Abstract
Objectives
Platelets and low-density neutrophils (LDNs) are major players in the immunopathogenesis of SLE. Despite evidence showing the importance of platelet–neutrophil complexes (PNCs) in inflammation, little is known about the relationship between LDNs and platelets in SLE. We sought to characterize the role of LDNs and Toll-like receptor 7 (TLR7) in clinical disease.
Methods
Flow cytometry was used to immunophenotype LDNs from SLE patients and controls. The association of LDNs with organ damage was investigated in a cohort of 290 SLE patients. TLR7 mRNA expression was assessed in LDNs and high-density neutrophils (HDNs) using publicly available mRNA sequencing datasets and our own cohort using RT-PCR. The role of TLR7 in platelet binding was evaluated in platelet–HDN mixing studies using TLR7-deficient mice and Klinefelter syndrome patients.
Results
SLE patients with active disease have more LDNs, which are heterogeneous and more immature in patients with evidence of kidney dysfunction. LDNs are platelet bound, in contrast to HDNs. LDNs settle in the peripheral blood mononuclear cell (PBMC) layer due to the increased buoyancy and neutrophil degranulation from platelet binding. Mixing studies demonstrated that this PNC formation was dependent on platelet–TLR7 and that the association results in increased NETosis. The neutrophil:platelet ratio is a useful clinical correlate for LDNs, and a higher NPR is associated with past and current flares of LN.
Conclusions
LDNs sediment in the upper PBMC fraction due to PNC formation, which is dependent on the expression of TLR7 in platelets. Collectively, our results reveal a novel TLR7-dependent crosstalk between platelets and neutrophils that may be an important therapeutic opportunity for LN.
Keywords: SLE, nephritis, neutrophils, platelets, TLR7
Rheumatology key messages.
Platelets and low-density neutrophils (LDNs) play an important role in inflammation and autoimmune diseases.
We show that LDNs are associated with nephritis and form PNCs in a platelet–TLR7-dependent manner.
This knowledge of platelet and neutrophil interactions may contribute to developing new therapeutics for nephritis.
Introduction
SLE is a multisystem autoimmune disorder that is characterized by an initial loss of tolerance in B cells and T cells and the production of ANAs. These associate as immune complexes (ICs) with nuclear material such as RNA and DNA and deposit in the tissues to activate leucocytes. This results in the stimulation of multiple immune pathways, together with disruption of regulatory processes, and a chronic cycle of inflammation leading to tissue destruction [1, 2].
Activation of platelets and thrombocytopenia are common in SLE and are associated with greater disease activity and worse prognosis [3–5]. Activation induces CD62P, enabling binding to its ligand, P-selectin glycoprotein ligand-1 (PSGL-1), on neutrophils and the formation of platelet–neutrophil complexes (PNCs) [6]. Neutrophils (polymorphonuclear cells; PMNs) contribute to both ongoing autoimmune responses and the dysregulation of mechanisms central to the resolution of inflammation [7–9]. Low-density neutrophils (LDNs) are a key feature of the disease and are identified through density centrifugation cell separation [10]. They are detected with lymphocytes and monocytes in the peripheral blood mononuclear cell (PBMC) layer, whereas ‘high-density’ neutrophils (HDNs) and other granulocytes sediment below. SLE LDNs demonstrate increased NETosis compared with autologous HDNs. NETosis is a process in which nuclear material studded with antimicrobial proteins is released into the extracellular environment as neutrophil extracellular traps (NETs) [11]. In SLE, this results in more self-reactive products, such as double-stranded DNA (dsDNA), RNA and LL-37, and higher levels of type I IFNs and TNF-α following activation [7, 8, 12, 13].
We have focused efforts on understanding the role of TLR7, the receptor that recognizes single-stranded RNA (ssRNA) in the development of SLE. Although essential for host defence against viruses, TLR7 and its downstream MyD88-dependent signalling pathway are critical for the initiation of systemic autoimmunity [14–16]. Genetic studies have shown an association of TLR7 with clinical disease, and a TLR7Y264H gain-of-function variant has been recently described to cause human lupus [17, 18]. Therefore, in this study we sought to characterize the role of LDNs and TLR7 in clinical disease.
Methods
Clinical samples
Studies were completed with written informed consent in accordance with the Declaration of Helsinki and approved by the National Healthcare Group Domain Specific Review Board (2013/00504 and 2014/01419). SLE diagnosis was according to 1997 ACR and 2012 SLICC classification criteria [3, 19]. Further details on patients are provided in the supplementary information, including demographic and clinical characteristics (Supplementary Tables S1–S4, available at Rheumatology online).
Blood sample collection and stimulation
Blood was collected in EDTA or sodium citrate tubes where indicated and PBMCs and HDNs were purified using Histopaque-1077/1119 (Sigma-Aldrich, St. Louis, MO, USA). Cells were washed in PBS (Gibco, Waltham, MA, USA) and resuspended in 10% foetal bovine serum/Roswell Park Memorial Institute (RPMI) 1640 medium. Blood, HDNs or PBMCs (LDNs) were stimulated with 10 µg/ml R837 or R848 (TLR7/8), 10 µM CpG-A (TLR9) (all from InvivoGen, San Diego, CA, USA), adenosine diphosphate (ADP; 200 µM; Sigma-Aldrich), heat-aggregated IgG (50 µg/ml; Sigma-Aldrich) or phorbol-12-myristate-13-acetate (PMA; 30 nM; Sigma-Aldrich) for the times indicated. Heat-aggregated IgG was used to induce reactive oxygen species generation via FcγRIIa and FcγRIIIb crosslinking (FcγRX) and NETosis [23]. PMA was used as a positive control [24]. NET formation was quantified using flow cytometry, as described previously [25].
Platelet isolation
Blood was centrifuged (150 g for 10 min at room temperature (RT)/break-off) and plasma was transferred to a new tube containing an equal volume of 2 µM prostaglandin E1/Tyrodes (Cayman Chemical, Ann Arbor, MI, USA) and centrifuged (50 g for 10 min at room temperature/break-off). The supernatant was centrifuged (800 g for 10 min at room temperature/break-off) and the platelet pellet was resuspended in complete RPMI before mixing with neutrophils at the indicated ratios for 3 h before flow cytometry analysis.
Flow cytometry and cell sorting
Cells were resuspended in staining buffer consisting of PBS with 1% foetal calf serum (FCS), blocked with FcR antibody (Thermo Fisher Scientific, Waltham, MA, USA) and incubated on ice for 30 min with the indicated antibodies (Supplementary Table S5, available at Rheumatology online). HDN and platelet purities after gradient centrifugation were determined by flow cytometry. Contamination of platelets in the HDN fractions was typically <0.5%, and <2% for neutrophils in mice and human preparations. Further details on specific staining have been included in Supplementary Data S1, available at Rheumatology online. Acquisition and sorting were performed using a BD LSR II, BD FACSymphony-A5, BD FACSAria II (BD, Franklin Lakes, NJ, USA) and Cytek Aurora (Cytek Biosciences, Fremont, CA, USA). FlowJo version 10.6 (BD) with Downsample (version 3.0), tSNE and PhenoGraph (version 2.4) and ClusterExplorer (version 1.4.9) plugins was used for analysis with clustering and heatmaps.
Murine HDN and platelet preparation
All procedures conformed to National Institutes of Health (NIH) guidelines and according to an Institutional Animal Care and Use Committee approved protocol (161176). C57BL/6J (B6) and TLR7 knockout (TLR7KO) mice were housed at the Biological Resource Centre, A*STAR [26]. HDNs were harvested from mouse femurs and tibias from 16- to 18-week-old females and purified using Histopaque-1077/1119, as described above [25]. Platelets were isolated as described above.
Microscopy and immunohistochemical analysis of kidney tissue
Cells were incubated with 3% goat serum for 30 min at room temperature, then stained with CD66b-APC and CD41-FITC for 1 h at room temperature. DNA was counterstained with Hoechst-33342 (Thermo Fisher Scientific) for 15 min at room temperature. Images were acquired using the Opera Phenix high-content confocal microscope screening system (Perkin Elmer, Waltham, MA, USA) and analysed using ImageJ (NIH, Bethesda, MD, USA). Immunohistochemistry was performed on formalin-fixed, paraffin-embedded renal biopsy sections from SLE patients and from disease control tissue using a Bond Max autostainer (Leica Biosystems, Wetzlar, Germany) with the indicated antibodies (Supplementary Table S5, available at Rheumatology online) and 4′,6-diamidino-2-phenylindole (DAPI) as the nuclear counterstain. Images were acquired using the Vectra 3 pathology imaging system using inForm (version 2.6.0; Akoya Biosciences, Marlborough, MA, USA) and HALO (version 3.5.3; Indica Labs, Albuquerque, NM, USA) software. Further details are available in Supplementary Data S1, available at Rheumatology online.
Meta-analysis of microarray and RNA-seq gene expression datasets
Queries of the GEO and PubMed databases for expression studies of LDNs compared with HDNs identified two datasets in addition to our previously published RNA-seq dataset [8, 27, 28]. Microarray data (GSE26975 and GSE79404) were processed using Bioconductor (R version 3.3.3; R Foundation for Statistical Computing, Vienna, Austria) using the Affy and Lumi packages [8, 27]. Logarithmically transformed data were used to evaluate the standardized mean difference. A meta-analysis was performed using a fixed effects model via the rma.uni function and forest plots were completed using the Metafor package in R.
TLR7 expression analysis using RNA-seq and quantitative RT-PCR
Data were extracted from a published RNA-seq dataset to assess comparative expression of TLR7 mRNA in healthy donor HDNs [29]. Known expressors of TLR7 [B cells and plasmacytoid dendritic cells (pDCs)] were selected, as well as unstimulated Th1, Th2 and Th17 that do not [30]. RNA was also isolated from human LDNs, HDNs and platelets using TRIzol chloroform precipitation (Invitrogen, Waltham, MA, USA) using the RNeasy Mini kit (Qiagen, Venlo, The Netherlands). Expression of TLR7 mRNA was determined using TaqMan Gene Expression Assays; TLR7 Hs00152971_m1 with reference gene, gapdh Hs99999905_m1 using TaqMan 1-Step kit on a 7900HT Fast (Applied Biosystems, Waltham, MA, USA) or CFX96 Touch (Bio-Rad Laboratories, Hercules, CA, USA) Real-Time PCR System. Target gene expression was quantified using mean normalized expression against gapdh as a housekeeping gene. Gapdh is considered stable for human neutrophils with a M-value of 0.556 using geNorm analysis, which is below the commonly accepted maximum of 1.5 [31, 32].
Assessment of endothelium-dependent flow-mediated dilation (FMD)
Endothelium-dependent FMD and arterial stiffness were assessed using the Prosound Alpha-10 ultrasound system (Aloka, Wallingford, CT, USA). SLE patients and control volunteers were asked to abstain from food and exercise, caffeine and alcohol for 12, 24 and 48 h, respectively, before scanning.
Statistical analysis
Data were assessed for Gaussian distribution and analysed using the appropriate statistical test using Prism 9.2 (GraphPad Software, San Diego, CA, USA). Further details are described in Supplementary Data S1, available at Rheumatology online. A linear mixed model was used to assess the association of neutrophil:platelet ratio (NPR) and renal disease activity in 290 patients from the National University Hospital (NUH) for the follow-up period from 9 November 2013 to 4 June 2020 (SPSS version 25.0; IBM, Armonk, NY, USA). Unsupervised Bayesian network analysis (maximum spanning tree) was performed to determine the associated network with NPR in SLE patients using BayesiaLab 8.0 (Bayesia, Changé, France). Statistical significance was defined as a two-tailed P-value <0.05.
Results
SLE patients with active disease have more LDNs
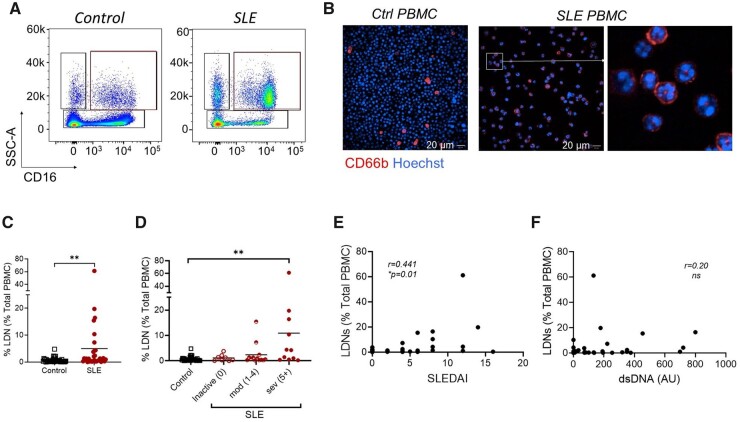
SLE patients demonstrate increased frequencies of LDNs, however, how this relates to disease activity is not fully characterized [7, 34]. Therefore, patients with SLE were recruited and LDNs were examined with respect to other leucocyte populations and clinical disease parameters (Supplementary Tables S1 and S2; Supplementary Fig. S1A–E, available at Rheumatology online). LDNs were identified in PBMCs as SSChiCD14loCD16+/hi and confirmed as CD66b+HLA-DR−CD15+, with polymorphonuclear morphology (Fig. 1A and B). We observed an expected increase in SLE patients compared with control donors (Fig. 1C). LDNs were higher in active disease and correlated with the SLEDAI score (Fig. 1D and E). However, the increase in LDNs in SLE patients was irrespective of anti-dsDNA levels in the sera, C3/C4 levels or prescribed doses of prednisolone or hydroxychloroquine (Fig. 1F, and data not shown).
Figure 1.
An increased prevalence of LDNs in SLE is associated with disease activity. PBMCs were isolated as described from SLE donors and healthy donors and LDNs were assessed using flow cytometry and confocal microscopy. (A) Representative analysis of lineage negative (LIN−; CD4−CD8−CD14−CD19−) PBMCs identifying SSChiCD14loCD16+/hi LDNs in SLE patients and controls. (B) Representative confocal microscopic images of CD66b+ cells with polymorphonuclear morphology within PBMCs in a patient with active SLE. (C) Cumulative data showing that LDNs are increased SLE patients (n = 34) compared with healthy controls (n = 41). (D, E) Cumulative data from healthy donors and SLE patients according to disease severity, as determined by SLEDAI score, showing that patients with severe disease have significantly higher frequencies of LDNs in their PBMC fraction. (E) Correlation analysis of the LDN frequency with the SLEDAI score. (F) Assessment of LDN frequency according to serum anti-dsDNA levels. Significance was determined using a Mann–Whitney U test, Kruskal–Wallis test or Spearman’s rank-order correlation, according to outcomes of Gaussian distribution tests. *P < 0.05, **P < 0.01
The SLE LDN population is heterogeneous
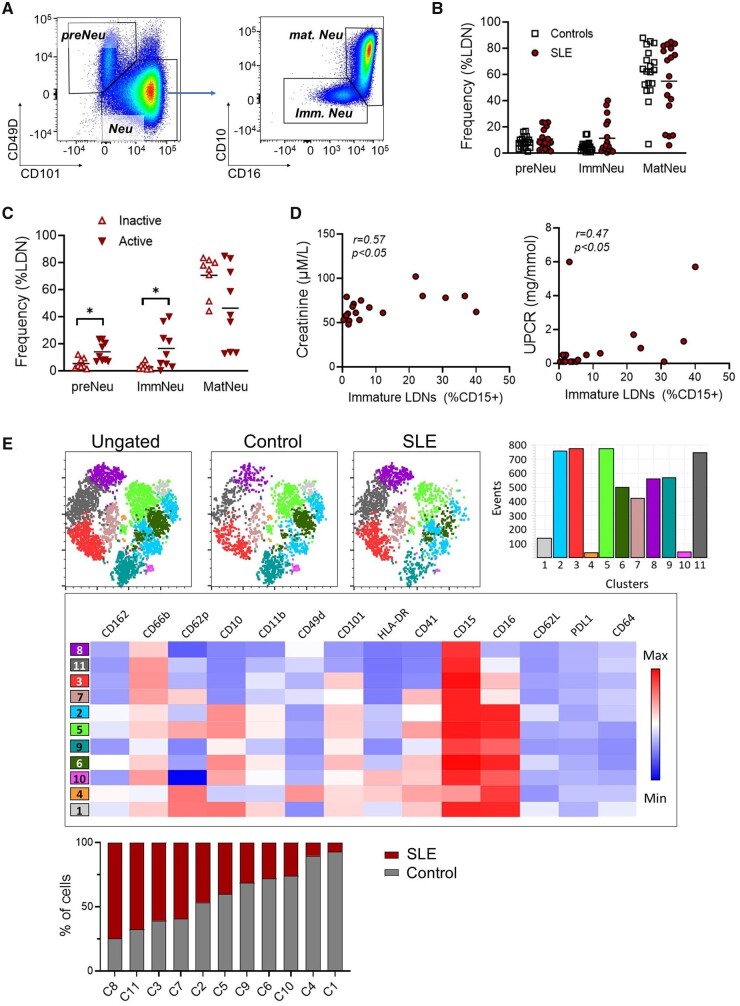
Studies characterizing SLE LDNs have assumed a homogeneous population, despite several subsets being described [7, 8]. We immunophenotyped PBMCs using flow cytometry where pre-neutrophils (preNeus) are CD49dhighCD101low and immature (ImmNeu) and mature (MatNeu) populations are both CD49d−/low and CD16lowCD10− and CD16highCD10high, respectively (Fig. 2A and Supplementary Fig. S2A and B, available at Rheumatology online) [35]. LDNs were primarily mature in healthy donors and SLE patients (Fig. 2B). However, patients with active disease had higher frequencies of preNeus and ImmNeus compared with those with inactive disease (Fig. 2C). This increase was associated with higher creatinine levels and urine protein:creatinine ratios (UPCRs; Fig. 2D), which suggest kidney dysfunction, and with the daily dose of prednisolone (Supplementary Fig. S2C, available at Rheumatology online).
Figure 2.
Immunophenotyping of LDNs reveals less mature phenotypes in active SLE. (A) Immunophenotyping of LIN−CD15+ LDN subsets using flow cytometry in SLE patients and healthy donors. Pre-neutrophils (preNeu) were identified as CD101−CD49+ and the remaining neutrophils were divided into mature (matNeu; CD10+CD16+/hi) and immature (ImmNeu; CD10−CD16low) subsets. (B) Cumulative data showing the distribution of neutrophil subsets in LDNs from SLE patients (n = 18) and healthy controls (n = 19). (C) Data from SLE patients were stratified according to disease activity. LDNs from patients with active disease (SLEDAI >4; n = 9) were less mature than LDNs from inactive (SLEDAI ≤4) SLE patients (n = 9). (D) Correlation analysis of renal disease with the frequency of immature LDNs. Left panel shows serum creatinine levels and right panel indicates the UPCR. (E) PhenoGraph revealed 11 clusters (C1–11). SLE LDNs compared with control LDNs clustered predominantly in C3, C7, C8 and C11 and expressed lower levels of CD16, CD10 and CD101. Significance was determined using two-way analysis of variance and Spearman’s rank-order correlation. *P < 0.05
UMAP and Phenograph analyses showed that preNeus and immature and mature LDNs clustered into 11 distinct groups (Supplementary Fig. S2D and E, available at Rheumatology online). Expanding populations in SLE patients (clusters C3, C7, C8 and C11) expressed reduced CD16, CD10 and CD101, suggesting reduced maturity, in agreement with the immature profile described above. Taken together, this suggests that the immature LDNs subsets may play an important part in immunopathogenesis.
LDNs are primarily platelet bound
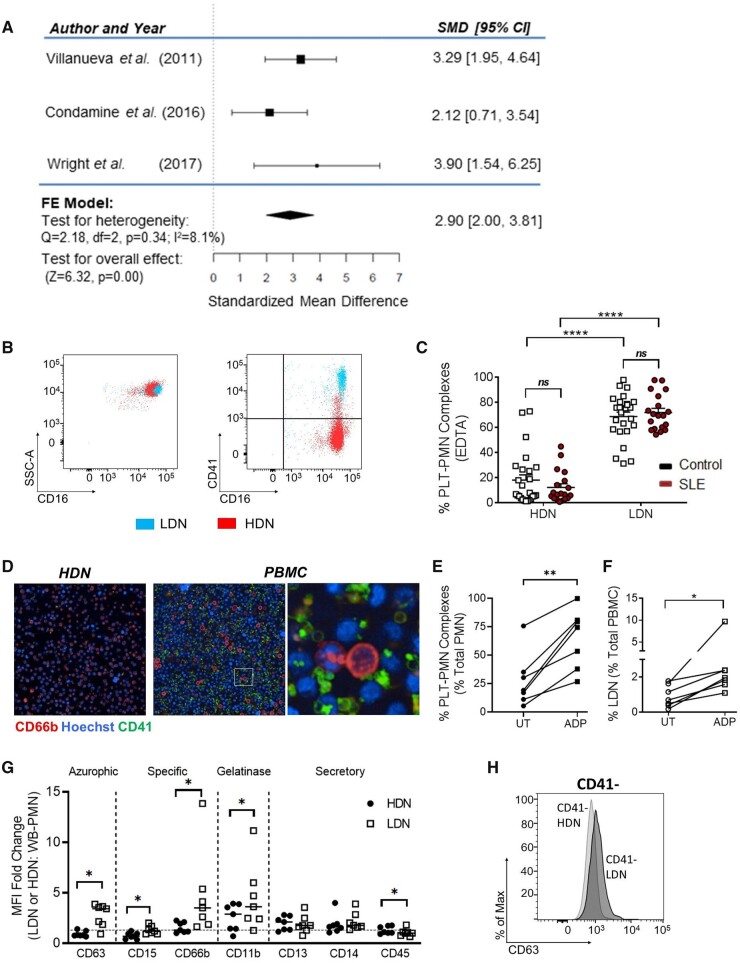
PNCs are increased in several autoimmune diseases and activated platelets have been implicated in NET formation [6, 36]. Given the differential expression of CD41 (itga2b), a platelet marker [37], in our immunophenotyping studies (Fig. 2E), we examined published transcriptome expression profiles. A meta-analysis revealed higher levels of ITGA2B mRNA in LDNs compared with HDNs (Fig. 3A), suggesting that LDNs were preferentially bound to platelets as PNCs or had internalized platelets (Fig. 3A) [8, 27, 28]. Flow cytometry and microscopy assessments in two separate cohorts demonstrated that LDNs were primarily platelet bound, in contrast to HDNs with no evidence of internalization (Fig. 3B–D; Supplementary Fig. S3A–C, available at Rheumatology online).
Figure 3.
PNC formation is upregulated in LDNs. (A) LDNs expressed more CD41 in a meta-analysis of three microarray and RNA-Seq gene expression datasets. (B) Representative flow cytometry plots illustrating PNCs in LDNs. (C) LDNs have more adherent platelets compared with HDNs on flow cytometry, using EDTA as an anticoagulant. (D) LDNs from an active SLE patient were adherent to platelets. (E) ADP treatment leads to increased PNC formation compared with untreated neutrophils in whole blood (n = 7 healthy donors). (F) ADP treatment leads to increased LDNs within the PBMC fraction (n = 7 healthy donors). (G) LDNs expressed higher levels of degranulation markers such as CD63, CD15, CD66b and CD11b compared with HDNs. (H) Overlay of histograms from CD41− neutrophils revealed that LDNs had increased expression of CD63, independent of platelet adhesion. Significance was determined using paired t-test and two-way analysis of variance. *P < 0.05 and ****P < 0.0001
We then examined whether neutrophils settled in the upper PBMC fraction due to increased buoyancy or neutrophil degranulation from platelet binding and activation. The platelet activator, ADP, was added to whole blood from control individuals and density centrifugation was performed. PNCs in whole blood increased 30 min following the addition of ADP and the frequency of LDNs in the PBMC fraction was higher, suggesting that platelet binding contributed to buoyancy (Fig. 3E and F). Immunophenotyping showed that LDNs expressed higher surface levels of CD63, CD15, CD66b and CD11b compared with HDNs, suggesting degranulation (Fig. 3G, Supplementary Fig. S3D, available at Rheumatology online) [38]. A comparison of CD41+ and CD41− LDNs revealed no differences in expression of CD15, CD66b or CD11b, eliminating platelets as the cause of this increase (Supplementary Fig. S3E, available at Rheumatology online). The CD63 expression increase was due to platelet binding (CD41+vs CD41− LDNs) and degranulation (CD41− LDNs vs HDNs) (Fig. 3H and Supplementary Fig. S3F, available at Rheumatology online).
The NPR correlates with LDN frequency and LN
Our immunological data showed associations of platelets with neutrophils with LDNs and disease. This association was then examined using routine diagnostic tests used in the clinic and disease manifestations. Increased LDNs were positively associated with NPR and LDN frequency, which was due to increased neutrophils, not platelets (Fig. 4A and B, Supplementary Fig. S4A and B, available at Rheumatology online). This suggested that we could use the NPR as a predictive factor of LDNs.
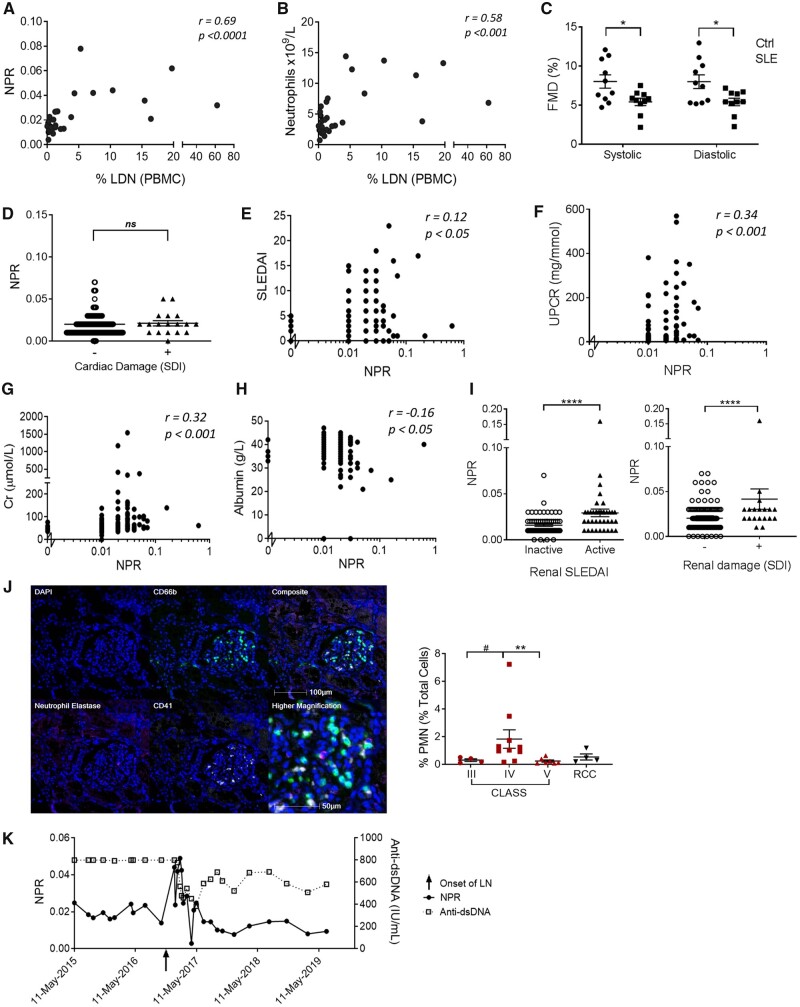
Figure 4.
The NPR precedes LN flares. (A, B) Correlation of LDN frequency measured using flow cytometry, with the NPR or neutrophil count obtained in the clinic (n = 32). (C) SLE patients and age- and gender-matched healthy donors (n = 10) without known cardiovascular risk factors were analysed for endothelial function using FMD at the brachial artery. (D–I) NPR values from SLE patients in the NUH database (n = 241) and associations (D) with or without cardiovascular damage, (E) SLEDAI, (F) UPCR, (G) creatine (Cr), (H) albumin and (I) in SLE patients with or without renal activity (n = 108, left panel) or renal damage (n = 247, right panel). (J) Representative multiplex immunofluorescent images on formalin-fixed, paraffin-embedded kidney section from SLE class IV nephritis, with region of interest at the glomerulus, with cumulative data of neutrophil infiltration from SLE patients with class III (n = 4), class IV (n = 10) and class V (n = 7) LN or renal cell carcinoma (n = 4). CD66b: green; CD41: white; neutrophil elastase: magenta; and DAPI: blue. (K) NPR, and not anti-dsDNA, was associated with the onset of LN in an SLE patient. Significance was determined using two-way analysis of variance, Spearman’s rank-order correlation and Mann–Whitney U test. ns: not significant, *P < 0.05 and ****P < 0.0001
Cardiac assessments revealed that SLE patients had significantly lower systolic and diastolic FMD compared with controls, suggesting endothelial function impairment, but this was not associated with LDNs (Supplementary Table S3, available at Rheumatology online, Fig. 4C and D) [39]. However, increased LDNs were associated with reduced arterial stiffness, as implicated by higher arterial compliance and reduced pulse wave velocity β with pressure strain elastic modulus (Supplementary Fig. S4E–G, available at Rheumatology online).
We next extracted clinical data from 290 individuals from the NUH database and used the NPR as a measure of LDNs. We did not determine any association between NPR and cardiovascular damage, measured as a composite of any cerebrovascular accident and cardiovascular and peripheral vascular disease (Fig. 4D), consistent with our small study findings. Our analysis also showed no association of NPR with active skin inflammation or prior damage from cutaneous lupus erythematosus (Supplementary Fig. S4H, available at Rheumatology online). However, there was a correlation between the NPR and SLEDAI (Fig. 4F). An evaluation of the larger SLE cohort revealed a positive association of the NPR with UPCR and serum creatinine and a negative association with serum albumin, which are all indicators of renal disease (Fig. 4F–H). Furthermore, the NPR was elevated in patients with active LN and in patients with prior damage from LN assessed using the SLEDAI-2K and SLICC/ACR Damage Index (Fig. 4I).
We used immunohistochemistry to examine cellular infiltrate in renal biopsies from SLE patients, comparing results to normal adjacent tissue from renal cell carcinoma patients (Supplementary Table S4, available at Rheumatology online, Fig. 4J). We observed more CD66b+ neutrophils in samples with class IV nephritis than in class III or V, suggesting that they are recruited in the later stages of damage and their recruitment is prevented with higher doses of immunosuppressives. An unsupervised Bayesian network analysis assessing all data variables revealed associations between the NPR node and renal manifestations, neutrophils, platelets and daily prednisolone dose (Supplementary Fig. S4I, available at Rheumatology online). A prospective analysis of an individual LN patient showed that the development of renal disease coincided with the peak of the NPR and was independent of anti-dsDNA antibody titres (Fig. 4K). Therefore, 290 SLE patients were prospectively followed up for a median duration of 4.7 years to analyse renal activity (renal SLEDAI) over time. NPR was positively associated with renal activity after adjustment for immunosuppressive therapy (Supplementary Table S6, available at Rheumatology online).
In summary, our data suggest that LDNs may play a critical role in the development of nephritis and that the NPR could be used to predict the development of renal disease.
LDNs express functional levels of TLR7
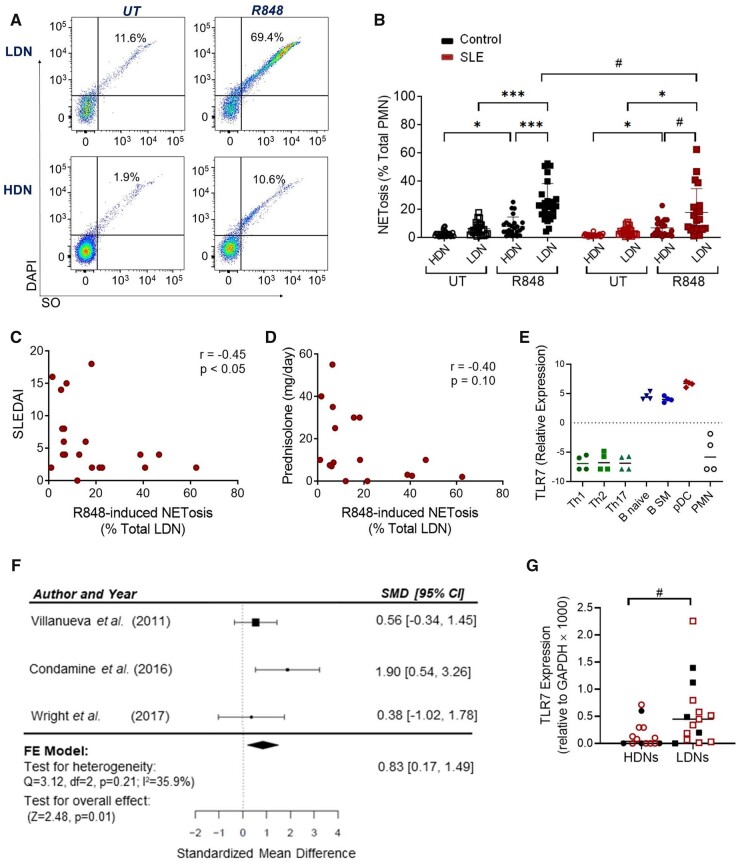
TLR7 plays a fundamental role in the initiation and progression of autoimmunity, yet data regarding neutrophil TLR7 expression and function is inconsistent. Earlier work showed RNA-containing ICs stimulate NETosis in paediatric HDNs in a TLR7-dependent manner [40]. Therefore, we stimulated LDNs and HDNs from our adult cohort with a TLR7 ligand, R848, and assessed NETosis using our single cell flow cytometry assay (Supplementary Fig. S5A, available at Rheumatology online, Fig. 5A and B) [25]. Activation resulted in an increase in NET-appendant neutrophils, in contrast to HDNs. SLE LDNs were less responsive than controls and the reduced TLR7-dependent NETosis response was associated with increasing SLEDAI disease activity (Fig. 5B and C, Supplementary Fig. S5B, available at Rheumatology online). Analogous observations were detected following stimulation with FcγR cross-linking, R837 and PMA (Supplementary Fig. S5B, available at Rheumatology online), suggesting that medication may play a role in the dampening of the functional response. Analysis of hydroxychloroquine, a known TLR inhibitor that is often used as the first-line therapy, was not associated with R848-induced NETosis in SLE LDNs [41] (Supplementary Fig. S5C, available at Rheumatology online). In contrast, higher doses of prednisolone corresponded with reduced NETosis, following R848, R837 and FcRX, and a higher disease activity (SLEDAI), as expected (Fig. 5D, Supplementary Fig. S5D and E, available at Rheumatology online).
Figure 5.
LDNs express detectable levels of TLR7 and undergo TLR7-induced NETosis. (A) Gating strategy showing that NET-appendant neutrophils detected by Sytox Orange and DAPI positivity were increased following R848 treatment in LDNs but not HDNs. (B) Cumulative data showing R848-induced NETosis in LDNs and HDNs from 10 SLE patients and 16 healthy controls. (C, D) Correlative assessment of R848-induced LDN NETosis with SLE disease activity and treatment. (E) RNA-seq analysis of TLR7 mRNA expression in circulating HDNs and T CD4+ Th1, CD4+ Th2 and CD4+ Th17 T cells (green), naïve and switched memory (SM) B cells (blue), and pDCs (red). (F) Meta-analysis of TLR7 mRNA expression in LDNs and HDNs from three microarray and RNA-seq gene expression datasets. (G) Quantitative RT-PCR analysis of TLR7 mRNA expression in HDNs and LDNs in healthy controls (n = 5) and SLE patients (n = 10). Significance was determined using two-way analysis of variance, Spearman’s rank-order correlation and Wilcoxon signed-rank test (#). */#P < 0.05 and ***P < 0.001
Given the conflicting reports on TLR7 and our NETosis responses in LDNs and HDNs, we went on to assess TLR7 mRNA expression. We used RNA-seq data from previous studies to assess levels in purified HDNs compared with other peripheral blood leucocytes from healthy donors [29]. HDNs did not express detectable levels of TLR7 mRNA, in contrast to pDCs and B cells, which are known expressors (Fig. 5E). In addition, a meta-analysis using the datasets described in Fig. 3A, demonstrated that TLR7 mRNA expression was significantly higher in LDNs compared with HDNs (Fig. 5F). We confirmed these findings using RT-PCR and demonstrated that isolated LDNs had detectable levels of TLR7 in 14 of 15 samples, in contrast to HDNs, where only 9 of 15 had significantly lower but detectable expression (Fig. 5G). There were no differences due to the presence of SLE or disease activity (e.g. C3, CD4, SLEDAI; data not shown). Consistent with previous reports, platelets expressed TLR7 mRNA, but the increase observed in SLE patients was not significant with the number of samples tested (Supplementary Fig. S5F, available at Rheumatology online) [42, 43].
R848, stimulates both TLR7 and TLR8, however, a meta-analysis of mRNA expression using the previously described datasets showed that TLR8 was significantly lower (not higher) in LDNs compared with HDNs [SMD: −1.20 (95% CI −0.52, −1.87), P < 0.01] (Supplementary Fig. S5G, available at Rheumatology online). TLR8 levels were also higher in SLE HDNs compared with healthy donor HDNs (Supplementary Fig. S5H, available at Rheumatology online), yet R848 did not effectively induce NETosis, making it unlikely that TLR8 was contributing to the differences in LDNs (Fig. 5B).
Taken together, our findings demonstrate that LDNs from adults express TLR7 mRNA and undergo NETosis in response to TLR7 ligands, in contrast to HDNs.
Platelet binding and NETosis by LDNs is dependent on platelet TLR7
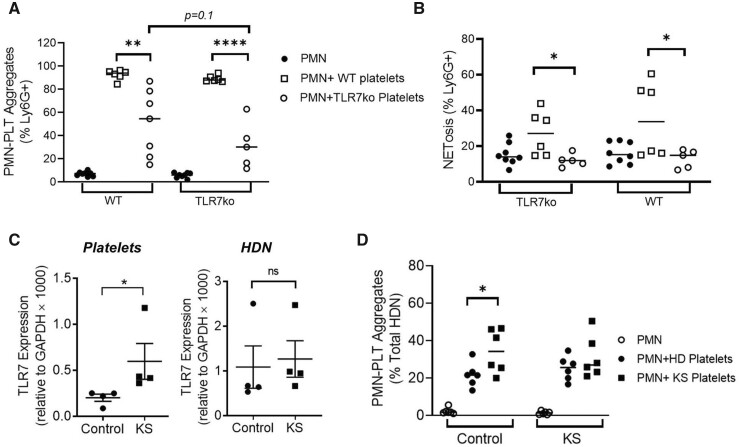
The role of TLR7 in platelet formation was then examined using mice, since platelets and neutrophils express TLR7 and retrieving sufficient numbers of TLR7-expressing LDNs from healthy donors is not feasible [43, 44]. Platelets and HDNs from TLR7KO mice and wild-type (WT) mice were mixed and PNC formation was assessed using flow cytometry (Supplementary Fig. S6A and B, available at Rheumatology online). While WT platelets were efficient at PNC formation with TLRKO or WT HDNs, TLR7KO platelets failed to efficiently bind HDNs from either strain (Fig. 6A). Furthermore, the addition of WT platelets resulted in increased NET-appendant neutrophils, which was absent with TLR7KO platelets (Fig. 6B).
Figure 6.
PNC formation depends on platelet activation and TLR7 expression. (A, B) Murine platelets and HDNs (PMNs) were isolated from WT and TLR7KO mice and mixed as indicated for 3 h. Cumulative data identifying (A) CD41+Ly6G+ PNCs and (B) neutrophil-appendant NET DNA, measured using flow cytometry (n = 5–8 per group). (C) TLR7 mRNA levels in human platelets and HDNs from KS patients compared with healthy donor males (n = 4 per group). (D) Platelets and HDNs were isolated from KS patients (n = 6) and healthy controls (n = 6) and mixed as indicated and CD41+CD66b+ PNCs measured using flow cytometry. Significance was determined using paired t-test and two-way analysis of variance. ns: not significant, *P < 0.05, **P < 0.01 and ***P < 0.001
TLR7 is on the X chromosome and expression in XX individuals is controlled through X chromosome inactivation (XCI) [45]. Klinefelter syndrome (KS) is a chromosomal variation where XY individuals have one extra X chromosome. We used RT-PCR and determined that TLR7 mRNA expression was higher in platelets from KS patients compared with healthy donors, in contrast to HDNs, which did not differ (Fig. 6C). Moreover, the addition of KS platelets to healthy donor HDNs resulted in more PNCs (Fig. 6D, Supplementary Fig. S6C and D, available at Rheumatology online). We also detected increased NETosis after mixing control HDNs with KS platelets, although this was not significant, which may be due to sample size (Supplementary Fig. S6E, available at Rheumatology online).
Discussion
The contribution of platelets and neutrophils to SLE immunopathogenesis have been recognized through genetics, transcriptomics and functional studies, however, little has been done to assess their intersecting roles. In the current study we show for the first time that LDNs are platelet bound and that this is dependent on platelet TLR7 expression. This PNC formation enables NETosis, which has been accredited with the ongoing generation of self-ligands and increased inflammation in SLE. Supporting this, the NPR, which correlates with the LDN frequency, was associated with renal damage, and there were increased kidney-infiltrating neutrophils in class IV LN. Through immunophenotyping, we demonstrated that LDNs are a heterogeneous population with a higher frequency of immature neutrophils, which are associated with disease activity and kidney dysfunction.
Earlier reports have shown neutrophil gene expression signatures in whole blood from SLE patients with active disease and nephritis, and researchers have shown an immature neutrophil RNA profile, in agreement with our studies [9, 46–48]. We propose that the inflammation in SLE patients increases circulating immature neutrophils, which then fractionate with PBMCs upon density centrifugation [35, 49]. Our binding studies and analysis of public transcriptome data showed that LDNs were preferentially platelet bound. Interestingly, this was due to the presence of TLR7 within the platelets themselves. We demonstrated this using TLR7-deficient murine systems and clinical samples from KS patients who have one1 or more additional X chromosome [22]. Our work indicates that the increased TLR7 expression that we detected in LDNs was due to platelets. Our analysis of platelet TLR7 mRNA in SLE patients showed an increase in expression; however, this was not significant. Further studies are warranted that examine expression and disease in a larger cohort.
There are differing reports regarding TLR7 expression and function in neutrophils and this issue is confounded by non-specificity of commercial anti-human TLR7 antibodies and neutrophil purification methods that result in differential LDN elution [49]. Lood et al. [50] showed that TLR7 activation cleaves the N-terminal of FcγRIIa, resulting in reduced RNP-IC-induced phagocytosis and increased NETosis. However, it is unclear if the neutrophils, isolated using Polymorphprep, were platelet bound. Other studies have shown that HDN activation by RNA-containing ICs is TLR7 independent, suggesting a purely structural role for small nuclear ribonucleoproteins [51]. In vivo, the net outcome for neutrophils will arise from the complex inflammatory milieu of ICs inflammatory mediators and cellular components, including activated platelets.
Platelet inhibitors, such as clopidogrel, reduce kidney disease and mortality in lupus-prone mice [52]. Furthermore, clopidogrel has shown reduced platelet activation in SLE patients with no safety concerns in recent phase 1/2 clinical trials, suggesting that platelet inhibitors may be potential therapeutics for SLE in the near future [53].
In summary, we have shown that TLR7-expressing platelets bind to neutrophils forming PNCs, which settle with PBMCs as LDNs. The increase of this LDN population, which correlates with the NPR, is associated with nephritis in lupus patients. We propose that these platelet-bound LDNs infiltrate the kidneys and play a key role in inflammation and tissue destruction, partly through their increased capacity for NETosis. The improved understanding of platelet and neutrophil interactions in SLE may potentially contribute to developing new therapeutic pathways and strategies.
Supplementary data
Supplementary material is available at Rheumatology online.
Supplementary Material
Acknowledgements
The authors would also like to express their appreciation to the patients in the study, to Kok Onn Lee and Shao Feng Mok (NUHS) for clinical study contributions and to Immanuel Kwok and Lai Guan Ng from SIgN for guidance in assessing neutrophil maturation. We also thank the flow cytometry facility at the Institute of Molecular and Cell Biology at A*STAR, Singapore.
Contributor Information
Sen Hee Tay, Division of Rheumatology, Department of Medicine, National University Hospital, Singapore; Department of Medicine, Yong Loo Lin School of Medicine, National University of Singapore, Singapore.
Olga Zharkova, Singapore Immunology Network (SIgN), Agency for Science, Technology and Research (A*STAR), Singapore.
Hui Yin Lee, Singapore Immunology Network (SIgN), Agency for Science, Technology and Research (A*STAR), Singapore; Institute of Molecular and Cell Biology (IMCB), Agency for Science, Technology and Research (A*STAR), Singapore.
Michelle Min Xuan Toh, Department of Medicine, Yong Loo Lin School of Medicine, National University of Singapore, Singapore.
Eshele Anak Libau, Department of Medicine, Yong Loo Lin School of Medicine, National University of Singapore, Singapore.
Teja Celhar, Singapore Immunology Network (SIgN), Agency for Science, Technology and Research (A*STAR), Singapore.
Sriram Narayanan, Institute of Molecular and Cell Biology (IMCB), Agency for Science, Technology and Research (A*STAR), Singapore.
Patricia Jennifer Ahl, Institute of Molecular and Cell Biology (IMCB), Agency for Science, Technology and Research (A*STAR), Singapore.
Wei Yee Ong, Institute of Molecular and Cell Biology (IMCB), Agency for Science, Technology and Research (A*STAR), Singapore.
Craig Joseph, Institute of Molecular and Cell Biology (IMCB), Agency for Science, Technology and Research (A*STAR), Singapore.
Jeffrey Chun Tatt Lim, Institute of Molecular and Cell Biology (IMCB), Agency for Science, Technology and Research (A*STAR), Singapore.
Lingzhi Wang, Cancer Science Institute of Singapore, Yong Loo Lin School of Medicine, National University of Singapore, Singapore.
Anis Larbi, Singapore Immunology Network (SIgN), Agency for Science, Technology and Research (A*STAR), Singapore.
Shen Liang, Biostatistics Unit, Yong Loo Lin School of Medicine, National University of Singapore, Singapore.
Aisha Lateef, Division of Rheumatology, Department of Medicine, National University Hospital, Singapore.
Shizuo Akira, Host Defence, Osaka University, Osaka, Japan.
Lieng Hsi Ling, Department of Medicine, Yong Loo Lin School of Medicine, National University of Singapore, Singapore; Department of Cardiology, National University Hospital, Singapore.
Thomas Paulraj Thamboo, Department of Pathology, National University Hospital, Singapore.
Joe Poh Seng Yeong, Institute of Molecular and Cell Biology (IMCB), Agency for Science, Technology and Research (A*STAR), Singapore; Department of Anatomical Pathology, Division of Pathology, Singapore General Hospital, Singapore.
Bernett Teck Kwong Lee, Singapore Immunology Network (SIgN), Agency for Science, Technology and Research (A*STAR), Singapore.
Steven W Edwards, Institute of Infection, Veterinary and Ecological Sciences, University of Liverpool, Liverpool, UK.
Helen L Wright, Institute of Life Course and Medical Sciences, University of Liverpool, Liverpool, UK.
Paul Anthony MacAry, Department of Microbiology and Immunology, Yong Loo Lin School of Medicine, National University of Singapore, Singapore.
John E Connolly, Singapore Immunology Network (SIgN), Agency for Science, Technology and Research (A*STAR), Singapore; Institute of Molecular and Cell Biology (IMCB), Agency for Science, Technology and Research (A*STAR), Singapore; Department of Microbiology and Immunology, Yong Loo Lin School of Medicine, National University of Singapore, Singapore; Institute of Biomedical Studies, Baylor University, Waco, TX, USA.
Anna-Marie Fairhurst, Singapore Immunology Network (SIgN), Agency for Science, Technology and Research (A*STAR), Singapore; Institute of Molecular and Cell Biology (IMCB), Agency for Science, Technology and Research (A*STAR), Singapore; Department of Microbiology and Immunology, Yong Loo Lin School of Medicine, National University of Singapore, Singapore.
Data availability
The data that support the meta-analyses on microarray and RNA sequencing have been previously published before as indicated in the text. The authors also confirm that the remaining data supporting the findings of this study are available within the article and/or its supplementary materials.
Authors’ contributions
S.H.T. designed the study, performed experiments and analysed the data. O.Z., M.M.X.T., E.A.L., H.Y.L, P.J.A. and T.C. assisted with the experiments. S.N. performed the Bayesian network analysis. W.Y.O. performed the confocal microscopy. C.J., J.C.T.L. and J.P.S.Y. performed multiplex immunohistochemistry. L.W. performed LC-MS/MS for measurement of hydroxychloroquine. B.T.K.L. performed the meta-analysis. L.H.L. performed ED-FMD for measurement of endothelial function. S.F.M., A.L., T.P.T., P.A.M. and J.E.C. provided material and participated in discussions. S.H.T., S.L. and A.-M.F. performed data analysis and interpretation. S.H.T. and A.-M.F. prepared the manuscript. A.-M.F. conceptualized and oversaw the project.
Funding
This work was supported by A*STAR-JJSI (joint grant 1218226002) and core funding from A*STAR Research Entities through the Institute of Molecular and Cell Biology and Singapore Immunology Network (AMF) and grants from the National University Health System (NUHS) Seed Fund (NUHSRO/2019/052/RO5 + 5/Seed-Mar/05) and National Medical Research Council (NMRC) Clinician-Scientist Individual Research Grant (NMRC/CNIG/1174/2017). S.H.T. was also supported by the NMRC Research Training Fellowship (NMRC/Fellowship/0020/2015).
Disclosure statement: The authors have declared no conflicts of interest.
References
- 1. Marshak-Rothstein A. Toll-like receptors in systemic autoimmune disease. Nat Rev Immunol 2006;6:823–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Podolska MJ, Biermann MH, Maueroder C, Hahn J, Herrmann M.. Inflammatory etiopathogenesis of systemic lupus erythematosus: an update. J Inflamm Res 2015;8:161–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum 1997;40:1725. [DOI] [PubMed] [Google Scholar]
- 4. Aringer M, Costenbader K, Daikh D. et al. 2019 European League Against Rheumatism/American College of Rheumatology classification criteria for systemic lupus erythematosus. Ann Rheum Dis 2019;78:1151–9. [DOI] [PubMed] [Google Scholar]
- 5. Scherlinger M, Guillotin V, Truchetet ME. et al. Systemic lupus erythematosus and systemic sclerosis: all roads lead to platelets. Autoimmun Rev 2018;17:625–35. [DOI] [PubMed] [Google Scholar]
- 6. Page C, Pitchford S.. Neutrophil and platelet complexes and their relevance to neutrophil recruitment and activation. Int Immunopharmacol 2013;17:1176–84. [DOI] [PubMed] [Google Scholar]
- 7. Denny MF, Yalavarthi S, Zhao W. et al. A distinct subset of proinflammatory neutrophils isolated from patients with systemic lupus erythematosus induces vascular damage and synthesizes type I IFNs. J Immunol 2010;184:3284–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Villanueva E, Yalavarthi S, Berthier CC. et al. Netting neutrophils induce endothelial damage, infiltrate tissues, and expose immunostimulatory molecules in systemic lupus erythematosus. J Immunol 2011;187:538–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Banchereau R, Hong S, Cantarel B. et al. Personalized immunomonitoring uncovers molecular networks that stratify lupus patients. Cell 2016;165:551–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tay SH, Celhar T, Fairhurst AM.. Low-density neutrophils in systemic lupus erythematosus. Arthritis Rheumatol 2020;72:1587–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brinkmann V, Reichard U, Goosmann C. et al. Neutrophil extracellular traps kill bacteria. Science 2004;303:1532–5. [DOI] [PubMed] [Google Scholar]
- 12. Lood C, Blanco LP, Purmalek MM. et al. Neutrophil extracellular traps enriched in oxidized mitochondrial DNA are interferogenic and contribute to lupus-like disease. Nat Med 2016;22:146–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kahlenberg JM, Carmona-Rivera C, Smith CK, Kaplan MJ.. Neutrophil extracellular trap-associated protein activation of the NLRP3 inflammasome is enhanced in lupus macrophages. J Immunol 2013;190:1217–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nickerson KM, Christensen SR, Shupe J. et al. TLR9 regulates TLR7- and MyD88-dependent autoantibody production and disease in a murine model of lupus. J Immunol 2010;184:1840–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Santiago-Raber ML, Dunand-Sauthier I, Wu T. et al. Critical role of TLR7 in the acceleration of systemic lupus erythematosus in TLR9-deficient mice. J Autoimmun 2010;34:339–48. [DOI] [PubMed] [Google Scholar]
- 16. Teichmann LL, Schenten D, Medzhitov R, Kashgarian M, Shlomchik MJ.. Signals via the adaptor MyD88 in B cells and DCs make distinct and synergistic contributions to immune activation and tissue damage in lupus. Immunity 2013;38:528–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Celhar T, Fairhurst A-M.. Toll-like receptors in systemic lupus erythematosus: potential for personalized treatment. Front Pharmacol 2014;5:265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brown GJ, Cañete PF, Wang H. et al. TLR7 gain-of-function genetic variation causes human lupus. Nature 2022;605:349–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Petri M, Orbai AM, Alarcon GS. et al. Derivation and validation of the Systemic Lupus International Collaborating Clinics classification criteria for systemic lupus erythematosus. Arthritis Rheum 2012;64:2677–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gladman D, Ginzler E, Goldsmith C. et al. The development and initial validation of the Systemic Lupus International Collaborating Clinics/American College of Rheumatology damage index for systemic lupus erythematosus. Arthritis Rheum 1996;39:363–9. [DOI] [PubMed] [Google Scholar]
- 21. Gladman DD, Ibanez D, Urowitz MB.. Systemic Lupus Erythematosus Disease Activity Index 2000. J Rheumatol 2002;29:288–91. [PubMed] [Google Scholar]
- 22. Scofield RH, Bruner GR, Namjou B. et al. Klinefelter's syndrome (47,XXY) in male systemic lupus erythematosus patients: support for the notion of a gene-dose effect from the X chromosome. Arthritis Rheum 2008;58:2511–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fairhurst AM, Wallace PK, Jawad AS, Goulding NJ.. Rheumatoid peripheral blood phagocytes are primed for activation but have impaired Fc-mediated generation of reactive oxygen species. Arthritis Res Ther 2007;9:R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Carmona-Rivera C, Zhao W, Yalavarthi S, Kaplan MJ.. Neutrophil extracellular traps induce endothelial dysfunction in systemic lupus erythematosus through the activation of matrix metalloproteinase-2. Ann Rheum Dis 2015;74:1417–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zharkova O, Tay SH, Lee HY. et al. A flow cytometry-based assay for high-throughput detection and quantification of neutrophil extracellular traps in mixed cell populations. Cytometry A 2019;95:268–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hemmi H, Kaisho T, Takeuchi O. et al. Small anti-viral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nat Immunol 2002;3:196–200. [DOI] [PubMed] [Google Scholar]
- 27. Condamine T, Dominguez GA, Youn JI. et al. Lectin-type oxidized LDL receptor-1 distinguishes population of human polymorphonuclear myeloid-derived suppressor cells in cancer patients. Sci Immunol 2016;1:aaf8943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wright HL, Makki FA, Moots RJ, Edwards SW.. Low-density granulocytes: functionally distinct, immature neutrophils in rheumatoid arthritis with altered properties and defective TNF signalling. J Leukocyte Biol 2017;101:599–611. [DOI] [PubMed] [Google Scholar]
- 29. Monaco G, Lee B, Xu W. et al. RNA-seq signatures normalized by mRNA abundance allow absolute deconvolution of human immune cell types. Cell Rep 2019;26:1627–40.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Petes C, Odoardi N, Gee K.. The toll for trafficking: toll-like receptor 7 delivery to the endosome. Front Immunol 2017;8:1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang X, Ding L, Sandford AJ.. Selection of reference genes for gene expression studies in human neutrophils by real-time PCR. BMC Mol Biol 2005;6:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vandesompele J, De Preter K, Pattyn F. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002;3:research0034.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang LZ, Ong RY, Chin TM. et al. Method development and validation for rapid quantification of hydroxychloroquine in human blood using liquid chromatography-tandem mass spectrometry. J Pharm Biomed Anal 2012;61:86–92. [DOI] [PubMed] [Google Scholar]
- 34. Rahman S, Sagar D, Hanna RN. et al. Low-density granulocytes activate T cells and demonstrate a non-suppressive role in systemic lupus erythematosus. Ann Rheum Dis 2019;78:957–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Evrard M, Kwok IWH, Chong SZ. et al. Developmental analysis of bone marrow neutrophils reveals populations specialized in expansion, trafficking, and effector functions. Immunity 2018;48:364–79.e8. [DOI] [PubMed] [Google Scholar]
- 36. Carestia A, Kaufman T, Rivadeneyra L. et al. Mediators and molecular pathways involved in the regulation of neutrophil extracellular trap formation mediated by activated platelets. J Leukocyte Biol 2016;99:153–62. [DOI] [PubMed] [Google Scholar]
- 37. van Velzen JF, Laros-van Gorkom BA, Pop GA, van Heerde WL.. Multicolor flow cytometry for evaluation of platelet surface antigens and activation markers. Thromb Res 2012;130:92–8. [DOI] [PubMed] [Google Scholar]
- 38. Naegelen I, Plançon S, Nicot N. et al. An essential role of syntaxin 3 protein for granule exocytosis and secretion of IL-1α, IL-1β, IL-12b, and CCL4 from differentiated HL-60 cells. J Leukocyte Biol 2015;97:557–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pirillo A, Norata GD, Catapano AL.. High-density lipoprotein subfractions—what the clinicians need to know. Cardiology 2013;124:116–25. [DOI] [PubMed] [Google Scholar]
- 40. Garcia-Romo GS, Caielli S, Vega B. et al. Netting neutrophils are major inducers of type I IFN production in pediatric systemic lupus erythematosus. Sci Transl Med 2011;3:73ra20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gardet A, Pellerin A, McCarl CA. et al. Effect of in vivo hydroxychloroquine and ex vivo anti-BDCA2 mAb treatment on pDC IFNα production from patients affected with cutaneous lupus erythematosus. Front Immunol 2019;10:275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Koupenova M, Mick E, Mikhalev E. et al. Sex differences in platelet toll-like receptors and their association with cardiovascular risk factors. Arterioscler Thromb Vasc Biol 2015;35:1030–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Koupenova M, Vitseva O, MacKay CR. et al. Platelet-TLR7 mediates host survival and platelet count during viral infection in the absence of platelet-dependent thrombosis. Blood 2014;124:791–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang JP, Bowen GN, Padden C. et al. Toll-like receptor-mediated activation of neutrophils by influenza A virus. Blood 2008;112:2028–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Odhams CA, Roberts AL, Vester SK. et al. Interferon inducible X-linked gene CXorf21 may contribute to sexual dimorphism in systemic lupus erythematosus. Nat Commun 2019;10:2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jourde-Chiche N, Whalen E, Gondouin B. et al. Modular transcriptional repertoire analyses identify a blood neutrophil signature as a candidate biomarker for lupus nephritis. Rheumatology (Oxford) 2017;56:477–87. [DOI] [PubMed] [Google Scholar]
- 47. Toro-Dominguez D, Martorell-Marugan J, Goldman D. et al. Stratification of systemic lupus erythematosus patients into three groups of disease activity progression according to longitudinal gene expression. Arthritis Rheumatol 2018;70:2025–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bennett L, Palucka AK, Arce E. et al. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med 2003;197:711–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hassani M, Hellebrekers P, Chen N. et al. On the origin of low-density neutrophils. J Leukocyte Biol 2020;107:809–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lood C, Arve S, Ledbetter J, Elkon KB.. TLR7/8 activation in neutrophils impairs immune complex phagocytosis through shedding of FcgRIIA. J Exp Med 2017;214:2103–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bonegio RG, Lin JD, Beaudette-Zlatanova B. et al. Lupus-associated immune complexes activate human neutrophils in an FcγRIIA-dependent but TLR-independent response. J Immunol 2019;202:675–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Duffau P, Seneschal J, Nicco C. et al. Platelet CD154 potentiates interferon-alpha secretion by plasmacytoid dendritic cells in systemic lupus erythematosus. Sci Transl Med 2010;2:47ra63. [DOI] [PubMed] [Google Scholar]
- 53. Vial G, Gensous N, Savel H. et al. The impact of clopidogrel on plasma-soluble CD40 ligand levels in systemic lupus erythematosus patients: the CLOPUS phase I/II pilot study. Joint Bone Spine 2021;88:105097. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the meta-analyses on microarray and RNA sequencing have been previously published before as indicated in the text. The authors also confirm that the remaining data supporting the findings of this study are available within the article and/or its supplementary materials.