Highlights
-
•
Identified all odorant receptor gene family members in the vine mealybug, Planococcus ficus, from genomic and transcriptomic resources.
-
•
Evaluated sex-specific expression patterns of odorant receptor genes.
-
•
Tested several receptors for responsiveness to mealybug sex pheromones and plant volatiles using a cell-based expression system.
-
•
Experimentally determined that one receptor is highly sensitive and specific to the sex pheromone used by this species.
Keywords: Vine mealybug, Insect odorant receptor, Sex pheromone, HEK293 cells, Lavandulyl senecioate, Planococcus ficus
Abstract
The vine mealybug, Planococcus ficus, is a significant pest of vineyards in all major grape growing regions of the world. This pest causes significant aesthetic damage to berry clusters through its feeding behavior and secretion of "honeydew", which leads to significant decreases in crop marketability. More importantly, the vine mealybug is a vector of several grapevine viruses which are the causal agent of grapevine leafroll disease, one of the most destructive and economically devastating diseases of the grape industry worldwide. As there is no cure for grapevine leafroll disease, the only control measures available to reduce its spread are to remove infected vines whilst simultaneously controlling mealybug populations. Using transcriptomic libraries prepared from male and female mealybugs and a draft genome, we identified and evaluated expression levels of members of the odorant receptor gene family. Interestingly, of the 50 odorant receptors identified from these P. ficus genetic resources, only 23 were found to be expressed in females, suggesting this flightless life stage has a decreased reliance on the olfactory system. In contrast, 46 odorant receptors were found to be expressed in the alate male life stage. Heterologous expression of eight of these receptors, along with the obligate co-receptor, Orco, in HEK293 cells allowed for the identification of two receptors that respond to lavandulyl senecioate, the sole constituent of the sex pheromone used by this species. Interestingly, one of these receptors, PficOR8, also responded to the sex pheromone used by the Japanese mealybug, Planococcus kraunhiae. The data presented here represent the first report of odorant receptor gene family expression levels, as well as the identification of the first sex pheromone receptor, in soft-scale insects. The identification of a receptor for the vine mealybug sex pheromone will allow for the development of novel, species-specific pest control tools and monitoring devices.
Introduction
Grapevine leafroll disease (GLD) is one of the most destructive and economically devastating diseases of the grape industry, worldwide (Almeida et al., 2013). GLD has been shown to disrupt grapevine growth rates and yields, especially in Vitis vinifera, and current economically based guidance directs growers to replant vineyards when infection rates reach 25 % or greater (Atallah et al., 2012; Ricketts et al., 2015). The disease is caused by several closely related RNA viruses known as grape leafroll-associated viruses (GLRaVs) belonging to the family Closteroviridae which can be transmitted between grapevines via grafting or by soft-scale insects, including several mealybug species (Maree et al., 2013; Naidu et al., 2014). Grapevines serve as the bona fide host for GLRaVs where they live and replicate for the lifespan of the grapevine. Female mealybugs are sap-sucking insects (males do not feed), having highly specialized mouthparts that they use to pierce and suck nutrients from the phloem of grapevines (Pitino et al., 2014; Alliaume et al., 2018). In doing so, mealybugs inadvertently suck up the viruses that cause the disease and carry them throughout the vineyard, injecting them into uninfected plants as they feed. To date there remains no cure or treatment for GLD, so the best strategy to protect a vineyard from GLD is to plant or graft only from certified virus-free rootstock and scion, rogue virus-infected vines, and vigorously attempt to control insect vector populations.
There currently exists over 2000 species of mealybugs in the world (Garcia Morales et al., 2016), with many being considered significant agricultural pests to various crops due to their ability to transmit plant pathogens. With respect to grapes, only a handful of mealybug species are considered significant pests due to their ability to transmit GLRaVs, with most being geographically restricted to certain grape growing regions around the world. For example, the grape mealybug, Pseudococcus maritimus, and the vine mealybug, Planococcus ficus, are the most common mealybug species in vineyards in the U.S., yet they are currently not found in New Zealand; instead, New Zealand's vineyards are dominated by the citrophilus mealybug, Pseudococcus calceolariae, and the long-tailed mealybug, Pseudococcus longispinus (reviewed in Daane et al., 2012). To date, several studies have investigated the vector specificity of various mealybug species with various GLRaVs, while only few have focused on understanding the transmission biology of the vector-virus relationship (Cid et al., 2007; Douglas and Kruger, 2008; Tsai et al., 2010; Kruger et al., 2015). These studies have shown that most mealybugs can transmit multiple GLRaV species, that GLRaVs exist in mealybugs semi-persistently (i.e., only for a few days), and that the transmission rates of virus from an individual mealybug to a grapevine are consistently low across species.
Historically, vineyard mealybugs have been controlled using cultural practices, biological control agents, and insecticides, with varying degrees of effectiveness depending on factors such as geographic location, degree of infestation, and resources available. However much of the difficulty in controlling mealybugs in vineyards results from inherent biological characteristics such as the fact that mealybugs are naturally coated with a waxy covering and that they spend most of their lives underneath the bark of grapevines (Geiger and Daane, 2001; Cocco et al., 2021). As a result, much of the standard insecticide arsenal is rendered inefficient and ineffective, necessitating the use of alternative, systemic chemistries (e.g., imidacloprid and spirotetramat) that can be delivered to the mealybugs through the plants while feeding. These chemistries are much more effective at controlling mealybugs in vineyards, however much like other insecticides, they too are facing increased regulation and use restrictions due to valid concerns regarding resistance development (Wen et al., 2009; Fournier-Level et al., 2019) and impacts on pollinator health (Chen et al., 2021; Zhang et al., 2022). There exists a need to develop more tools to control mealybugs in vineyards to stop the spread of GLD.
Like all insects, mealybugs rely heavily upon their sense of smell for foraging and mating purposes. Grapevines emit unique blends of odorant molecules into the environment that mealybugs detect and use to differentiate grapevines from other plants. More specifically, grapevine volatiles enter the mealybug antennae where they bind to odorant receptors (ORs) that, upon activation, cause neuronal firing that leads to attractive behavior towards the source of the odorant molecule. Similarly, female mealybugs emit sex pheromone molecules into the environment that are detected by males of the same species using ORs that are highly specific and sensitive to the mating cues. The reliance of mealybugs on using sex pheromones to communicate has led to the development of the practice of mating disruption, in which the mealybug's own sex pheromone is released in high concentrations in vineyards during reproductive cycles to disrupt the ability of males to find females. While this tactic has proven effective for some species as an augmentation to existing IPM practices, it has its own limitations primarily due to the requirement of large quantities of mealybug pheromones which can be very expensive or difficult to synthesize. Nevertheless, mealybug sex pheromones are incredibly unique, which offers an opportunity for exploitation (Franco et al., 2022). For example, the molecule that is produced and used by the vine mealybug as a sex pheromone, lavandulyl senecioate, is not found anywhere else in nature (Hinkens et al., 2001). The fact that mealybugs produce distinct chemicals and emit them into the environment presents an opportunity to revolutionize our ability to detect these cryptic species in the field through the development of sex pheromone-detecting biosensors.
In insects, there are three main types of proteins that facilitate the detection of odorant molecules: the odorant binding proteins (OBPs), that transport the hydrophobic odorant molecules through the aqueous sensillar lymph to the ORs; the ORs, which function as ligand-gated ion channels causing neuronal depolarization upon activation; and odorant degrading enzymes (ODEs), which are responsible for degrading odorant molecules after OR activation, thereby resetting the system for subsequent reactivation. Interestingly, these molecules have evolved independently in insects and are not related to the molecules used in the olfactory systems by all other animals (Robertson et al., 2003; Benton et al., 2006; Missbach et al., 2014). Moreover, insect olfactory proteins are highly divergent, even between closely related insects, with many of these proteins being unique to a single species. The uniqueness of insect olfactory proteins makes them ideal targets in next-generation “insecticide” development programs, as there are likely to be significantly less off-target effects compared to traditional neurotoxic insecticides that target highly conserved proteins, such as acetylcholine receptors. Functional studies have revealed that insect OBPs and ODEs tend to be broadly “tuned” (Hamiaux et al., 2020; Corcoran et al., 2023), in that they are not highly specific for certain odorants, but rather capable of transporting or degrading, respectively, a wide range of chemically related molecules. In contrast, insect ORs have generally been found to be highly specific and sensitive towards a certain odorant molecule that is biologically relevant to the organism in question (Mansourian et al., 2016; Yuvaraj et al., 2022). These properties of ORs make them ideal candidates as the core constituent of recombinant protein-based biosensors. For example, the vine mealybug possesses an OR that is highly specific and sensitive to its sex pheromone which could theoretically function as the core element in a field-based sensor designed to detect the presence of the insect in vineyards.
The aim of the current study is to identify a receptor for the vine mealybug sex pheromone to, 1) facilitate the identification of receptor agonists and/or antagonists that can ultimately be used as tools in mating disruption programs in a more efficient and effective manner than using the sex pheromone itself, and 2) to facilitate the development of a vine mealybug sex pheromone-detecting biosensor. Towards this end, we undertook a bioinformatic approach to identify the vine mealybug OR genes and identify putative sex pheromone receptor genes based on their expression levels in male and female adults. We then cloned the open reading frames of eight OR genes, expressed them in mammalian cell lines, and tested their ability to respond to the vine mealybug sex pheromone, lavandulyl senecioate, as well as to various odorant molecules known to be emitted by grapevines.
Materials & methods
Insects and extraction of genetic material
Male and female adult vine mealybugs were obtained from an insectary maintained by Dr. Kent Daane at the University of California's Kearney Agricultural Research and Extension Center in Parlier, California. Heads and antennae were removed from 50 male and 50 female vine mealybugs and immediately placed into separate 1.5 mL Eppendorf tubes on dry ice. This process was repeated on three separate occasions, comprising three male and three female replicate samples. Total RNA was then extracted from each sample using Trizol (ThermoFisher, Waltham, MA, USA) reagent following the manufacturer's protocol. Genomic DNA was purified from three adult P. ficus females using DNAzol (ThermoFisher) following the manufacturer's instructions.
Genetic sequencing and bioinformatic analyses
The six RNA samples were then shipped to Novogene, Inc (Sacramento, California, USA) for sample integrity analysis, sequencing, and transcriptome assembly. Briefly, 250–300 bp cDNA libraries were prepared for each sample and sequenced on Illumina's Novaseq platform (PE150), after which low-quality reads and adapter sequences were removed from a minimum of 154,000,000 raw reads per sample prior to assembly. Trinity software was used for de novo transcriptome assembly (Grabherr et al., 2011), RSEM software was used to map clean reads onto the transcriptome (Li and Dewey, 2011), and DESeq2 software was used to normalize readcounts amongst biological replicates for gene expression analyses (Love et al., 2014). A minkercov value of 1 was chosen for transcript assembly which allowed for the identification of rare and lowly expressed transcript variants. Purified genomic DNA was sent to Novogene, Inc., for sample integrity analysis, sequencing, and genome assembly. Briefly, a 350 bp insert library was prepared from the DNA sample and sequenced on Illumina's Novaseq platform (PE150), after which low-quality reads and adapter sequences were removed from 52,804,232 raw reads prior to assembly into a preliminary genome using SOAPdenovo software (Luo et al., 2012).
Odorant receptors were identified from the transcriptome using published OR sequences from other insects (e.g., Drosophila melanogaster) as queries in tBLASTn analyses using Geneious software (Biomatters, Auckland, New Zealand). Once P. ficus ORs (PficORs) were identified from the transcriptome, these sequences were used as queries in further, iterative searches to ensure all ORs present in the transcriptome had been identified. The open reading frame sequences (ORFs) of PficORs obtained from the transcriptome were verified, and partial sequences were extended where possible, using the draft P. ficus genome. Vine mealybug ORs were deemed to be expressed if their transcript readcounts were 1 or higher, and differentially expressed between males and females if the log2(fold change) was greater than 1 and the adjusted p-value was less than 0.05.
Phylogenetics
The ORF sequences obtained from the vine mealybug transcriptome were used in phylogenetic analyses to evaluate orthologous relationships between the PficORs and those of other hemipteran insects in the same suborder (Sternorrhyncha) as P. ficus. Amino acid sequences encoding ORs from the pea aphid, Acyrthosiphon pisum were obtained from the literature (Smadja et al., 2009), and ORs from the giant scale insect, Drosicha corpulenta, grape phylloxera, Daktulosphaira vitifoliae, and the Asian citrus psyllid, Diaphorina citri, were obtained from the National Center for Biotechnology Information (NCBI.gov). The OR sequences were used to produce a multiple sequence alignment using the MUSCLE (version 3.8.425) application embedded in Geneious software (Biomatters Ltd). The resulting sequence alignment was then used to construct a phylogenetic tree using PhyML software (version 3.3.20180621) based on 1000 bootstrap replicates. Accession numbers of ORs obtained from NCBI for phylogenetic analyses are listed in Supplementary File 1.
Molecular cloning of PficORs
Eight odorant receptors that displayed relatively high expression in male samples in transcriptomic analyses were selected for functional testing in HEK293 cells. PficORs 1A, 2, 3, 4, 5, 6, 7 and 8, plus the obligate OR-coreceptor, Orco, were amplified from cDNA as previously described (Corcoran et al., 2014). Briefly, full-length coding sequences of PficOrco and each PficOR were amplified from cDNA and ligated into the entry vector pJET1.2 (ThermoFisher). Escherichia coli was transformed with each ligation, spread on LB plates, incubated overnight at 37 °C, and resulting colonies were tested for the presence of the plasmid containing the correct inserts by PCR. Positive colonies were scaled up in LB broth overnight and plasmids were harvested using a GeneJet plasmid miniprep kit (ThermoFisher). Purified plasmids were Sanger sequenced (Eurofins, Louisville, KY, USA) and those without errors were used as templates for subsequent PCRs in which 5′ and 3′ restrictions sites were added to the DNA with a second set of primers. Purified DNA encoding each OR gene was then digested using NotI/ApaI (PficORs) or NotI/XhoI (PficOrco) restriction enzymes (New England Bio, Ipswich, Massachusets) and ligated into the expression vectors pcDNA5TO or pcDNA4TO (ThermoFisher), respectively. Expression vectors containing each PficOR or Orco were purified and sequenced and those without errors were used for transfection into HEK293 cells. Large quantities of high-quality pcDNA5TO/PficOR and pcDNA4TO/PficOrco plasmids were produced using a PureLink HiPure Plasmid Midiprep Kit (ThermoFisher) following the manufacturer's protocol.
Heterologous expression of PficORs in HEK293 cells
The methods used to generate HEK293 cell lines stably expressing combinations of insect Orco/ORs have been described in detail previously (Corcoran et al., 2014). Briefly, pcDNA4TO/PficOrco was transfected into a previously reported isogenic HEK293 cell line (HEK293/TRc14; Corcoran et al., 2014) stably expressing pcDNA6TR (ThermoFisher). This cell line constitutively expresses a tetracycline repressor (TR) protein that represses transcription from pcDNA4TO and pcDNA5TO until a TR inhibitor is added to the cell culture media. For transfections, five micrograms of pcDNA4TO/PficOrco and 15 μL of Lipofectamine 2000 (ThermoFisher) transfection reagent were each diluted into 500 μL of Optimem medium (ThermoFisher) and incubated at room temperature for 10 min, after which they were mixed together and incubated for an additional 60 min. The plasmid/Lipofectamine 2000 mixture was then added to a T-25 cell culture flask containing HEK293/TRc14 cells at approximately 70 % confluency and incubated overnight (37 °C, 5 % CO2). The next day the culture medium was removed and replaced with fresh cell culture medium containing 400 μg/mL of Zeocin (ThermoFisher). The cells were cultured for four weeks in the presence of Zeocin until an antibiotic-resistant cell line was established, then the Zeocin concentration was reduced to 200 μg/mL and blasticidin (10 μg/mL) was added to the cell culture medium. The resulting HEK293/TRc14/PficOrco (TREx/PficOrco) cell line was passaged three times and frozen prior to further use. Each pcDNA5TO/PficOR plasmid was transfected into a fresh thaw of the TREx/PficOrco cell line using the methods described above, except that cells were cultured for approximately three weeks in the presence of 200 µg/mL hygromycin. Once antibiotic-resistant cell lines were established, the hygromycin concentration was reduced to 100 μg/mL and zeocin (200 μg/mL) and blasticidin (10 μg/mL) were added to the cell culture medium. All TREx/PficOrco/PficOR'X’ cell lines were passaged three times, frozen at −80°C and thawed prior to functional testing.
Functional testing
The materials and methods used to test cell lines expressing PficORs for response to pheromones and odorant compounds are similar to those described in detail previously (Corcoran et al., 2014). Briefly, each cell line was lifted from culture flasks and 25,000 cells were plated into wells of black-walled, poly-d-lysine coated 96-well plates. Cells were incubated overnight at 37 °C with 5 % CO2. On day two, the cell culture medium was removed from the plates, fresh medium was added to the top four rows, and fresh medium containing 1 μg/mL doxycycline induction reagent was added to the bottom four rows to induce expression of exogenous genes from pcDNA4TO and pcDNA5TO. Cells were incubated again overnight at 37 °C with 5 % CO2. The following morning, cells were rinsed and loaded with the calcium-sensitive indicator Fluo4-AM. After incubation and rinsing, cells were tested for response to test compounds using a FlexStation-3 platereader. The baseline fluorescence of cells was monitored for 30 s prior to injection with 11 μL of controls or test compounds using the automated injection system of the Flexstation-3. After compound injection, changes in well fluorescence were monitored every 5 s for 2 min. The mean response of four wells (technical replicates) that received the same treatment was calculated as a percent increase in fluorescence between baseline and 15 s post-treatment (when the response peak occurred). The response of TREx/PficOrco/PficOR'X’ cell lines were tested on three independent occasions (biological replicates) and the mean response of cells from each experiment was used to generate concentration response curves using the non-linear regression function embedded in GraphPad Prism software (version 10.1.1, GraphPad Software, Boston, MA, USA).
For screening experiments cells were tested for a response to a single dose (30 μM) of each of the test compounds. Odorant compounds used in screening experiments were purchased from commercial suppliers and all had purities ≥ 95 %. Mealybug pheromones were synthesized and provided by Dr. Jocelyn Millar and had purities ≥ 97 %. Test compounds were diluted to a 200X concentration (6 mM) in 100 % DMSO, then diluted to 10X (300 μM) in assay buffer. Compounds were diluted to 1X (30 μM) by adding 11 μL of compound to 99 μL of assay buffer in wells. In addition, four wells of non-induced and four wells of induced cells on each plate were treated with a negative (vehicle, 0.5 % DMSO in assay buffer) and positive (50 μM VUAA1) control. VUAA1 directly agonizes Orco in most insect species (Jones et al., 2011; Andersson et al., 2016; Corcoran et al., 2018) and its use provides confidence that PficOrco was present and functional in each cell line and that the reporter assay was operating properly. Each well of cells was treated with only a single compound, or dose of compound, in all cases. Where possible, compounds that elicited responses during screening experiments were tested further in dose-response studies. Serial dilutions of each compound were performed in 100 % DMSO starting at 200X, followed by dilutions to 10X in assay buffer. Each dose of each compound was delivered to four induced and four non-induced wells of cells using the FlexStation injector system as described above.
Results
Odorant receptor identification and expression analysis
A total of 47 OR genes plus the OR coreceptor, Orco, were found to be expressed at an mRNA level in either male or female P. ficus antennae/head samples. Three receptors, PficORs 9, 14 and 49, were not detected in the transcriptome and were identified using the draft genome only. Alternative splicing variants were found in the transcriptome and verified in the genome for PficORs 1 and 7. PficOR1A and 1B share the same first 387 amino acids, however there are an additional 77 amino acids comprising two additional exons (exons 5 and 6) present on the C-terminus of PficOR1A which are not present in PficOR1B; instead, an additional 22 amino acid extension of exon 4 is present on the C-terminus of PficOR1B. PficORs 7A and 7B differ in their first 299 amino acids (exons 1 and 2), yet share the same C-terminal 108 amino acids (exons 3 and 4). Of the 47 PficOR genes present in the transcriptome, only one, PficOR1B, was not expressed (readcount < 1) in male samples. The odorant receptor co-receptor Orco displayed the highest normalized readcounts (NR) in male samples (NR = 1595) followed by ORs 6, 8, 2, 1A and 5 (NRs of 830, 331, 297, 186 and 109, respectively). The remaining PficORs that were expressed in males displayed varying readcounts of less than 100. In contrast, only 23 of the 47 PficORs were found to be expressed in female samples, however 22 of these, as well as Orco, displayed readcounts of less than 13. The remaining OR detected in females, PficOR1A, had relatively high expression (NR = 127). The OR coreceptor Orco, PficOR6 and PficOR8 were the only OR genes found to have statistically significant differences (padj ≤ 0.05) in expression levels between male and female samples (Fig. 1; Supplementary Table 1), with all being significantly male-biased.
Fig. 1.
Relative odorant receptor (OR) gene expression in male and female Planococcus ficus as determined by RNASeq analyses. For heatmap visualization, readcount data were rounded to integers before normalizing in DESEQ2 by “regularized” log (R-log) transforming each transcript count (log2 (x + 0.5)) (Love et al., 2014).
Phylogenetics
The evolutionary relatedness of PficORs to other hemipteran odorant receptors was evaluated through phylogenetic analyses. Roughly one-third of the PficORs were interspersed throughout the resulting phylogenetic tree, likely indicating orthologous relationships between the proteins and those from other related insects (Fig. 2). Interestingly, those ORs that display clear orthologous relationships (e.g., PficOR1A and DcorOR17) are clustered towards the base of the phylogeny suggesting these ORs are more ancestral; the remaining ORs (with few exceptions) form large, species-specific expansions of paralogous genes that have likely evolved more recently, coinciding with the evolution and specialization of the insects over time. Because so much biodiversity exists within the suborder Sternorrhyncha (e.g., aphids, psyllids, mealybugs, phylloxera), it is not surprising that only a small portion of the PficORs demonstrated orthologous relationships with other sternorrhynchan OR genes. To date, there is very little genetic information available regarding ORs from soft-scale insects, such as other mealybugs, which limits the ability to draw inferences regarding orthologous relationships of the PficORs identified in this study.
Fig. 2.
Evolutionary relationships of odorant receptors (OR) of five hemipteran insects within the suborder Sternorrhynca, including the vine mealybug, Planococcus ficus (Pfic; black font), the giant scale insect, Drosicha corpulenta (Dcor; green font), grape phylloxera, Daktulosphaira vitifoliae (Dvit; red font), the pea aphid, Acyrthosiphon pisum (Apis; blue font), and the Asian citrus psyllid, Diaphorina citri (Dcit; orange font). Node values represent bootstrap values based on 1000 replicates.
Functional testing
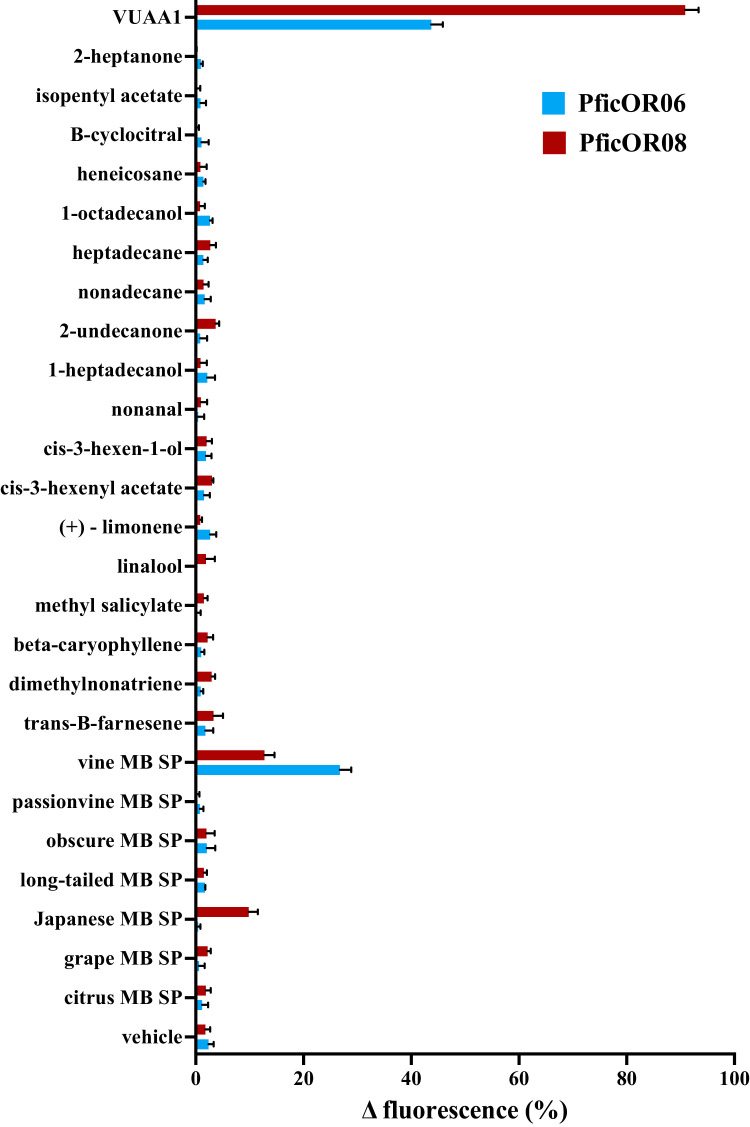
Of the eight PficORs that were tested for their ability to respond to grapevine volatiles and mealybug sex pheromones in cell-based assays, only two, PficORs 6 and 8, responded to any of the compounds tested. All eight cell lines responded to the Orco agonist VUAA1 and did not respond to the negative control (vehicle) when Orco/OR expression was induced, indicating that the Orco/ORs were expressed in each cell line in a regulated fashion and that the functional assay was operating properly. The TREx/PficOrco/PficOR6 cell line responded to the Orco agonist VUAA1 and to the vine mealybug sex pheromone component, lavandulyl senecioate, in screening assays (Fig. 3) and dose-dependently to the pheromone with an EC50 of 4.6 μM (Fig. 4). The TREx/PficOrco/PficOR8 cell line responded to VUAA1, lavandulyl senecioate, and the Japanese mealybug sex pheromone component, 2-isopropyliden-5-methyl-4-hexen-1-yl butyrate, in screening assays (Fig. 3). This cell line responded dose-dependently to both the vine mealybug and Japanese mealybug sex pheromone components with EC50s of 52.6 μM and 8.6 μM, respectively (Fig. 4). While both PficOR6 and PficOR8 responded to the vine mealybug sex pheromone component, PficOR6 was ∼13x more sensitive to the compound than PficOR8, and PficOR6 was specifically tuned to the compound compared to PficOR8, which together suggests that PficOR6 is the receptor used by the insect to detect the sex pheromone in the environment. Since the Japanese mealybug, Planococcus kraunhiae, and the vine mealybug are relatively closely related evolutionarily, it is possible that PficOR8 represents an OR possessed by the last common ancestor of the vine and Japanese mealybugs from which PficOR6 has evolved to be specifically tuned to the vine mealybug sex pheromone.
Fig. 3.
Response of TREx/HEK293/PficOrco/PficOR6 and TREx/HEK293/PficOrco/PficOR8 cell lines to various grapevine volatiles or mealybug sex pheromone (MB SP) compounds. Data represent the mean (± SEM) response of cells from three independent experiments.
Fig. 4.
Response of TREx/HEK293/PficOrco/PficOR6 and TREx/HEK293/PficOrco/PficOR8 cell lines to various concentrations of active compounds identified in screening experiments. Response of induced and non-induced cells expressing PficOrco/OR6 (A) or PficOrco/OR8 (B) to the vine mealybug sex pheromone, lavandulyl senecioate. Response of induced and non-induced cells expressing PficOrco/OR8 (C) to the Japanese mealybug sex pheromone, 2-isopropyliden-5-methyl-4-hexen-1-yl butyrate. Data represent the mean (± SEM) response of cells from three independent experiments. All figures are displaying error bars, some are too small to be visible.
Discussion
The sense of smell is, by far, the chemosensory modality that insects use the most as they navigate their environment. In some cases, certain insects have evolved to depend heavily on, for example, vision (e.g., dragonflies) or hearing (e.g., crickets), however these are exceptions and not the rule. For most insects, it is all about how things smell; food sources, mates, oviposition sites, congenitors, and caste members are all recognized and differentiated based on the unique volatile chemical signatures that they possess. These olfactory based “decisions” are critical to an insect's survival, and they are directly influenced by underlying peripheral olfactory proteins, such as OBPs, ODEs and ORs. Therefore, insect behavior can be directly influenced by interfering with the function of these proteins. One can envision a whole new suite of insect repellant, attractant, or mating disruption products that could be developed by taking a pharmacological approach to target insect olfactory proteins. The obvious big hurdle to pursuing this approach, and perhaps why there are no synthetic commercial products on the market that target specific insect olfactory proteins, is that not a lot is known about insect olfactory proteins, relatively speaking. The incredible uniqueness of insect olfactory proteins has only been recognized in the last decade or so due to dramatic advances in genetic and molecular biology technologies, such as next-generation sequencing platforms. Two prerequisites to targeting insect olfactory proteins, having the ability to identify them in an organism of interest and the ability to study their function, have only become tangible relatively recently.
This study fulfills the prerequisites of developing olfactory-based commercial products that can be used to help control vine mealybug populations in vineyards. Towards this end, we undertook a relatively simple approach to identify the ideal target olfactory protein from the vine mealybug, the receptor for the sole constituent of its sex pheromone. The bioinformatic approach we deployed here allowed us to identify the entire repertoire of odorant receptors that are possessed by the vine mealybug and to determine which are expressed in the insect's primary olfactory organ. In our analyses, we were able to identify a total of 50 OR genes, which represents a complement that is lower than the total typically found in lepidopteran (e.g., 70 in Epiphyas postvittana; Corcoran et al., 2015), coleopteran (e.g., 259 in Tribolium castaneum; Engsontia et al., 2008), dipteran (e.g., 131 in Aedes aegypti; Bohbot et al., 2007), or even other hemipteran (e.g., 79 in Acyrthosiphon pisum; Smadja et al., 2009) insects. Interestingly, despite having 50 OR genes present in its genome, female vine mealybugs were found to express less than half in their antennae. Moreover, with one exception, those that were expressed in the female antennae were expressed at very low levels compared to male vine mealybugs. It has been proposed that the insect olfactory system rapidly evolved with the capability of flight (Missbach et al., 2014), so it is possible the reduced expression of OR genes in female vine mealybugs – and therefore reduced reliance on the olfactory system – is a result of their inability to fly. This theory is supported by our findings that male mealybugs, which do fly, displayed some level of expression of most of the ORs encoded in the genome.
Given that female mealybugs emit a sex pheromone that is detected by an OR in male mealybugs and given the importance of the male's ability to detect these cues on its reproductive fitness, it stood to reason that the receptor for the sex pheromone would be one of the most highly expressed receptors in male antennae. Indeed, this reasoning allowed us to identify a receptor that was highly specific and sensitive to the vine mealybug sex pheromone. Interestingly, in our functional studies, we found that only 2 of the 8 receptors tested responded to any of the test compounds studied, which included several important grapevine volatiles. As mentioned above, it is quite likely that because female mealybugs do not fly there is significantly less selective pressure to maintain a functioning olfactory system capable of detecting grapevine volatiles. With males, it is a little less clear because they do fly and there would likely exist selective pressure to maintain ORs that detect grapevine volatiles because they would increase their reproductive fitness by allowing them to locate females more efficiently, as has been shown to be the case in other insects (Gonzalez et al., 2020). However, in the case of mealybugs, while the males fly, they do not feed and only live for a few days after emerging from eggs laid by the females. These biological characteristics of male mealybugs may alleviate the selection pressure to maintain ORs capable of detecting grapevine volatiles as well. It is also quite possible, and likely, that the various larval stages of P. ficus display varying OR expression patterns compared to adult males and females. The first instar (or “crawler”) stage is the most mobile, feeding mealybug life stage, so it could be that ORs tuned to grapevine volatiles are primarily expressed and used during this developmental period. Finally, it is possible that our heterologous expression system is not perfect, and responses of ORs to grapevine volatiles were not observed for various reasons, such as a lack of other required olfactory proteins (e.g., OBPs) or improper OR protein folding or trafficking within the mammalian cell lines. In the absence of any behavioral or electrophysiological data regarding the biological relevance of the various odorants tested in our cell-based assays (with the exception of the sex pheromone used by this species), it is impossible to know what compounds there “should” be receptors for.
Previous studies have reported the presence of a second component, (S)-lavandulyl isovalerate, in the pheromone blends of certain Israeli vine mealybug populations (Zada et al., 2003; Kol-Maimon et al., 2010). Interestingly, subsequent genetic studies confirmed the presence of a cryptic, sympatric species (Planococcus vitis) within Eastern Mediterranean vineyards (Correa et al., 2023), and that North American P. ficus populations originated from this region (Daane et al., 2018). It is possible that this “new” species, P. vitis, utilizes (S)-lavandulyl isovalerate as a sex pheromone (and not P. ficus), however, to date there have been no studies that have evaluated sex pheromone production in P. vitis in conjunction with DNA barcoding genetic analyses. Based on what has been learned about sex pheromone communication in other sympatric insects, it would be expected that these two Planococcus species are each capable of detecting both pheromone compounds (De Pasqual et al., 2021; Khallaf et al., 2021). The difference would be that in both species, one compound serves as an attractant and the other serves as an inhibitor of attraction. Therefore, with respect to the current study, some of the remaining non-responsive PficORs tested in our assays may in fact be tuned to (S)-lavandulyl isovalerate, however this compound was not available for use during functional characterization studies. Future studies are needed to evaluate and compare olfactory gene expression levels and function between these two closely related species.
We chose to focus on the vine mealybug because it is a very serious agricultural pest to the worldwide grape industry, current control methods are inadequate, and the geographic range of this pest is likely to spread with impending global warming (Ji et al., 2020). We chose to focus on the sex pheromone receptor used by this species not only because of its biological uniqueness and critical function (characteristics that most olfactory proteins possess), but also because its identification would allow for the pursual of two different types of novel pest control tools: synthetic mating disruptants and species-specific biosensors. The vine mealybug sex pheromone receptor responded robustly and consistently in our mammalian cell-based assay system, which are characteristics desirable for high-throughput screening campaigns. For years, scientists have experimented with modifying the natural version of insect pheromones through modifications such as halogenation or isoteric replacements and found altered activity towards ORs, showing promise in the development of novel, behavior modifying pest control tools (Renou and Guerrero, 2000; Pesenti and Viani, 2004). However, these studies historically have had to rely on behavioral or electrophysiological experiments to evaluate the activity of the molecules, approaches that are both far from being high-throughput. The cell-based approach used here will allow for the evaluation of the activity of these types of molecules against pheromone receptors much more efficiently and effectively, which may speed up the development of novel, vine mealybug-specific mating disruptants. Finally, the identification of the vine mealybug sex pheromone receptor facilitates the development of a vine mealybug specific biosensor. A biosensor capable of detecting vine mealybugs in the field would have various applications that would benefit the viticulture industry. Individual growers would be able to know exactly how exposed they were to the risk of GLD spreading over time if they had infected vines, or detect a mealybug infestation early enough to prevent significant losses from transmission of GLD. In addition to simply improving our ability to detect and monitor mealybug populations in vineyards, these sensors could be spread over larger geographical regions and linked to automated reporting systems that could monitor the spread of these pests throughout a region or across a border.
Funding
This work was supported by Oregon State University's Agricultural Research Foundation (W.F.M.), and the U.S. Department of Agriculture, Agricultural Research Service under CRIS project: 5070-22000-038-00D (J.A.C.), and 2072-22000-045-00D (W.F.M.).
CRediT authorship contribution statement
Jacob A. Corcoran: Conceptualization, Funding acquisition, Methodology, Investigation, Formal analysis, Writing – original draft, Writing – review & editing. Walter F. Mahaffee: Conceptualization, Project administration, Funding acquisition, Writing – review & editing.
Declaration of competing interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgements
We are very grateful to Kent Daane for providing vine mealybug specimens and to Jocelyn Millar for providing various mealybug sex pheromone components. The findings and conclusions in this publication are those of the authors and should not be construed to represent any official USDA or U.S. Government determination or policy. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.cris.2024.100072.
Appendix. Supplementary materials
Data availability
Data will be made available on request.
References
- Alliaume A., Reinbold C., Uzest M., Lemaire O., Herrbach E. Mouthparts morphology of the mealybug Phenacoccus aceris. Bull. Insectol. 2018;71(1):1–9. [Google Scholar]
- Almeida R.P.P., Daane K.M., Bell V.A., Blaisdell G.K., Cooper M.L., Herrbach E., Pietersen G. Ecology and management of grapevine leafroll disease. Front. Microbiol. 2013;4 doi: 10.3389/fmicb.2013.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson M.N., Corcoran J.A., Zhang D.D., Hillbur Y., Newcomb R.D., Lofstedt C. A sex pheromone receptor in the hessian fly Mayetiola destructor (Diptera, Cecidomyiidae) Front. Cell Neurosci. 2016;10:212. doi: 10.3389/fncel.2016.00212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atallah S.S., Gomez M.I., Fuchs M.F., Martinson T.E. Economic impact of grapevine leafroll disease on Vitis vinifera cv. Cabernet franc in Finger Lakes vineyards of New York. Am. J. Enol. Vitic. 2012;63(1):73–79. doi: 10.5344/ajev.2011.11055. [DOI] [Google Scholar]
- Benton R., Sachse S., Michnick S.W., Vosshall L.B. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006;4(2):e20. doi: 10.1371/journal.pbio.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohbot J., Pitts R.J., Kwon H.W., Rutzler M., Robertson H.M., Zwiebel L.J. Molecular characterization of the Aedes aegypti odorant receptor gene family. Insect Mol. Biol. 2007;16(5):525–537. doi: 10.1111/j.1365-2583.2007.00748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.R., Tzeng D.T.W., Yang E.C. Chronic effects of imidacloprid on honey bee worker development-molecular pathway perspectives. Int. J. Mol. Sci. 2021;22(21) doi: 10.3390/ijms222111835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cid M., Pereira S., Cabaleiro C., Faoro F., Segura A. Presence of Grapevine leafroll-associated virus 3 in primary salivary glands of the mealybug vector Planococcus citri suggests a circulative transmission mechanism. Eur. J. Plant Pathol. 2007;118(1):23–30. doi: 10.1007/s10658-006-9090-8. [DOI] [Google Scholar]
- Cocco A., da Silva V.C.P., Benelli G., Botton M., Lucchi A., Lentini A. Sustainable management of the vine mealybug in organic vineyards. J. Pest. Sci. 2021;94(2):153–185. doi: 10.1007/s10340-020-01305-8. [DOI] [Google Scholar]
- Corcoran J.A., Hamiaux C., Faraone N., Lofstedt C., Carraher C. Structure of an antennally-expressed carboxylesterase suggests lepidopteran odorant degrading enzymes are broadly tuned. Curr. Res. Insect Sci. 2023;3 doi: 10.1016/j.cris.2023.100062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran J.A., Jordan M.D., Carraher C., Newcomb R.D. A novel method to study insect olfactory receptor function using HEK293 cells. Insect Biochem. Mol. Biol. 2014;54:22–32. doi: 10.1016/j.ibmb.2014.08.005. [DOI] [PubMed] [Google Scholar]
- Corcoran J.A., Jordan M.D., Thrimawithana A.H., Crowhurst R.N., Newcomb R.D. The peripheral olfactory repertoire of the lightbrown apple moth, Epiphyas postvittana. PLoS. One. 2015;10(5) doi: 10.1371/journal.pone.0128596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran J.A., Sonntag Y., Andersson M.N., Johanson U., Lofstedt C. Endogenous insensitivity to the Orco agonist VUAA1 reveals novel olfactory receptor complex properties in the specialist fly Mayetiola destructor. Sci. Rep. 2018;8(1):3489. doi: 10.1038/s41598-018-21631-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa M.C.G., Palero F., da Silva V.C.P., Kaydan M.B., Germain J.F., Abd-Rabou S., Daane K.M., Cocco A., Poulin E., Malausa T. Vol. 96. 2023. Identifying cryptic species of Planococcus infesting vineyards to improve control efforts; pp. 573–586. (J. Pest. Sci.). [DOI] [Google Scholar]
- Daane K.M., Almeida R.P., Bell V.A., Walker J.T., Botton M., Fallahzadeh, Mani M., Miano J.L., Sforza R., Walton V.M., Zaviezo T. Chap. 12 in Arthropod Management in Vineyards : Pests, Approaches, and Future Directions. Springer; New York: 2012. Biology and management of mealybugs. [Google Scholar]
- Daane K.M., Middleton M.C., Sforza R.F.H., Kamps-Hughes N., Watson G.W., Almeida R.P.P., Correa M.C.G., Downie D.A., Walton V.M. Determining the geographic origin of invasive populations of the mealybug Planococcus ficus based on molecular genetic analysis. PLoS. One. 2018;13(3) doi: 10.1371/journal.pone.0193852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pasqual C., Groot A.T., Mappes J., Burdfield-Steel E. Evolutionary importance of intraspecific variation in sex pheromones. Trends. Ecol. Evol. 2021;36(9):848–859. doi: 10.1016/j.tree.2021.05.005. [DOI] [PubMed] [Google Scholar]
- Douglas N., Kruger K. Transmission efficiency of Grapevine leafroll-associated virus 3 (GLRaV-3) by the mealybugs Planococcus ficus and Pseudococcus longispinus (Hemiptera : pseudococcidae) Eur. J. Plant Pathol. 2008;122(2):207–212. doi: 10.1007/s10658-008-9269-2. [DOI] [Google Scholar]
- Engsontia P., Sanderson A.P., Cobb M., Walden K.K.O., Robertson H.M., Brown S. The red flour beetle's large nose: an expanded odorant receptor gene family in Tribolium castaneum. Insect Biochem. Mol. Biol. 2008;38(4):387–397. doi: 10.1016/j.ibmb.2007.10.005. [DOI] [PubMed] [Google Scholar]
- Fournier-Level A., Good R.T., Wilcox S.A., Rane R.V., Schiffer M., Chen W.…Robin C. The spread of resistance to imidacloprid is restricted by thermotolerance in natural populations of Drosophila melanogaster. Nat. Ecol. Evol. 2019;3(4):647–656. doi: 10.1038/s41559-019-0837-y. [DOI] [PubMed] [Google Scholar]
- Franco J.C., Cocco A., Lucchi A., Mendel Z., Suma P., Vacas S., Mansour R., Navarro-Llopis V. Scientific and technological developments in mating disruption of scale insects. Entomol. Gener. 2022;42(2):251–273. doi: 10.1127/entomologia/2021/1220. [DOI] [Google Scholar]
- Garcia Morales M., Denno B.D., Miller D.R., Miller G.L., Ben-Dov Y., Hardy N.B. ScaleNet: a literature-based model of scale insect biology and systematics. Database (Oxford) 2016;2016 doi: 10.1093/database/bav118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger C.A., Daane K.M. Seasonal movement and distribution of the grape mealybug (Homoptera: pseudococcidae):: Developing a sampling program for San Joaquin Valley vineyards. J. Econ. Entomol. 2001;94(1):291–301. doi: 10.1603/0022-0493-94.1.291. [DOI] [PubMed] [Google Scholar]
- Gonzalez F., Borrero-Echeverry F., Josvai J.K., Strandh M., Unelius C.R., Toth M.…Walker W.B. Odorant receptor phylogeny confirms conserved channels for sex pheromone and host plant signals in tortricid moths. Ecol. Evol. 2020;10(14):7334–7348. doi: 10.1002/ece3.6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I.…Regev A. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29(7):644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamiaux C., Carraher C., Lofstedt C., Corcoran J.A. Crystal structure of Epiphyas postvittana pheromone binding protein 3. Sci. Rep. 2020;10(1):16366. doi: 10.1038/s41598-020-73294-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinkens D.M., McElfresh J.S., Millar J.G. Identification and synthesis of the sex pheromone of the vine mealybug, Planococcus ficus. Tetrahedron Lett. 2001;42(9):1619–1621. doi: 10.1016/S0040-4039(00)02347-9. [DOI] [Google Scholar]
- Ji W., Han K., Lu Y.Y., Wei J.F. Predicting the potential distribution of the vine mealybug, under climate change by MaxEnt. Crop Protect. 2020;137 doi: 10.1016/j.cropro.2020.105268. [DOI] [Google Scholar]
- Jones P.L., Pask G.M., Rinker D.C., Zwiebel L.J. Functional agonism of insect odorant receptor ion channels. Proc. Natl. Acad. Sci. USA. 2011;108(21):8821–8825. doi: 10.1073/pnas.1102425108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khallaf M.A., Cui R., Weissflog J., Erdogmus M., Svatos A., Dweck H.K.M., Valenzano D.R, Hansson B.S., Knaden M. Large-scale characterization of sex pheromone communication systems in drosophila. Nat. Commun. 2021;12(1):4165. doi: 10.1038/s41467-021-24395-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kol-Maimon H., Levi-Zada A., Franco J.C., Dunkelblum E., Protasov A., Eliyaho M., Mendel Z. Male behaviors reveal multiple pherotypes within vine mealybug Planococcus ficus (Signoret) (Hemiptera; Pseudococcidae) populations. Naturwissenschaften. 2010;97(12):1047–1057. doi: 10.1007/s00114-010-0726-3. [DOI] [PubMed] [Google Scholar]
- Kruger K., Saccaggi D.L., van der Merwe M., Kasdorf G.G.F. Transmission of grapevine leafroll-associated virus 3 (GLRAV-3): acquisition, inoculation and retention by the mealybugs Planococcus ficus and Pseudococcus longispinus (Hemiptera: pseudococcidae) S. Afr. J. Enol. Viticulture. 2015;36(2):223–230. [Google Scholar]
- Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R.B., Liu B.H., Xie Y.L., Li Z.Y., Huang W.H., Yuan J.Y., He G.Z., Chen Y.X., Pan Q., Liu Y.J., Tang J.B., Wu G.X., Zhang H., Shi Y.J., Liu Y., Yu C., Wang B., Lu Y., Han C.L., Cheung D.W., Yiu S.M., Peng S.L., Zhu X.Q., Liu G.M., Liao X.K., Li Y.R., Yang H.M., Wang J., Lam T.W., Wang J. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1 doi: 10.1186/2047-217x-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansourian S., Corcoran J., Enjin A., Lofstedt C., Dacke M., Stensmyr M.C. Fecal-derived phenol induces egg-laying aversion in drosophila. Curr. Biol. 2016;26(20):2762–2769. doi: 10.1016/j.cub.2016.07.065. [DOI] [PubMed] [Google Scholar]
- Maree H.J., Almeida R.P.P., Bester R., Chooi K.M., Cohen D., Dolja V.V., Fuchs M.F., Golino D.A., Jooste A.E.C., Martelli G.P., Naidu R.A., Rowhani A., Saldarelli P., Burger J.T. Grapevine leafroll-associated virus 3. Front. Microbiol. 2013;4 doi: 10.3389/fmicb.2013.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missbach C., Dweck H.K.M., Vogel H., Vilcinskas A., Stensmyr M.C., Hansson B.S., Grosse-Wilde E. Evolution of insect olfactory receptors. Elife. 2014;3 doi: 10.7554/eLife.02115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidu R., Rowhani A., Fuchs M., Golino D., Martelli G.P. Grapevine leafroll: a complex viral disease affecting a high-value fruit crop. Plant Dis. 2014;98(9):1172–1185. doi: 10.1094/PDIS-08-13-0880-FE. [DOI] [PubMed] [Google Scholar]
- Pesenti C., Viani F. The influence of fluorinated molecules (semiochemicals and enzyme substrate analogues) on the insect communication system. Chembiochem. 2004;5(5):590–613. doi: 10.1002/cbic.200300829. [DOI] [PubMed] [Google Scholar]
- Pitino M., Hoffman M.T., Zhou L., Hall D.G., Stocks I.C. The phloem-sap feeding mealybug (Ferrisia virgata) Carries 'Candidatus liberibacter asiaticus' populations that do not cause disease in host plants (vol 9, e85503, 2014) PLoS. One. 2014;9(3) doi: 10.1371/journal.pone.0085503.g002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renou M., Guerrero A. Insect parapheromones in olfaction research and semiochemical-based pest control strategies. Annu. Rev. Entomol. 2000;45:605–630. doi: 10.1146/annurev.ento.45.1.605. [DOI] [PubMed] [Google Scholar]
- Ricketts K.D., Gomez M.I., Atallah S.S., Fuchs M.F., Martinson T.E., Battany M.C., Bettiga L.J., Cooper M.L., Verdegaal P.S., Smith R.J. Reducing the economic impact of grapevine leafroll disease in California: identifying optimal disease management strategies. Am. J. Enol. Vitic. 2015;66(2):138–147. doi: 10.5344/ajev.2014.14106. [DOI] [Google Scholar]
- Robertson H.M., Warr C.G., Carlson J.R. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 2003;100(Suppl 2):14537–14542. doi: 10.1073/pnas.2335847100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smadja C., Shi P., Butlin R.K., Robertson H.M. Large gene family expansions and adaptive evolution for odorant and gustatory receptors in the Pea aphid, Acyrthosiphon pisum. Mol. Biol. Evol. 2009;26(9):2073–2086. doi: 10.1093/molbev/msp116. [DOI] [PubMed] [Google Scholar]
- Tsai C.W., Rowhani A., Golino D.A., Daane K.M., Almeida R.P.P. Mealybug transmission of grapevine leafroll viruses: an analysis of virus-vector specificity. Phytopathology. 2010;100(8):830–834. doi: 10.1094/Phyto-100-8-0830. [DOI] [PubMed] [Google Scholar]
- Wen Y.C., Liu Z.W., Bao H.B., Han Z.J. Imidacloprid resistance and its mechanisms in field populations of brown planthopper, Nilaparvata lugens Stal in China. Pestic. Biochem. Physiol. 2009;94(1):36–42. doi: 10.1016/j.pestbp.2009.02.009. [DOI] [Google Scholar]
- Yuvaraj J.K., Jordan M.D., Zhang D.D., Andersson M.N., Lofstedt C., Newcomb R.D., Corcoran J.A. Sex pheromone receptors of the light brown apple moth, Epiphyas postvittana, support a second major pheromone receptor clade within the Lepidoptera. Insect Biochem. Mol. Biol. 2022;141 doi: 10.1016/j.ibmb.2021.103708. [DOI] [PubMed] [Google Scholar]
- Zada A., Dunkelblum E., Assael F., Harel M., Cojocaru M., Mendel Z. Sex pheromone of the vine mealybug, in Israel: occurrence of a second component in a mass-reared population. J. Chem. Ecol. 2003;29(4):977–988. doi: 10.1023/A:1022944119077. [DOI] [PubMed] [Google Scholar]
- Zhang Y.K., Zeng D.Q., Li L., Hong X.C., Li-Byarlay H., Luo S.D. Assessing the toxicological interaction effects of imidacloprid, thiamethoxam, and chlorpyrifos on Bombus terrestris based on the combination index. Sci. Rep. 2022;12(1) doi: 10.1038/s41598-022-09808-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.