Abstract
SAP18, a polypeptide associated with the Sin3–HDAC co-repressor complex, was identified in a yeast two-hybrid screen as capable of interacting with the Drosophila GAGA factor. The interaction was confirmed in vitro by glutathione S-transferase pull-down assays using recombinant proteins and crude SL2 nuclear extracts. The first 245 residues of GAGA, including the POZ domain, are necessary and sufficient to bind dSAP18. In polytene chromosomes, dSAP18 and GAGA co-localize at a few discrete sites and, in particular, at the bithorax complex where GAGA binds some silenced polycomb response elements. When the dSAP18 dose is reduced, flies heterozygous for the GAGA mutation Trl67 show the homeotic transformation of segment A6 into A5, indicating that GAGA–dSAP18 interaction contributes to the functional regulation of the iab-6 element of the bithorax complex. These results suggest that, through recruitment of the Sin3–HDAC complex, GAGA might contribute to the regulation of homeotic gene expression.
INTRODUCTION
The GAGA factor of Drosophila is a sequence-specific DNA binding protein that is involved in a variety of different nuclear processes. Many genes in Drosophila contain GAGA binding sites in their regulatory regions and GAGA has been shown to regulate the expression of some developmentally regulated and stress-induced genes in vivo (Wilkins and Lis, 1997). GAGA is encoded by the trithorax-like (Trl) gene, which is required for the normal expression of the homeotic genes (Farkas et al., 1994). The homeotic transformations observed in some Trl alleles suggest a role for GAGA in transcription activation. Consistent with these results, GAGA acts as a transcriptional activator in vitro, or upon transient transfection in cultured SL2 cells (Soeller et al., 1993; Benyajati et al., 1997; Vaquero et al., 2000). However, GAGA is also found associated with some silenced polycomb response elements (PREs) of the bithorax and antennapedia complexes (Strutt et al., 1997). Moreover, GAGA also associates with some heterochromatin regions (Raff et al., 1994; Platero et al., 1998; Deuring et al., 2000) and the Trl gene is an enhancer of position effect variegation (PEV), which affects chromosome condensation and segregation (Farkas et al., 1994; Bhat et al., 1996). Finally, it was proposed that GAGA can cooperate with chromatin remodeling factors, such as NURF, to modify chromatin structure (Tsukiyama et al., 1994). All these results indicate that GAGA is a multifunctional protein that plays fundamental roles in chromosome structure and function.
GAGA is organized into several functionally distinct domains. A single zinc finger is involved in nucleic acid recognition (Pedone et al., 1996). In addition to this central DNA binding domain (DBD), GAGA carries a C-terminal glutamine-rich domain (Q-domain), which is involved in transcription activation (Vaquero et al., 2000), and a highly conserved N-terminal POZ domain, which mediates protein–protein interactions (Bardwell and Treisman, 1994; Huynh and Bardwell, 1998; Espinás et al., 1999; Katsani et al., 1999). A relatively long (140 amino acids) region of unknown function(s) links the POZ and DBD domains. Little is known about the interaction of GAGA with other nuclear proteins. It was shown that the POZ domain of GAGA supports homomeric as well as heteromeric interactions with other POZ-containing proteins, such as tramtrack (ttk) for instance (Bardwell and Treisman, 1994; Espinás et al., 1999; Katsani et al., 1999). Here, we report that GAGA interacts with SAP18, a polypeptide that is found associated with the Sin3–HDAC co-repressor complex (Zhang et al., 1997).
RESULTS AND DISCUSSION
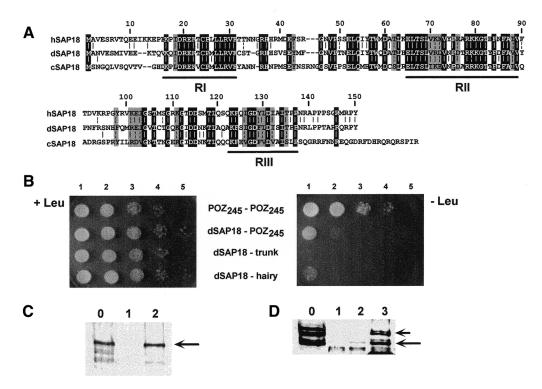
A yeast two-hybrid screen was performed to search for Drosophila proteins capable of interacting with the POZ domain of GAGA. For this screen, the first 245 amino acids of GAGA, which include the POZ domain (Figure 2A), were used. One of the 20 different clones isolated in this screen showed high homology to SAP18, a component of the Sin3 co-repressor complex (Zhang et al., 1997). The identity of dSAP18 with either human (hSAP18) or Caenorhabditis elegans (cSAP18) SAP18 is high, ∼60 and 47%, respectively (Figure 1A). The three polypeptides show high homology throughout their sequences, except for the most N- (1–15) and C-terminal (138–150) residues, and a central region (residues 32–45). Two specific regions, RI (16–31) and RII (65–89), show a very high degree of conservation with a similarity >80% (Figure 1A). A third region, RIII (123–137), also shows significant similarity (80%), but in this case the identity is lower (47%) than for regions RI (81%) and RII (67%).
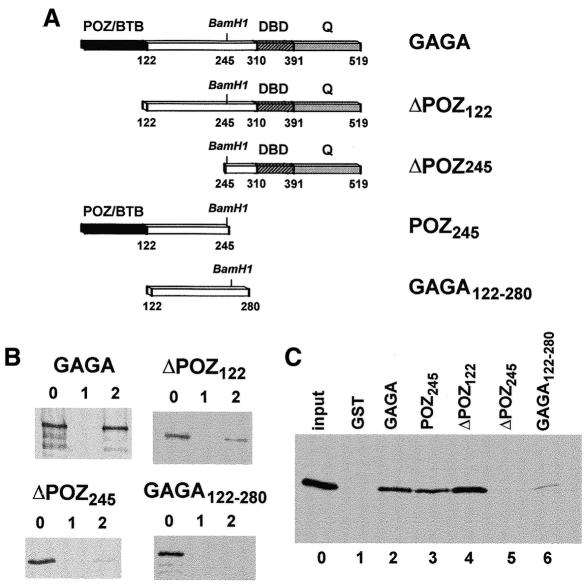
Fig. 2. Determination of the GAGA domains involved in the interaction with dSAP18. (A) Constructs used in these experiments. (B) GST pull-down assay of the interaction of the GAGA constructs indicated, with GST (lanes 1) and GST–dSAP18 (lanes 2). Lanes 0 show 10% of the input proteins. (C) GST pull-down assay of the interaction of dSAP18 with GST (lane 1) and the GST fusion proteins indicated (lanes 2–6). Lane 0 shows 10% of the input protein.
Fig. 1. The interaction of GAGA with dSAP18. (A) Sequence comparison of dSAP18, hSAP18 and cSAP18. Residues present in all three sequences are shaded black. Residues present in two sequences with a similar residue in the third or being similar in all three sequences are shaded gray. Similarity groups: IVL, FY, NQ, DE, RK and ST. Numbers indicate the amino acid position corresponding to the dSAP18 sequence. (B) Yeast two-hybrid assay of the POZ245–POZ245, dSAP18–POZ245, dSAP18–trunk and dSAP18–hairy interactions. Twenty microliters of the culture were plated on Leu+ (left) and Leu– (right) plates at a cell density of 2 × 106 cells/ml (lanes 1) and serial 10-fold dilutions (lanes 2–5). (C) GST pull-down assay of the interaction of recombinant GAGA with GST (lane 1) and GST–dSAP18 (lane 2). Lane 0 shows 10% of the input protein. The arrow indicates the position of the recombinant GAGA. (D) As in C but crude SL2 nuclear extracts were used as a source of GAGA proteins. Lane 3 shows the proteins retained by GST–POZ245. Arrows indicate the position of the GAGA519 and GAGA581 forms (see text).
As judged by yeast two-hybrid assay (Figure 1B), the interaction of dSAP18 with POZ245 is strong. Confluent growth is obtained when the culture is plated on selective medium at the highest density, and significant growth is observed when plated at a 10-fold dilution (Figure 1B, dSAP18–POZ245, lanes 1 and 2). No interaction at all was observed between dSAP18 and the unrelated fusion protein lexA–trunk. Similarly, a much weaker interaction was detected with the sequence-specific transcriptional repressor hairy. In this case, when the culture is plated at the highest density, only a few colonies are observed, similar in number to those obtained at a 10-fold dilution for dSAP18–POZ245. However, the dSAP18–POZ245 interaction is weaker than the very strong homomeric POZ245–POZ245 interaction, for which significant growth is detected even at a 103-fold dilution. The interaction of GAGA with dSAP18 was confirmed in vitro through glutathione S-transferase (GST) pull-down assays. A GST–dSAP18 fusion protein was shown to interact very efficiently with recombinant GAGA (Figure 1C, lane 2) and, vice versa, recombinant dSAP18 was strongly bound by a GST–GAGA fusion (Figure 2C, lane 2). GST–dSAP18 was also found specifically to bind GAGA from crude SL2 nuclear extracts (Figure 1D, lane 2), though to a much lower extent than when recombinant GAGA was used (Figure 1C). In SL2 nuclear extracts, the majority of GAGA appears as two duplets (Figure 1D, lane 0). The duplet of faster electrophoretic mobility arises from the GAGA519 form, while the species of slow mobility are derived from the GAGA581 form (Benyajati et al., 1997; C. Bonet and F. Azorín, unpublished results). As seen in Figure 1D, only the GAGA519 species appears to bind to GST–dSAP18 with some efficiency. Furthermore, a GST–POZ245 fusion was found to bind both GAGA forms indistinctly and much more efficiently than GST–dSAP18 (Figure 1D, lane 3). These results strongly suggest that only a minor, though still significant, subpopulation of the GAGA complexes present in SL2 nuclear extracts can actually support dSAP18 binding. Though little is known about the different types of complexes that GAGA forms in vivo, its potential interaction with various nuclear proteins (Bardwell and Treisman, 1994; Huynh and Bardwell, 1998; Espinás et al., 1999; Katsani et al., 1999) suggests that the interaction with dSAP18 may only account for the formation of part of such complexes.
The GAGA domains involved in binding dSAP18 were determined by GST pull-down assays (Figure 2). The first 245 amino acids of GAGA are essential for this interaction since deletion of this region results in a lack of binding to GST–dSAP18 (Figure 2B, panel ΔPOZ245) and GST–ΔPOZ245 was unable to bind dSAP18 (Figure 2C, lane 5). Furthermore, GST–POZ245 was capable of binding dSAP18 (Figure 2C, lane 3). The POZ domain of GAGA, which extends for the first 120 amino acids (Zollman et al., 1994) and is sufficient for self-oligomerization (Espinás et al., 1999), importantly contributes to the interaction with dSAP18. Deletion of the first 122 amino acids of GAGA results in a much weaker binding to GST–dSAP18 (Figure 2B, panel ΔPOZ122). Similarly, the GAGA122–280 construct is not bound efficiently by GST–dSAP18 (Figure 2B, panel GAGA122–280) and GST–GAGA122–280 binds dSAP18 only very weakly (Figure 2C, lane 6). However, ΔPOZ122 binds to GST–dSAP18 more efficiently than ΔPOZ245 (Figure 2B), suggesting that the GAGA122–245 region also contributes to the interaction with dSAP18. Actually, GST–ΔPOZ122 binds dSAP18 efficiently (Figure 2C, lane 4), reflecting the contribution of the linking domain to this interaction. The inability of GAGA122–280 to support the interaction with dSAP18 might reflect that, in the absence of the C-terminal part of GAGA, the linking domain is not fully folded. Altogether, these results indicate that the first 245 residues of GAGA are necessary and sufficient for binding dSAP18, and that efficient GAGA–dSAP18 interaction requires the contribution of both the POZ and linking domains of GAGA.
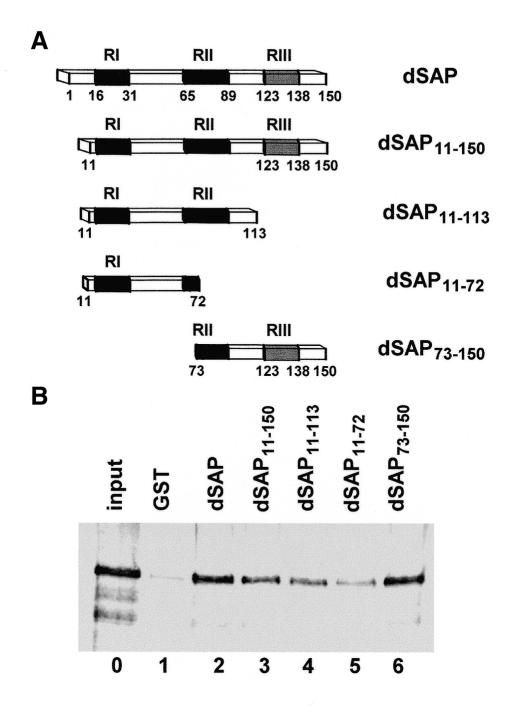
A similar approach was followed to analyze the contribution of the different regions of dSAP18 to its interaction with GAGA (Figure 3). The N- and C-terminal parts of SAP18 are not conserved and the deletion of the first 11 residues of dSAP18 or the last 38 residues of SAP11–150, does not result in a significant decrease in GAGA binding (Figure 3B, lanes 3 and 4). A larger N-terminal deletion, up to residue 72, does not show a significant effect either (Figure 3B, lane 6), indicating that the N-terminal half of dSAP18, including the highly conserved RI region, is dispensable for binding to GAGA. Consistent with these results, the N-terminal half of dSAP18 binds GAGA significantly worse than full dSAP18 (Figure 3B, lane 5). These results indicate that not all regions of dSAP18 contribute equally to its interaction with GAGA and that residues 73–113, which include most of the highly conserved RII region, are mainly responsible for binding to GAGA.
Fig. 3. Determination of the region of dSAP18 involved in the interaction with GAGA. (A) Constructs used in these experiments. (B) GST pull-down assay of the interaction of GAGA with GST (lane 1) and the GST fusion proteins indicated (lane 2–6). Lane 0 shows 10% of the input protein.
SAP18 was identified as a polypeptide associated with the mammalian transcriptional repressor Sin3 (Zhang et al., 1997). The core mSin3 complex contains a total of seven polypeptides, which include the histone deacetylases HDAC1 and HDAC2, RbAp48 and RbAp46, and SAP30 and SAP18. Recruitment of the Sin3–HDAC complex to specific target genes appears to rely on its interaction with sequence-specific DNA binding proteins, since none of the known components of the complex are capable of binding DNA. SAP30 and SAP18 could mediate some of these interactions. It was shown that mSAP30 binds both mSin3 and N-CoR, and is required for N-CoR-mediated repression by a set of sequence-specific DNA-binding transcription factors (Laherty et al., 1998). SAP18 could also be involved in interactions with sequence-specific DNA binding proteins. It is known that SAP18 interacts directly with mSin3 (Zhang et al., 1997). Here, we have shown that GAGA interacts with dSAP18 and that the POZ domain of GAGA contributes importantly to this interaction. Since POZ is a highly conserved structural domain (Bardwell and Treisman, 1994; Zollman et al., 1994), it is likely that similar interactions would be observed with other POZ domains. Actually, several sequence-specific transcriptional repressors carry POZ domains (Brown and Wu, 1993; Chang et al., 1996; Aoki et al., 1998; Huynh and Bardwell, 1998; Wong and Privalsky, 1998), some of which were also found to interact with N-CoR and SMRT (Huynh and Bardwell, 1998; Wong and Privalsky, 1998). Most likely, formation of a stable complex requires multiple interactions between its various components.
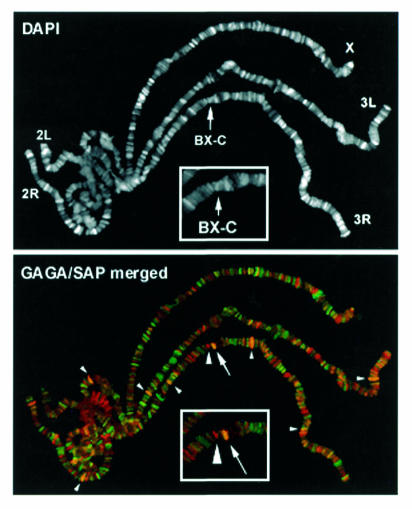
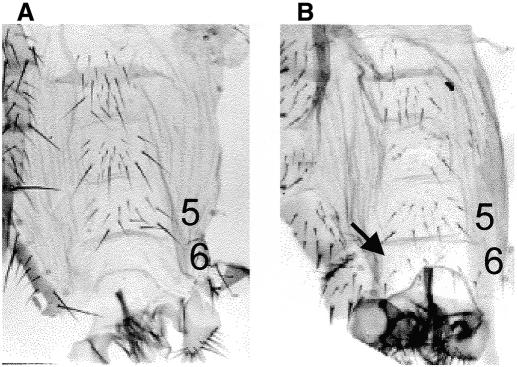
Contrary to most POZ-containing proteins, the Drosophila GAGA factor acts as a transcriptional activator. Its interaction with a component of the Sin3 co-repressor complex indicates that GAGA could also act as a repressor in some cases. In this respect, the presence of GAGA at some silenced PREs of the bithorax and antennapedia complexes might be especially revealing (Strutt et al., 1997; Horard et al., 2000). Interestingly, though the immunostaining patterns of GAGA and dSAP18 show only a limited general overlapping in polytene chromosomes, the two proteins co-localize at the region of the bithorax complex (BX-C) (Figure 4), suggesting a possible contribution of GAGA–dSAP18 interaction to BX-C regulation. Consistent with this possibility, we observe a genetic interaction between Trl and a deficiency that uncovers dSAP18 (Figure 5). Flies heterozygous for the Trl67 mutation and hemizygous for Df(3R)sbd26 show a homeotic transformation of the sixth abdominal segment into the fifth as indicated by the presence in the sixth sternite of several bristles (6–8) (Figure 5B) in the vast majority of the individuals (Table I). Such homeotic transformation is very infrequent in Trl67/+ and Df(3R)sbd26/+ heterozygotes where no males were detected to have more than two bristles and many contained no bristles at all (Table I). A similar interaction was observed with the Trl62 allele (not shown). It is most probable that this interaction is due to a reduced dose of dSAP18 and not to other genetic elements in the Df(3R)sbd26, since we also observed the homeotic transformation associated with a P-element insertion in the 5′ UTR of dSAP18 [see EP(3)3462/Df(3R)sbd26 in Table I]. Preliminary results suggest that this insertion might correspond to a hypomorphic allele of dSAP18 (not shown). No homeotic phenotype is detected in EP(3)3462/Trl trans-heterozygous flies (not shown), probably because of the hypomorphic nature of EP(3)3462. All of these results strongly indicate that GAGA–dSAP18 interaction has a significant contribution to the functional regulation of the iab-6 element of BX-C.
Fig. 4. The immunostaining of polytene chromosomes with αSAP18 (pseudocolored in red) and αGAGA (pseudocolored in green) antibodies. The region of the bithorax complex (BX-C) is shown enlarged. The two proteins colocalize at few sites (small arrowheads). The arrow indicates the overlapped SAP/GAGA signal at BX-C and the big arrowhead indicates an individual SAP signal in the vicinity.
Fig. 5. Whole mount of abdominal cuticles of (A) wild-type and (B) Trl67/Df(3R)sbd26 males. In wild-type males the sixth sternite is devoid of bristles while in Trl67/Df(3R)sbd26 males it contains several hairs (6–8) (arrow).
Table I. Frequency of the homeotic transformation of segment A6 into A5 in different genetic backgrounds.
| |
|
No. of bristlesb |
||
|---|---|---|---|---|
| Genotype | Na | 0 | ≤2 | >2 |
| Trl67/Df(3R)sbd26 | 12 | 0% | 16% | 84% |
| Trl67/+ | 35 | 94% | 6% | 0% |
| Df(3R)sbd26/+ | 51 | 29% | 71% | 0% |
| Df(3R)sbd26/EP(3)3462 | 14 | 0% | 29% | 71% |
aN = number of males scored.
bPercentage of males carrying none (0), 2 or less (≤2) and more than 2 (>2) bristles in the sixth sternite.
The concurrent presence of GAGA and polycomb at some silenced PREs is surprising since, as derived from genetic analysis, these two proteins are expected to have opposing functions on the regulation of the expression of the homeotic genes. Acting at the level of the core promoter elements, GAGA is likely to activate transcription of the homeotic genes (Strutt et al., 1997; Orlando et al., 1998). However, functional trithorax response elements (TREs) are frequently found in the vicinity of PREs (Orlando et al., 1998; Tillib et al., 1999) and a contribution of GAGA to the functional regulation of several segment-specific cis-regulatory regions of the bithorax complex has been reported (Hagstrom et al., 1997; Cavalli and Paro, 1998; Horard et al., 2000). Whether GAGA helps to establish the repressed or the active state of these elements is still uncertain. GAGA has been shown to contribute to the relief of repression at the Fab-7 element (Strutt et al., 1997), and the homeotic transformations described here and elsewhere (Farkas et al., 1994) are also consistent with a role in activation. On the other hand, in the case of the iab-7 and bxd PREs, GAGA was shown to contribute to silencing (Hagstrom et al., 1997; Horard et al., 2000) and the genetic interactions observed between some Pc and Trl alleles also suggest a contribution to repression (Strutt et al., 1997). Our results indicate that GAGA might participate in the recruitment of the Sin3–HDAC co-repressor complex to some PREs, but that contrary to what would be anticipated for such an interaction, it contributes to the relief of repression at the iab-6 element. The same phenotype is observed in flies homozygous for the hypomorph Trl13C allele (Farkas et al., 1994). Interestingly, some rpd3 alleles behave as enhancers of PEV, also leading to an increase in repression (De Rubertis et al., 1996). It is possible that by modifying chromatin structure, GAGA–SAP18 interaction could contribute to the establishment of the domain boundaries that insulate different cis-regulatory elements, rather than to the formation of the repressed or active states themselves.
METHODS
Recombinant proteins and extracts. GAGA and dSAP18 constructs used in these experiments (Figures 2A and 3A) were derived from GAGA519 (Soeller et al., 1993) and the dSAP18 cDNA described below. His6-tagged recombinant proteins were expressed in BL21(DE3) Escherichia coli cells (Espinás et al., 1999) and either purified on a Ni2+–NTA column, for the GAGA constructs, or used as crude extracts, for dSAP18. GST fusion proteins were obtained in pGEX4T-3, for the dSAP18 constructs, or in pGEX-KG expression vectors (Amersham), for the GAGA constructs. GST fusion proteins were expressed in BL21(DE3) cells. Crude SL2 nuclear extracts were prepared from ∼107 cells according to (Andrews and Faller, 1991).
Yeast two-hybrid assays. The yeast two-hybrid screen was performed in EGY48 cells (MATα, ura3, his3, trp1, LexAopx6–LEU2) co-transformed with a plexA202+PL plasmid expressing a lexA–POZ245 fusion protein and the Drosophila (0–12 h) embryonic cDNA library RFLY1 fused to the B42 activation domain of plasmid pJG4-5 (Finley et al., 1996), in which expression of the fused protein is driven by the GAL1 promoter. Transformants were selected for growth on minimal medium lacking leucine (Leu–) and supplemented with 2% galactose and 1% raffinose. Colonies showing galactose-dependent growth on Leu– plates were analyzed. One of the positive clones carried most of the cDNA coding for dSAP18, from residues 11 to 150. The first 10 residues of dSAP18 were obtained by PCR according to DDBJ/EMBL/GenBank AC019750, which contains the ORF for dSAP18.
When the specificity of the dSAP18–POZ245 interaction was analyzed, EGY48 cells were co-transformed with the B42–dSAP18 fusion and either the lexA–POZ245 fusion or similar lexA fusion proteins carrying the full cDNAs of hairy (lexA–hairy) or trunk (lexA–trunk). Cells were grown to a density of 2 × 106 cells/ml and 20 µl were then plated on Leu+ and Leu– plates at the original cell density of the culture and at serial 10-fold dilutions.
GST pull-down assays. GST fusion proteins were bound to glutathione Sepharose-4B beads. Protein concentrations, determined by gel electrophoresis, were adjusted by dilution with unbound beads. Agarose-bound GST fusion proteins were equilibrated in binding buffer (20 mM HEPES pH 7.9, 20% glycerol, 0.2 mM EDTA, 150 mM NaCl, 0.1% NP-40, 0.5 mM dithiothreitol, 0.1 mM phenylmethylsulfonyl fluoride) and incubated in 450 µl for 1 h at 4°C with either purified recombinant proteins, bacterial cell extracts or crude SL2 nuclear extracts. When purified recombinant proteins were used, the beads were blocked with 0.2% BSA for 2 h at 4°C before use. Beads were boiled directly in SDS–PAGE loading buffer and bound proteins were analyzed by western blotting with rabbit αGAGA or αdSAP18 polyclonal antibodies and detected by ECL (Amersham).
Fly stocks and immunostaining of polytene chromosomes. EP(3)3462 and Df(3R)sbd26 stocks were obtained from the Bloomington Center. For the genetic interactions they were crossed to the Trl alleles, Trl62 and Trl67 (Farkas et al., 1994). Double immunostaining of polytene chromosomes with rat αGAGA and rabbit αdSAP18 polyclonal antibodies was performed as described (James et al., 1989).
Acknowledgments
ACKNOWLEDGEMENTS
We are thankful to R. González for mounting the cuticles, to Dr Finley for the RFLY1 library, to Dr F. Karch for the Trl alleles, and to Drs J. Bernués, M. Martínez-Balbás and B. Piña for helpful discussions. This work was financed by grants from the DGES (PB96-812), the CIRIT (SGR97-55) and the Istituto Pasteur Fondazione Cenci Bolognetti (Università di Roma ‘La Sapienza’), and was carried out within the framework of the ‘Centre de Referència en Biotecnologia’ (Generalitat de Catalunya). S.C. is a fellow from the DGES.
REFERENCES
- Andrews N.C. and Faller, D.V. (1991) A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells Nucleic Acids Res., 19, 2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K., Meng, G., Suzuki, K., Takashi, T., Kameoka, Y., Nakahara, K., Ishida, R. and Kasai, M. (1998) RP58 associates with condensed chromatin and mediates a sequence-specific transcriptional repression. J. Biol. Chem., 273, 26698–26704. [DOI] [PubMed] [Google Scholar]
- Bardwell V.J. and Treisman, R. (1994) The POZ domain: a conserved protein–protein interaction motif. Genes Dev., 8, 1664–1677. [DOI] [PubMed] [Google Scholar]
- Benyajati C. et al. (1997) Multiple isoforms of GAGA factor, a critical component of chromatin structure. Nucleic Acids Res., 25, 3345–3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat K.M., Farkas, G., Karch, F., Gyurkovics, H., Gausz, J. and Schedl, P. (1996) The GAGA factor is required in the early Drosophila embryo not only for transcriptional regulation but also for nuclear division. Development, 122, 1113–1124. [DOI] [PubMed] [Google Scholar]
- Brown J.L. and Wu, C. (1993) Repression of Drosophila pair-rule segmentation genes by ectopic expression of tramtrack. Development, 117, 45–58. [DOI] [PubMed] [Google Scholar]
- Cavalli G. and Paro, R. (1998) The Drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis. Cell, 93, 505–518. [DOI] [PubMed] [Google Scholar]
- Chang C.-C., Ye, B.H., Chaganti, R.S.K. and Dalla-Favera, R. (1996) BCL-6, a POZ/zinc finger protein, is a sequence-specific transcriptional repressor. Proc. Natl Acad. Sci. USA, 93, 6947–6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubertis F., Kadosh, D., Henchoz, D., Pauli, D., Reuter, G., Struhl, K. and Spierer, P. (1996) The histone deacetylase RPD3 counteracts genomic silencing in Drosophila and yeast. Nature 384, 589–591. [DOI] [PubMed] [Google Scholar]
- Deuring R. et al. (2000) The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol. Cell, 5, 355–365. [DOI] [PubMed] [Google Scholar]
- Espinás M.L., Jiménez-García, E., Vaquero, A., Canudas, S., Bernués, J. and Azorín, F. (1999) The N-terminal POZ domain of GAGA mediates the formation of oligomers that bind DNA with high affinity and specificity J. Biol. Chem., 274, 16461–16469. [DOI] [PubMed] [Google Scholar]
- Farkas G., Janos, G., Galloni, M., Reuter, G., Gyurkovics, H. and Karch, F. (1994) The Trithorax-like gene encodes the Drosophila GAGA factor. Nature, 371, 806–808. [DOI] [PubMed] [Google Scholar]
- Finley R.L. Jr, Thomas, B.J., Zipursky, S.L. and Brent, R. (1996) Isolation of Drosophila cyclin D, a protein expressed in the morphogenetic furrow before entry into S phase. Proc. Natl Acad. Sci. USA, 93, 3011–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagstrom K., Muller, M. and Schedl, P. (1997) A Polycomb and GAGA dependent silencer adjoins the Fab-7 boundary in the Drosophila Bithorax complex. Genetics, 146, 1365–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horard B., Tatout, C., Poux, S. and Pirrotta, V. (2000) Structure of a polycomb response element and in vitro binding of polycomb group complexes containing GAGA factor. Mol. Cell. Biol., 20, 3187–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh K.D. and Bardwell, V.J. (1998) The BCL-6 POZ domain and other POZ domains interact with the co-represors N-CoR and SMRT. Oncogene, 17, 2473–2484. [DOI] [PubMed] [Google Scholar]
- James T.C., Eissenberg, J.C., Craig, C., Dietrich, V., Hobson, A. and Elgin, S.C.R. (1989) Distribution patterns of HP1, a heterochromatin-associated nonhistone chromosomal protein of Drosophila. Eur. J. Cell Biol., 50, 170–180. [PubMed] [Google Scholar]
- Katsani K.R., Hajibagheri, M.A.N. and Verrijzer, C.P. (1999) Co-operative DNA binding by GAGA transcription factor requires the conserved BTB/POZ domain and reorganizes promoter topology. EMBO J., 18, 698–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laherty C.D. et al. (1998) SAP30, a component of the mSin3 corepressor complex involved in N-CoR-mediated repression by specific transcription factors. Mol. Cell, 2, 33–42. [DOI] [PubMed] [Google Scholar]
- Orlando V., Jane, E.J., Chinwalla, V., Harte, P.J. and Paro, R. (1998) Binding of trithorax and polycomb proteins to the bithorax complex: dynamic changes during early Drosophila embryogenesis. EMBO J., 17, 5141–5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedone P.V., Ghirlando, R., Clore, G.M., Gronenborn, A.M., Felsenfeld, G. and Omichinski, J.G. (1996) The single Cys2-His2 zinc finger domain of the GAGA protein flanked by basic residues is sufficient for high-affinity specific DNA binding. Proc. Natl Acad. Sci. USA, 93, 2822–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platero J.S., Csink, A.K., Quintanilla, A. and Henikoff, S. (1998) Changes in chromosomal localization of heterochromatin binding proteins during the cell cycle in Drosophila. J. Cell Biol., 140, 1297–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff J.W., Kellum, R. and Alberts, B. (1994) The Drosophila GAGA transcription factor is associated with specific regions of heterochromatin throughout the cell cycle. EMBO J., 13, 5977–5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeller W.C., Oh, C.E. and Kornberg, T.B. (1993) Isolation of cDNAs encoding the Drosophila GAGA transcription factor. Mol. Cell. Biol., 13, 7961–7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strutt H., Cavalli, G. and Paro, R. (1997) Co-localization of Polycomb protein and GAGA factor on regulatory elements responsible for the maintenance of homeotic gene expression. EMBO J., 16, 3621–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillib S., Petruk, S., Sedkov, Y., Kuzin, A., Fujioka, M., Goto, T. and Mazo, A. (1999) Trithorax- and Polycomb-group response elements within an Ultrabithorax transcription maintenance unit consist of closely situated but separable sequences. Mol. Cell. Biol., 19, 5189–5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama T., Becker, P.B. and Wu, C. (1994) ATP-dependent nucleosome disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature, 367, 525–532. [DOI] [PubMed] [Google Scholar]
- Vaquero A., Espinás, M.L., Azorín, F. and Bernués, J. (2000) Functional mapping of the GAGA factor assigns its transcriptional activity to the C-terminal glutamine-rich domain. J. Biol. Chem., 275, 19461–19468. [DOI] [PubMed] [Google Scholar]
- Wilkins R.C. and Lis, J.T. (1997) Dynamics of potentiation and activation: GAGA factor and its role in heat shock gene regulation. Nucleic Acids Res., 25, 3963–3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C.-W. and Privalsky, M.L. (1998) Components of the SMRT corepressor complex exhibit distinctive interactions with the POZ domain oncoproteins PLZF, PLFZ-RARα. J. Biol. Chem., 273, 27695–27702. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Iratni, R., Erdjument-Bromage, H., Tempst, P. and Reinberg, D. (1997) Histone deacetylase and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell, 89, 357–364. [DOI] [PubMed] [Google Scholar]
- Zollman S., Godt, D., Privé, G.G., Couderc, J.-L. and Laski, F.A. (1994) The BTB domain, found primarily in zinc finger proteins, defines an evolutionarily conserved family that includes several developmentally regulated genes in Drosophila. Proc. Natl Acad. Sci. USA, 91, 10717–10721. [DOI] [PMC free article] [PubMed] [Google Scholar]