Abstract
Bacterial artificial chromosomes (BACs) offer many advantages for functional studies of large eukaryotic genes. To utilize the potential applications of BACs optimally, new approaches that allow rapid and precise engineering of these large molecules are required. Here, we describe a simple and flexible two-step approach based on ET recombination, which permits point mutations to be introduced into BACs without leaving any other residual change in the recombinant product. Introduction of other modifications, such as small insertions or deletions, is equally feasible. The use of ET recombination to achieve site-directed mutagenesis opens access to a powerful use of BACs and is extensible to DNA molecules of any size in Escherichia coli, including the E. coli chromosome.
INTRODUCTION
Functional genetic analysis in higher eukaryotes relies on the ability to engineer precise mutations in the gene of interest. Cloning vectors that carry large inserts, such as bacterial artificial chromosomes (BACs), PACs and P1s (Shizuya et al., 1992; Sternberg, 1992; Ioannou et al., 1994), offer great potential for these studies, because in most cases all of the regulatory regions of the gene can be included. Thus, these large constructs provide an environment in which the native expression pattern is preserved, without the need to identify all the relevant regulatory elements beforehand. Therefore, BACs, PACs and P1s provide an ideal basis upon which a gene can be studied in its native context. For many functional studies, it is necessary to manipulate the gene sequence precisely at a desired site. The ability to make very precise changes in the nucleotide sequence of large DNA molecules including BACs, which can be up to 300 kb in size, has therefore become critically necessary. Conventional cloning methods that rely on the occurrence of suitable restriction sites are impractical for this purpose. We have recently developed an alternative approach that enables precise modification of both small and large DNA molecules by homologous recombination in Escherichia coli. This strategy, called ET recombination, allows the manipulation of DNA molecules by recombination with linear DNA fragments (Zhang et al., 1998). These linear DNA molecules are synthesized to contain 50 nucleotide terminal homology arms that mediate recombination to the desired locus on the target molecule. Here, ET recombination, in combination with selection and counterselection using the sacB–neo fusion gene, is used for introducing precise subtle changes in the nucleotide sequence of a BAC, without the additional introduction of any operational sequences.
RESULTS
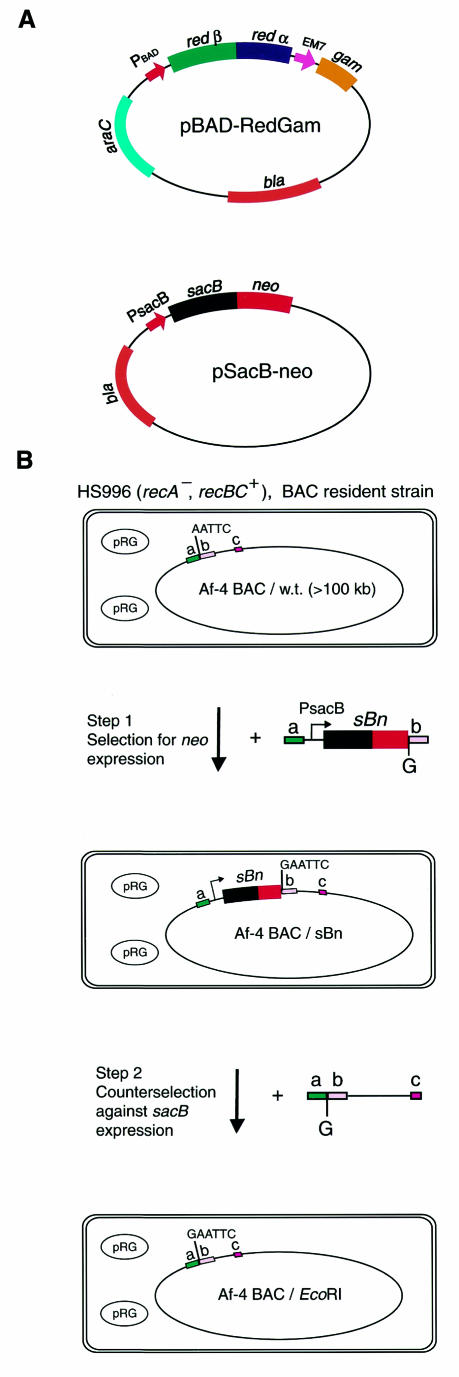
To make subtle changes in the nucleotide sequence of a BAC, such as the introduction of a point mutation, we developed a two-step use of ET recombination. Both steps were performed using the SacB–neo fusion protein present on pSacB-neo (Figure 1A). In the first step, selection pressure was applied for expression of the Neo part of the fusion protein, and in the second step, selection pressure was applied against the SacB part. Figure 1B shows an example of the strategy, in which a single G nucleotide was introduced directly upstream of an AATTC sequence present in a BAC of >100 kb, which carries the mouse Af-4 gene (Baskaran et al., 1997). As previously described (Zhang et al., 1998; Muyrers et al., 1999), the E. coli host carrying the Af-4 BAC was capacitated for ET recombination by transforming it with a plasmid carrying the ET recombination genes. Here we used pBAD-RedGam (Figure 1A), which contains redα/redβ under the l-arabinose-inducible promoter and from which gam is constitutively expressed to inhibit endogenous RecBC activity (Murphy, 1991).
Fig. 1. (A) Schematic presentation of the plasmids used. pBAD-RedGam is based on pBAD24 (Guzman et al., 1995). redα and redβ are expressed from the l-arabinose-inducible BAD promoter PBAD, and gam is expressed from the constitutive EM-7 promoter (Invitrogen). pSacB-neo contains the sacB–neo fusion gene under the control of the sacB promoter (PsacB). Both plasmids carry the ColE1 replication origin. (B) Strategy used to introduce a G nucleotide into the Af-4 BAC. First, pBAD-RedGam (pRG) was transformed into the HS996 strain harboring the BAC (Research Genetics). Subsequently, in step 1, Redα/Redβ expression was induced by l-arabinose, after which competent cells were prepared and electroporated with the linear targeting molecule, containing sacB–neo (sBn). pSacB-neo was used as a template for the PCR amplification of that linear molecule, using oligonucleotides that each contained a homology arm (a or b), followed by the region that primed the amplification of sBn. In addition, the 3′ oligonucleotide contained a C nucleotide, coding for the G to be introduced, between the homology arm b and the PCR priming region. Since the first 5 nucleotides of homology arm b were AATTC, introduction of the additional G by ET recombination created an additional EcoRI site. Correct recombinants were obtained after plating on LB plates containing kanamycin. Cells containing correct recombinants were grown, induced with l-arabinose and electroporated with the linear PCR product of step 2. The template for the PCR amplification of this linear DNA was the wild-type Af-4 BAC. The 5′ oligonucleotide consisted of homology arm a plus an additional G, followed by the first 18 nucleotides of homology arm b (starting with AATTC and priming PCR amplification). The 3′ oligonucleotide c consisted of 18 nucleotides of the Af-4 BAC ∼0.7 kb downstream of homology arm b. Thus, the entire sequence from b to c was used as one homology arm. Correct recombinants were obtained after plating on LB plates containing sucrose.
After induction of Redα/Redβ expression to open a recombinogenic window, electro-competent cells were prepared. These cells were transformed with a PCR fragment containing the sacB–neo fusion gene flanked by 50 nucleotide homology arms that were identical in sequence to the targeted locus on the BAC. Thereby, this linear fragment was introduced by homologous recombination into the chosen position on the BAC. The linear fragment contained the additional G nucleotide directly upstream of homology arm b. Correct recombinants of this first ET recombination step were selected via kanamycin resistance, conveyed by the Neo part of the SacB–neo fusion protein. Over 200 colonies were obtained in each of three independent experiments, and >75% of the colonies analyzed by restriction mapping and Southern blotting were correctly targeted recombinants (data not shown, but see Zhang et al., 1998; Angrand et al., 1999; Muyrers et al., 1999, 2000b; Narayanan et al., 1999; Datsenko and Wanner, 2000; Hill et al., 2000; Yu et al., 2000, for other examples of efficiency and fidelity in positively selected ET recombination exercises).
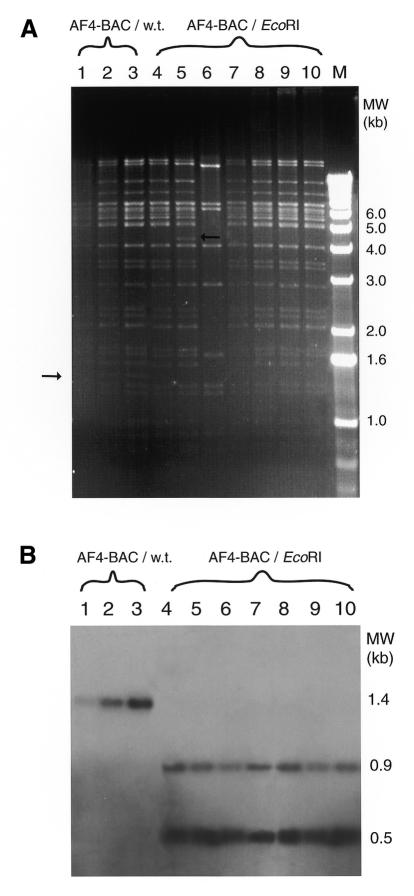
Colonies containing correct recombinants were checked for expression of sacB by streaking on Luria–Bertani (LB) plates containing sucrose. On these plates, selection pressure is applied against SacB expression, because sucrose is converted by SacB to a bacteriotoxin (Blomfield et al., 1991). The vast majority (>95%) failed to grow, indicating the expected expression of the SacB part of the SacB–neo fusion protein. These colonies were used for the second targeting step. Again, cells were induced to express Redα/Redβ and prepared for electroporation. They were then targeted with a 0.8 kb PCR product containing the original sequence of the targeted Af-4 BAC locus plus the additional G nucleotide, which was added to the 5′ oligonucleotide used for PCR amplification (see Methods). After targeting, candidate recombinants were identified by culturing on 7% sucrose plates. In four independent experiments, 700–2500 colonies were obtained. To check whether these colonies had indeed lost the sacB–neo gene, they were restreaked on LB plates containing kanamycin. Approximately 10–15% of these restreaked colonies failed to grow on plates containing kanamycin, indicating that the majority of mutational events that inactivate sacB expression are subtle and still permit functional expression of the neo part of the fusion gene. Since correct recombinants will have lost the neo gene, only kanamycin-sensitive colonies were taken for further analysis. Of these, seven were analyzed by EcoRI restriction digestion and Southern blot analysis. All of the colonies analyzed had acquired the additional G nucleotide directly upstream of the targeted AATTC locus, resulting in the creation of an additional EcoRI site (Figure 2). Of these clones, five did not display any undesired internal rearrangement (Figure 2 and data not shown). The correct introduction of the additional G was further confirmed by direct DNA sequencing from the BAC template of four independent clones (data not shown). Moreover, DNA sequencing revealed no extraneous changes in the sequence of the recombinant Af-4 BAC, in a total of 2500 nucleotides examined.
Fig. 2. Analysis of BAC DNA preparations. (A) The EcoRI digestion patterns of the wild-type Af-4 BAC (Af-4 BAC/w.t., lanes 1–3), and the Af-4 BAC containing the extra G (Af-4 BAC/EcoRI, lanes 4–10) are shown. The Af-4 BAC/w.t. lanes contain a specific band at ∼1.4 kb (left arrow), which results from the presence of EcoRI recognition sites on the wild-type BAC around the targeted locus, see (B). This band is not present in the clones that contain an additional EcoRI site due to the introduction of the additional G. Here, the two specific bands in the Af-4 BAC lanes are not visible, either due to the lowered gel resolution at shorter length or due to the presence of co-migrating bands in the same region. In lanes 5 and 6, the BAC has been rearranged internally (right arrow); the clones of lanes 4 and 7–10 all show the correct pattern. M, 1 kb DNA ladder (New England Biolabs). (B) Southern analysis using a probe that was made from the linear molecule used to target the BAC during step 2 (Figure 1B). Lane numbers correspond to (A). The Af-4 BAC/w.t. (lanes 1–3) displays hybridization to the 1.4 kb fragment marked by the left arrow in (A). In contrast, all clones containing the newly generated EcoRI site (lanes 4–10) display hybridization to two fragments, at 0.5 and 0.9 kb.
DISCUSSION
ET recombination is a new way to engineer DNA without the need to rely on the presence of suitable sites for restriction enzymes. Therefore, it permits modification of large DNA molecules, including BACs. ET recombination can be mediated either by the RecE/RecT protein pair (Zhang et al., 1998; Narayanan et al., 1999; Muyrers et al., 2000a,b) or by the Redα/Redβ protein pair (Muyrers et al., 1999, 2000a,b; Datsenko and Wanner, 2000; Hill et al., 2000; Yu et al., 2000). Here, we present a two-step procedure that combines ET recombination with selection and counterselection to allow subtle manipulation of BACs. The strategy used confers all of the advantages of other ET recombination applications, including the following. (i) A high degree of fidelity is preserved. As shown in Figure 2B, all of the clones examined contained the additional EcoRI site at the correct locus. (ii) Recombination occurs in a recA– background. The recombinogenic window is further limited by the transient nature of the l-arabinose-induced expression of Redα and Redβ, which are both strictly required for ET recombination to occur (Muyrers et al., 2000a). Thus, the risk of unwanted intramolecular rearrangement is minimized, allowing recombinants that contain no other unintended changes to be recovered efficiently (Figure 2 and data not shown). (iii) ET recombination is transferable to the host strain in which the BAC resides, thereby alleviating the need to retransform the BAC into a special strain. (iv) Like the pBADαβγ plasmid (Muyrers et al., 1999), the pBAD-RedGam plasmid is rapidly lost in the absence of continued selection pressure (most probably because of constitutive expression of the Gam protein). The functionally homologous plasmid pBAD-RecGam, in which redα and redβ have been replaced by their homologs from the Rac prophage, recE606 (Muyrers et al., 2000a) and recT, respectively, is also suitable for the strategy described here (data not shown).
By designing the oligonucleotide sequences used to amplify the linear targeting DNAs required for the two steps of the strategy decribed here, one or several nucleotides can be deleted from, added to, or changed in a BAC at a desired locus. Modifications can be designed freely either by adding or substituting nucleotides at the 3′ end of a 50 nucleotide homology arm, or by spacing the homology arms in such a way that the desired number of nucleotides is deleted from the BAC after the first targeting step. When adding or changing bases, it is important to include the desired change in the linear DNAs of both steps. If available, a template that contains the desired mutation(s) can also be used to amplify the linear DNA of step 2. We obtained evidence that suggests that the linear molecule of step 2 needs to be at least 500 bp long (data not shown). Since linear molecules containing a mutation in either homology arm are recombined at a very low efficiency (data not shown), introduction of secondary mutations, originating from errors in the PCR amplification of the linear product of step 2, is unlikely. In support of this, partial sequencing of four independent recombinant clones containing the additional EcoRI site revealed no secondary mutations.
An important virtue of the sacB–neo fusion gene is that selection for kanamycin resistance also selects for expression of SacB. This ensures that virtually all correct recombinants obtained at step 1 are sucrose sensitive. Moreover, it guarantees SacB expression during the competent cell preparation of step 2, since any mutation in the sacB region that inactivates the fusion protein cannot be amplified in the presence of kanamycin selection. After the BAC was targeted by the linear DNA of step 2, 85–90% of the obtained sucrose-resistant colonies retained kanamycin resistance. This indicates that the vast majority of mutational events are small ones affecting SacB protein function. Simply by restreaking for kanamycin sensitivity, these false candidates can easily be identified. Thereby, the most labor intensive step, in which putative recombinants are analyzed by restriction digestion and Southern hybridization, is done on a pre-screened subset of primary colonies, thus allowing efficient recovery of true recombinants (Figure 2 and data not shown).
By combining the advantages of ET recombination and selection/counterselection using the SacB–neo fusion protein, we have developed an efficient strategy that potentially enables the introduction of any deletions, insertions or mutations in large molecules such as BACs, PACs or the E.coli chromosome. Given the inherent practical simplicity of the two ET recombination steps involved, this strategy will greatly facilitate functional genetic studies, especially of large higher eukaryotic genes, for which the introduction of subtle modifications is often a requirement.
METHODS
pSacB-neo and pBAD-RedGam were constructed by conventional methods and by ET recombination. Oligonucleotides were synthesized by the EMBL oligonucleotide service. ET recombination steps were performed as described previously (Zhang et al., 1998; Muyrers et al., 1999); additional information is available (Muyrers et al., 2000b; see http://www.embl-heidelberg.de/ExternalInfo/stewart/index.html). Briefly, the strain containing the Af-4 BAC was transformed with pBAD-RedGam and grown on LB medium containing 20 µg/ml chloramphenicol and 50 µg/ml ampicillin. Prior to harvesting for competent cell preparation, the growing cells were induced with l-arabinose to 0.1% final concentration for 1 h. The linear product used in step 1 was PCR amplified from pSacB-neo using the following oligonucleotides (homology arms a and b are underlined; PCR primers are italicized; the additional C, coding for the additional G, is in bold type): 5′ oligonucleotide, containing homology arm a, TTAATGAATCGGCCAACGCGAACCCCTTGCGGCCGCCCGGGCCGTCGACCATCCTTTTATGATTTTCTATCA; 3′ oligonucleotide, containing homology arm b, AAGCGGATGAATGGCAGAAATTCGATGATAAGCTGTCAAACATGAGAATTCTCAGAAGAACTCGTCAAGAAGGCG.
All PCR amplifications were performed in triplicate using the Expand high fidelity PCR system according to the manufacturer’s instructions (Roche). The triplicate PCR products containing the sacB–neo fusion gene were pooled and 0.5 µg were digested with DpnI (New England Biolabs) and used to electroporate the cells containing the BAC, after which correct recombinants were obtained by selection on plates containing 20 µg/ml chloramphenicol, 20 µg/ml kanamycin and 50 µg/ml ampicillin. Although not required for obtaining recombinants, ampicillin selection was maintained to alleviate the need to retransform pBAD-RedGam for step 2. The linear product used in step 2 was PCR amplified using the unmodified, wild-type Af-4 BAC as the template. The following oligonucleotides were used (sequence from homology arm a is underlined; PCR primers are italicized; the additional G is in bold type): 5′ oligonucleotide, TTAATGAATCGGCCAACGCGAACCCCTTGCGGCCGCCCGGGCCGTCGACCGAATTCTCATGTTTGACAGCTTATC; 3′ oligonucleotide, ATGGAGAAAAAAATCACTGGATAT. Of the resulting 0.8 kb PCR product, 0.5 µg were electroporated into competent l-arabinose-induced cells containing pBAD-RedGam and the Af-4 BAC containing sacB–neo, which were grown on LB medium containing 20 µg/ml chloramphenicol, 20 µg/ml kanamycin and 50 µg/ml ampicillin. After electroporation, cells were incubated at 37°C for 1.5 h, spread onto plates containing 7% sucrose plus 20 µg/ml chloramphenicol, and grown at 25°C for ∼48–60 h. To identify true recombinants, the resulting primary colonies were restreaked in parallel on plates containing 20 µg/ml chloramphenicol plus 20 µg/ml kanamycin and on plates containing 20 µg/ml chloramphenicol plus 7% sucrose. Putative recombinants were grown overnight at 37°C in LB medium containing 20 µg/ml chloramphenicol, and characterized by restriction digestion and Southern analysis using standard methods.
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to thank Michelle Meredyth and Inhua Muyrers-Chen for critical reading of the manuscript. This work was supported in part by grants from the Volkswagen Foundation, Program on Conditional Mutagenesis and the NIH, National Institute for Aging.
REFERENCES
- Angrand P.O., Daigle, N., van der Hoeven, F., Schoeler, H.R. and Stewart, A.F. (1999) Simplified generation of targeting constructs using ET recombination. Nucleic Acids Res., 27, e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskaran K., Erfurth, F., Taborn, G., Copeland, N.G., Gilbert, D.J., Jenkins, N.A., Iannaccone, P.M. and Domer, P.H. (1997) Cloning and developmental expression of the murine homolog of the acute leukemia proto-oncogene AF4. Oncogene, 15, 1967–1978. [DOI] [PubMed] [Google Scholar]
- Blomfield I.C., Vaughn, V., Rest, R.F. and Eisenstein, B.I. (1991) Allelic exchange in Escherichia coli using the Bacillus subtilis sacB gene and a temperature-sensitive pSC101 replicon. Mol. Microbiol., 5, 1447–1457. [DOI] [PubMed] [Google Scholar]
- Datsenko K.A. and Wanner, B.L. (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA, 97, 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman L.M., Belin, D., Carson, M.J. and Beckwith, J. (1995) Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol., 177, 4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill F., Benes, V., Thomasova, D., Stewart, A.F., Kafatos, F.C. and Ansorge, W. (2000) BAC trimming: minimizing clone overlaps. Genomics, 64, 111–113. [DOI] [PubMed] [Google Scholar]
- Ioannou P.A., Amemiya, C.T., Garnes, J., Kroisel, P.M., Shizuya, H., Chen, C., Batzer, M.A. and de Jong, P.J. (1994) A new bacteriophage P1-derived vector for the propagation of large human DNA fragments. Nature Genet., 6, 84–89. [DOI] [PubMed] [Google Scholar]
- Murphy K.C. (1991) Lambda Gam protein inhibits the helicase and chi-stimulated recombination activities of Escherichia coli RecBCD enzyme. J. Bacteriol., 173, 5808–5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyrers J.P.P., Zhang, Y., Testa, G. and Stewart, A.F. (1999) Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic Acids Res., 27, 1555–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyrers J.P.P., Zhang, Y., Buchholz, F. and Stewart, A.F. (2000a) RecE/RecT and Redα/Redβ initiate double stranded break repair by specifically interacting with their respective partners. Genes Dev., 14, 1971–1982. [PMC free article] [PubMed] [Google Scholar]
- Muyrers J.P.P., Zhang, Y. and Stewart, A.F. (2000b) ET-cloning: Think recombination first. In Setlow, J.K. (ed.), Genetic Engineering, Principles and Methods. Volume 22. Kluwer Academic/Plenum Press, New York, NY, (in press). [DOI] [PubMed]
- Narayanan K., Williamson, R., Zhang, Y., Stewart, A.F. and Ioannou, P.A. (1999) Efficient and precise engineering of a 200 kb β-globin human/bacterial artificial chromosome in E.coli DH10B using an inducible homologous recombination system. Gene Ther., 6, 442–447. [DOI] [PubMed] [Google Scholar]
- Shizuya H., Birren, B., Kim, U.J., Mancino, V., Slepak, T., Tachiiri, Y. and Simon, M. (1992) Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc. Natl Acad. Sci. USA, 89, 8794–8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg N.L. (1992) Cloning high molecular weight DNA fragments by the bacteriophage P1 system. Trends Genet., 8, 11–16. [DOI] [PubMed] [Google Scholar]
- Yu D., Ellis, H.M., Lee, E.C., Jenkins, N.A., Copeland, N.G. and Court, D.L. (2000) An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl Acad. Sci. USA, 97, 5978–5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Buchholz, F., Muyrers, J.P.P. and Stewart, A.F. (1998) A new logic for DNA engineering using recombination in Escherichia coli. Nature Genet., 20, 123–128. [DOI] [PubMed] [Google Scholar]