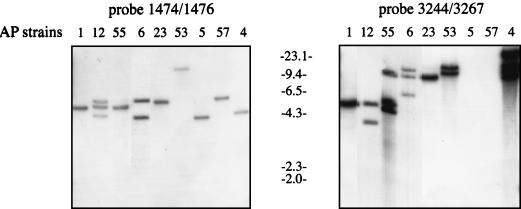
Some strains of S. pyogenes express an apolipoproteinase called the opacity factor (OF) (29). With primer pairs 3244-3267 and A16-3267 within the mga regulon (Fig. 1B), none of these OF-positive strains generated PCR products (Table 2). In contrast, most of the OF-negative strains gave rise to bands by PCR (Table 2), and in strains of M serotypes 1, 12, and 55, the ORF sequence was found immediately upstream the scpA gene. Southern blotting experiments, following digestion of chromosomal DNA with XbaI, EcoRI, ClaI, and HindIII and probing with scpA and the ORF sequence, verified these results (Fig. 2). In OF-positive strains several bands hybridized with the ORF probe, as exemplified by strain AP4 (Fig. 2), and multiple bands were also seen in several of the OF-negative strains. The results demonstrate that sequences homologous to the ORF are found, in variable numbers, in most S. pyogenes strains. As mentioned above, primer pair 3244-3267 failed to generate PCR products in OF-positive strains. However, primer pair 3266-3267 generated bands with expected sizes, showing that a region similar to the ORF sequence encoding the putative zinc finger motif is present in these strains. Whether these sequences are part of an IS also in OF-positive strains is under investigation.
TABLE 2.
Chromosomal DNA from different strains of S. pyogenes used as targets in PCR amplifications with oligonucleotides derived from the ORF sequence, the emm or emm-like genes, or scpA (see Fig. 1B)
| Straina | OF | Size of PCR product (bp) generated with primer pair indicated
|
|||||
|---|---|---|---|---|---|---|---|
| 3244-3267 | A16-3267 | A16-4419 | ES1-3244 | 3244-4419 | 3266-3267 | ||
| AP1 | − | 1,000 | 1,500 | 1,600 | 2,800 | 350 | |
| Umeå | − | 1,000 | 1,500 | 1,600 | 2,800 | 350 | |
| AP12 | − | 1,000 | 1,500 | 2,800 | 350 | ||
| AP55 | − | 1,000 | 1,500 | 2,800 | 350 | ||
| AP6 | − | 1,000 | 500 | 350 | |||
| AP23 | − | 1,000 | 500 | 350 | |||
| AP53 | − | 1,000 | 500 | 350 | |||
| AP5 | − | 500 | |||||
| AP57 | − | 500 | |||||
| AP2 | + | 500 | 350 | ||||
| AP4 | + | 500 | 350 | ||||
| AP25 | + | 500 | 350 | ||||
| AP58 | + | 500 | 350 | ||||
The number denotes the M protein serotype. Umeå refers to eight strains of the M1 serotype isolated from patients with septicemia (16).
FIG. 2.
Southern blotting experiments using scpA and the ORF as probes. Chromosomal material from different S. pyogenes strains were digested with XbaI, separated by agarose gel electrophoresis, and blotted to Hybond-N membranes. Probes were generated by radiolabeling of PCR products generated by the primer pair 1474-1476 to detect scpA and by the primer pairs 3244-3267 to detect the ORF sequence. AP1 chromosomal DNA was used as target to generate the PCR products.
Conclusions.
In the AP1 strain of the M1 serotype, the sequence between the genes encoding protein SIC and the C5a peptidase was determined. It was found to contain an ORF with homology to six proteins encoded by IS in other bacterial species. IS encode transposases responsible for the transposition of the IS (11). Three sets of inverted repeats flanking the ORF were found, but none was flanked by target site duplications. The homology to the promoter region of scpA in other strains ends at nt 380, suggesting that the repeats (marked IR1 in Fig. 1) are used by the IS. The sequence fulfills the criteria for an IS (4) and was designated IS1562.
One IS has previously been described in S. pyogenes, IS1239 (18). It is located 103 bp upstream from the gene encoding the streptococcal superantigen SSA (25) in a strain of the M15 serotype. IS can enhance the transcription of downstream genes (11), suggesting that the two IS thus far identified in S. pyogenes may contribute to virulence. IS can also form compound transposons by flanking other genes. It is possible that both IS1562 and sic are remains of a compound transposon inserted in the mga regulon. Finally, horizontal gene transfer has been implicated in playing an important role in the pathogenicity and epidemiology of S. pyogenes (19, 23, 34). Consequently, the property of IS to facilitate recombinational events (11), and thereby genetic flexibility and microbial adaptation, could also promote the virulence of this important human pathogen.
Nucleotide sequence accession number.
The GenBank accession number of the nucleotide sequence determined in this study is AF064540.
Acknowledgments
This work was supported by grants from the Swedish Medical Research Council (project 7480); the Medical Faculty, Lund University; the Foundations of Kock and Österlund; the Göran Gustavsson Foundation for Research in Natural Sciences and Medicine; and Actinova, Ltd.
We are indebted to Arne Olsén for kindly providing the modification of the method used for isolation of streptococcal DNA and to Kristin Persson for excellent technical assistance.
REFERENCES
- 1.Åkesson P, Sjöholm A G, Björck L. Protein SIC, a novel extracellular protein of Streptococcus pyogenes interfering with complement function. J Biol Chem. 1996;271:1081–1088. doi: 10.1074/jbc.271.2.1081. [DOI] [PubMed] [Google Scholar]
- 2.Archer G L, Niemeyer D M, Thanassi J A, Pucci M J. Dissemination among staphylococci of DNA sequences associated with methicillin resistance. Antimicrob Agents Chemother. 1994;38:447–454. doi: 10.1128/aac.38.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Archer G L, Thanassi J A, Niemeyer D M, Pucci M J. Characterization of IS1272, an insertional sequence-like element from Staphylococcus haemolyticus. Antimicrob Agents Chemother. 1996;40:924–929. doi: 10.1128/aac.40.4.924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell A, Berg D, Botstein D, Lederberg E, Novick R, Starlinger P, Szybalski W. Nomenclature of transposable elements in prokaryotes. In: Bukhari A I, Shapiro J A, Adhya S L, editors. DNA insertion elements, plasmids, and episomes. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1977. pp. 15–22. [Google Scholar]
- 5.Caparon M G, Scott J R. Identification of a gene that regulates expression of M protein, the major virulence determinant of group A streptococci. Proc Natl Acad Sci USA. 1987;84:8677–8681. doi: 10.1073/pnas.84.23.8677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandler M, Fayet O. Translational frameshift in the control of transposition in bacteria. Mol Microbiol. 1993;7:497–503. doi: 10.1111/j.1365-2958.1993.tb01140.x. [DOI] [PubMed] [Google Scholar]
- 7.Chen C, Cleary P P. Complete nucleotide sequence of the streptococcal C5a peptidase gene of Streptococcus pyogenes. J Biol Chem. 1990;265:3161–3167. [PubMed] [Google Scholar]
- 8.Cunningham R P, Ahern H, Xing D, Thayer M M, Tainer J A. Structure and function of Escherichia coli endonuclease III. Ann N Y Acad Sci. 1994;29:215–222. doi: 10.1111/j.1749-6632.1994.tb52818.x. [DOI] [PubMed] [Google Scholar]
- 9.Evans R M, Hollenberg S M. Zinc fingers: gilt by association. Cell. 1988;52:1–3. doi: 10.1016/0092-8674(88)90522-3. [DOI] [PubMed] [Google Scholar]
- 10.Fischetti V A. Streptococcal M protein: molecular design and biological behavior. Clin Microbiol Rev. 1989;2:285–314. doi: 10.1128/cmr.2.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galas D J, Chandler M. Bacterial insertion sequences. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C: American Society for Microbiology; 1989. pp. 109–162. [Google Scholar]
- 12.Hacker J, Blum-Oehler G, Müldorfer I, Tschäpe H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 13.Haggoud A, Reysset G, Azeddoug H, Sebald M. Nucleotide sequence analysis of two 5-nitroimidazole resistance determinants from Bacteroides strains and of a new insertion sequence upstream of the two genes. Antimicrob Agents Chemother. 1994;38:1047–1051. doi: 10.1128/aac.38.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammerschmidt S, Hilse R, van Putten J P, Gerardy-Schahn R, Unkmeir A, Frosch M. Modulation of cell surface sialic acid expression in Neisseria meningitidis via a transposable genetic element. EMBO J. 1996;15:192–198. [PMC free article] [PubMed] [Google Scholar]
- 15.Hollingshead S K, Arnold J, Readdy T L, Bessen D E. Molecular evolution of a multigene family in group A streptococci. Mol Biol Evol. 1994;11:208–219. doi: 10.1093/oxfordjournals.molbev.a040103. [DOI] [PubMed] [Google Scholar]
- 16.Holm S E, Norrby A, Bergholm A-M, Norgren M. Aspects of pathogenesis of serious group A streptococcal infections in Sweden 1988–1989. J Infect Dis. 1992;166:31–37. doi: 10.1093/infdis/166.1.31. [DOI] [PubMed] [Google Scholar]
- 17.Ji Y, McLandsborough L, Kondagunta A, Cleary P P. C5a peptidase alters clearance and trafficking of group A streptococci in infected mice. Infect Immun. 1996;64:503–510. doi: 10.1128/iai.64.2.503-510.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kapur V, Reda K B, Li L L, Ho L J, Rich R R, Musser J M. Characterization and distribution of insertion sequence IS1239 in Streptococcus pyogenes. Gene. 1994;150:135–140. doi: 10.1016/0378-1119(94)90872-9. [DOI] [PubMed] [Google Scholar]
- 19.Kehoe M A, Kapur V, Whatmore A M, Musser J M. Horizontal gene transfer among group A streptococci: implications for pathogenesis and epidemiology. Trends Microbiol. 1996;4:436–443. doi: 10.1016/0966-842x(96)10058-5. [DOI] [PubMed] [Google Scholar]
- 20.McIver K S, Heath A S, Green B D, Scott J R. Specific binding of the Mga to promotor sequences of the emm and scpA genes in the group A streptococcus. J Bacteriol. 1995;177:6619–6624. doi: 10.1128/jb.177.22.6619-6624.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Connor S P, Cleary P P. In vivo Streptococcus pyogenes C5a peptidase activity: analysis using transposon- and nitroguanidine-induced mutants. J Infect Dis. 1987;156:495–504. doi: 10.1093/infdis/156.3.495. [DOI] [PubMed] [Google Scholar]
- 22.Pitcher D G, Saunders N A, Owen R J. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett App Microbiol. 1989;8:151–156. [Google Scholar]
- 23.Podbielski A, Hawlitsky J, Pack T D, Flosdorff A, Boyle M D P. A group A streptococcal protein potentially resulting from intergenomic recombination exhibits atypical immunoglobulin-binding characteristics. Mol Microbiol. 1994;12:725–736. doi: 10.1111/j.1365-2958.1994.tb01060.x. [DOI] [PubMed] [Google Scholar]
- 24.Podglajen I, Breuil J, Collatz E. Insertion of a novel DNA sequence, IS1186, upstream of the silent carbapenemase gene cfiA, promotes expression of carbapenem resistance in clinical isolates of Bacteroides fragilis. Mol Microbiol. 1994;12:105–114. doi: 10.1111/j.1365-2958.1994.tb00999.x. [DOI] [PubMed] [Google Scholar]
- 25.Reda K B, Kapur V, Mollick J A, Lamphear J G, Musser J M, Rich R R. Molecular characterization and phylogenetic distribution of the streptococcal superantigen gene (ssa) from Streptococcus pyogenes. Infect Immun. 1994;62:1867–1874. doi: 10.1128/iai.62.5.1867-1874.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson J H, Kehoe M A. Group A streptococcal M proteins: virulence factors and protective antigens. Immunol Today. 1992;13:362–367. doi: 10.1016/0167-5699(92)90173-5. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 28.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saravani G A, Martin D R. Opacity factor from group A streptococci is an apoproteinase. FEMS Microbiol Lett. 1990;56:35–40. doi: 10.1111/j.1574-6968.1990.tb04118.x. [DOI] [PubMed] [Google Scholar]
- 30.Schneewind O, Mihaylova-Petkov D, Model P. Cell wall sorting signals in surface proteins of Gram-positive bacteria. EMBO J. 1993;12:4803–4811. doi: 10.1002/j.1460-2075.1993.tb06169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simpson W J, LaPenta D, Chen C, Cleary P P. Coregulation of type 12 M protein and streptococcal C5a peptidase genes in group A streptococci: evidence for a virulence regulon controlled by the virR locus. J Bacteriol. 1990;172:696–700. doi: 10.1128/jb.172.2.696-700.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trinh S, Haggoud A, Reysset G, Sebald M. Plasmids pIP419 and pIP421 from Bacteroides: 5-nitroimidazole resistance genes and their upstream insertion sequence elements. Microbiology. 1995;141:927–935. doi: 10.1099/13500872-141-4-927. [DOI] [PubMed] [Google Scholar]
- 33.Wexler D E, Nelson R D, Cleary P P. Human neutrophil chemotactic response to group A streptococci: bacteria-mediated interference with complement-derived chemotactic factors. Infect Immun. 1983;39:239–246. doi: 10.1128/iai.39.1.239-246.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whatmore A M, Kehoe M A. Horizontal gene transfer in the evolution of group A streptococcal emm-like genes: gene mosaics and variation in Vir regulons. Mol Microbiol. 1994;11:363–374. doi: 10.1111/j.1365-2958.1994.tb00316.x. [DOI] [PubMed] [Google Scholar]
- 35.Zhang L, Al-Hendry A, Toivonen P, Skurnik M. Genetic organization and sequence of the rfb gene cluster of Yersinia enterocolitica serotype O:3: similarities to the dTDP-l rhamnose biosynthesis pathway of Salmonella and to the bacterial polysaccharide transport systems. Mol Microbiol. 1993;9:309–321. doi: 10.1111/j.1365-2958.1993.tb01692.x. [DOI] [PubMed] [Google Scholar]