Abstract
As a first step towards a more comprehensive functional characterization of cDNAs than bioinformatic analysis, which can only make functional predictions for about half of the cDNAs sequenced, we have developed and tested a strategy that allows their systematic and fast subcellular localization. We have used a novel cloning technology to rapidly generate N- and C-terminal green fluorescent protein fusions of cDNAs to examine the intracellular localizations of >100 expressed fusion proteins in living cells. The entire analysis is suitable for automation, which will be important for scaling up throughput. For >80% of these new proteins a clear intracellular localization to known structures or organelles could be determined. For the cDNAs where bioinformatic analyses were able to predict possible identities, the localization was able to support these predictions in 75% of cases. For those cDNAs where no homologies could be predicted, the localization data represent the first information.
INTRODUCTION
Reconciling the large amount of DNA sequence information that is now available with more informative functional data presents itself as a fundamental issue in modern biology. The rapid increase in the productivity of both genomic and cDNA sequencing projects (Dunham et al., 1999; Kikuno et al., 2000; S. Wiemann et al., submitted) worldwide now provides an elementary base from which the next task is to relate sequence with function, in a sufficiently high-throughput manner. Since eukaryotic cells, and in particular mammalian cells are highly compartmentalized, a protein’s localization is in most cases tightly bound to its function. Extending this brings us to the notion that combining the knowledge of a protein’s intracellular localization with the DNA sequence from which it is derived and its bioinformatic analysis would give us a tangible feel for its function. Indeed, it has become clear that despite the rapid expansion of sequence databases, sequence comparison and bioinformatic analyses alone are still not strong enough to assign function assuredly to novel cDNAs (Eisenhaber and Bork, 1998). As a first step towards the systematic functional characterization of novel cDNAs identified by large scale cDNA sequencing projects, we have devised and tested a strategy to tag systematically the open reading frames (ORFs) of novel cDNAs with the coding sequence of spectral variants of green fluorescent protein (GFP), subsequently expressed the fusion proteins in cells and determined their subcellular localization.
RESULTS AND DISCUSSION
If any strategy for large scale functional analysis of ORFs and encoded proteins is to be initiated it is evident that the methods need to have the potential to be automated. Therefore, one of the major issues that must be resolved is the availability and application of a high-capacity cloning system. Cloning of ORFs needs to be rapid, efficient and directional, and compatible with a range of expression vectors, therefore any conventional approach attempting to analyse a large number of target ORFs cannot meet all these criteria. To this end, we have adapted and applied the Gateway™ cloning system, recently described by Walhout et al. (2000). This system allows the cloning of amplified ORFs by recombination, thereby circumventing the need for screening for restriction sites that are present within the cloning sites of the cloning vector(s) but not in the ORF in question.
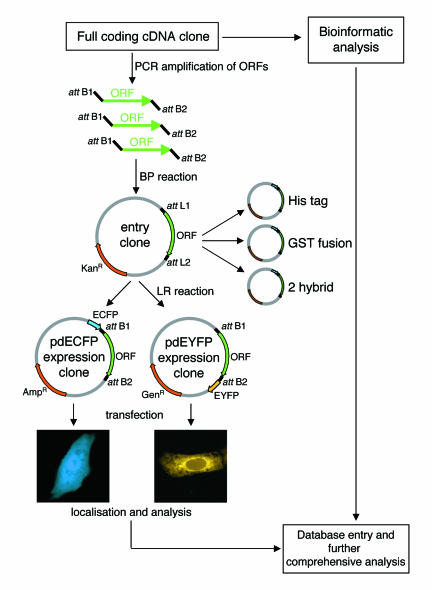
ORFs were amplified by PCR and the products were then cloned in a single tube (BP) reaction, thereby generating universal ‘entry clones’ compatible with any Gateway™ expression vector (Figure 1; see also Methods). Subsequently the inserts of the entry clones were cloned (LR reaction), again by recombination, into suitable GFP expression vectors (see Methods) allowing the expression of the ORFs as cyan fluorescent protein (CFP, N-terminal fusion) or yellow fluorescent protein (YFP, C-terminal fusion) fusion proteins (Figure 1; see also Methods). The ORF–GFP fusion plasmids generated were then transfected into mammalian cells and expression was analysed in living cells at various time points after transfection and the localizations recorded. Monitoring the cells at these multiple time points allowed for any effects of the increasing expression levels to be correlated with the expression time. After 48 h of expression, the cells were fixed and stored for further immunofluorescence analysis as appropriate. In order to test the reliability of the cloning and transfection strategy, we first took a selection of known proteins with already well characterized subcellular localizations. The ORFs representing these proteins were amplified, cloned into the CFP and YFP vectors using the Gateway™ cloning system, then transfected into cells. From these control experiments we found that seven out of nine of these GFP-tagged proteins localized as expected, representing structures such as the nucleus, Golgi complex and microtubules. (These data are available at the project website: http://www.dkfz-heidelberg.de/abt0840/GFP/) These results suggest that the approach described here, and in particular the presence of the Gateway™ recombination peptides on the fusion proteins, only causes interference with a protein’s localization in a minority of examples and should thus be suitable to determine the localizations of novel proteins with a high degree of certainty.
Fig. 1. Strategy for rapid systematic localization and functional characterization of proteins encoded by novel cDNAs. Individual full coding cDNAs were PCR amplified using automatically designed primers, which also added the recombination sequences attB1 and attB2 at the 5′ and 3′ ends, respectively. These products were then recombined into the entry clones, which then served as a universal source of material for all expression vectors. N- and C-terminal GFP (CFP/YFP) fusion expression vectors were both generated from the entry clone in a single recombination reaction. These clones were then transfected into cells and the localizations of the fusion proteins recorded. This information was then combined with the bioinformatic data generated from sequence analysis and additional immunostainings using compartment-specific antibodies when appropriate.
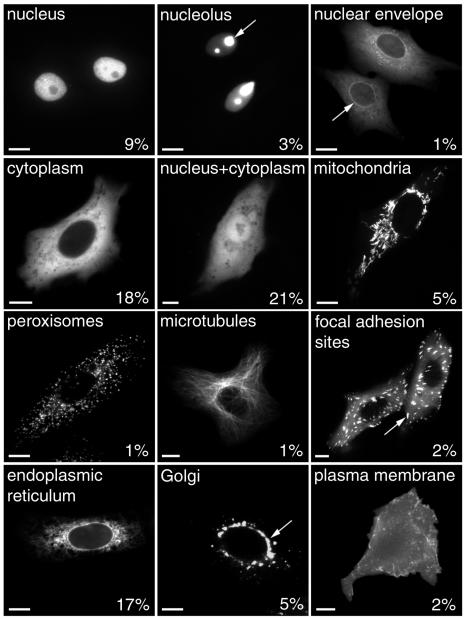
Figure 2 shows examples of the clear subcellular localizations observed in experiments with novel cDNAs, including the nucleolus, mitochondria, membranes of the secretory pathway and cytoskeletal elements. In total, 107 ORFs were tested, two independent N- and C-terminal fusions of each, representing a total of 428 transfections. Importantly, these results were also highly consistent between cell types. In control experiments, 14 examples of these different localizations were tested in human HeLa cells, and the localizations observed were found to be indistinguishable from the original data recorded in the monkey Vero cells. Again the full data are available at the project website.
Fig. 2. cDNA–GFP fusions express and localize to a wide variety of intracellular compartments. Vero cells were transfected with each cDNA–GFP in turn, and allowed to express the proteins for the times stated in Methods. The cells were imaged live and the localizations recorded. The figure shows examples of twelve of the localizations observed, with arrows used to point out particular structures as appropriate. Numbers indicate the percentage of individual cDNA molecules that express and localize in the categories shown. Other categories not depicted are Golgi and plasma membrane localization (6%), other unknown localizations (8%) and no expression (1%). Bar, 10 µm.
As expected, a significant percentage of the cDNA products localized to either the cytosol (18%) or the nucleus (12%); however, a large number of molecules were also found to be associated with the secretory pathway (28%). In addition, mitochondrial proteins (5%) and cytoskeletal structures (3%) were also identified. An additional category (other/unknown) was also denoted (8% of the cDNA products), and refers to structures that were not easily identifiable from visual analysis. In some examples these were likely to be either protein aggregates or proteins where the GFP tag was inhibiting membrane association (for example, Rab proteins). In other examples large globular structures could be seen that on subsequent immunostaining analysis did not co-localize with any known organelle markers. The remaining 21% of the cDNA products localized to both the cytosol and nucleus, and it is this population that provides the least information.
One frequent problem with the use of GFP is that it is critical to which end of the molecule of interest it should be fused (see e.g. Scales et al., 1997). The cloning approach used here however, has allowed us to generate both N- and C-terminal GFP fusions in a single reaction, thus we were immediately able to identify any aberrant effects that the GFP may have had on targeting signals of the protein. Clearly this appeared to be critical for the localization studies, as many of the patterns observed were only seen in a particular fusion order. The most striking example was in the case of the mitochondrial proteins identified. Of the five proteins to localize in this compartment, in every case the fusion order ORF–YFP gave specific mitochondrial localization, however the CFP–ORF order gave a nuclear and cytosolic localization (indicative of GFP targeting). Clearly the inference from this is that any perturbation of the N-terminal mitochondrial targeting signal abolished the ability of this protein to localize correctly. Further examples were also seen with the proteins of the secretory pathway; in the cases where bioinformatic analyses had predicted the presence of a signal peptide, only the orientation ORF–YFP correctly localized to the endoplasmic reticulum (ER). However, where a peripheral membrane protein was predicted for example, both ORF–YFP and CFP–ORF constructs localized in an ER pattern. The strategy presented here, therefore, largely circumvents the danger of missing entire classes of proteins due to misplacement of the GFP tag.
We next considered the usefulness of the bioinformatic analysis data on the cDNAs with respect to their actual localizations. For 47% of these novel cDNAs bioinformatics was able to make some predictions based on sequence similarities or predicted domains. The strengths of similarities and usefulness of the bioinformatic information were extremely variable. For example, one cDNA (DKFZp564J1864) had a 99% identity with the Canis familiaris microsomal signal peptidase 23 kDa subunit, and as such would be presumed to be the human homologue of this protein. The actual localization determined was the ER, thereby effectively confirming the prediction. Lower identity values from the bioinformatic prediction also often proved to be valuable. The highest similarity for one cDNA (DKFZp564I2482) was a 50% identity with an RNA helicase from Drosophila melanogaster. The observed localization of this cDNA product was the nucleus, therefore adding strength to this bioinformatic prediction. One other RNA helicase (DKFZp564C183) was also predicted from sequence analysis of the cDNA set. However, in contrast to the previous example, this cDNA product localized to the nuclear envelope. The possibility therefore exists that in this case the localization data have allowed us to discriminate between two helicases with different function: the first involved in RNA maturation, the second in RNA transport into the cytosol. This example clearly shows how the data here not only confirm or contradict bioinformatic predictions but also can extend them with important information that has implications for functional predictions. Overall, the results obtained indicated that the bioinformatic predictions for the cDNAs were supported in three times more cases than they were contradicted. Perhaps more importantly, however, bioinformatic analysis was unable to make any homology predictions for 53% of the cDNAs and encoded proteins. For these cDNA products therefore, their specific localization to a discrete compartment in living cells provides the first data at all. Due to space restrictions the complete data set (including the bioinformatic analysis results and images of the localizations for every cDNA) generated by this work cannot be presented here, however it is constantly updated and freely available at the project website (http://www.dkfz-heidelberg.de/abt0840/GFP/).
The tools generated here also represent a valuable source of material for subsequent analysis of interesting candidates. These clones are immediately useful to investigate the effects of overexpression of a protein, to study a protein’s trafficking, to reveal possible post-translational modifications or to identify interacting proteins using immunoprecipitation of the expressed GFP-tagged proteins with anti-GFP antibodies. In addition, the entry clone of each ORF generated by this work is ready to be transferred into other functionally useful destination vectors: for example, glutathione S-transferase (GST) or His6 fusion vectors to allow easy purification of the protein of interest, or yeast two-hybrid vectors to assay for interacting partners (Walhout et al., 2000).
In summary, we have identified the intracellular localization of ∼100 proteins encoded by novel human cDNAs. We have both adapted and proved the feasibility of a novel cloning system, which allows the rapid transfer of coding sequences between functionally useful vectors. This has allowed us to make better use of the large data and clone set generated by the German cDNA Consortium, adding the first in vivo data to the previously available bioinformatic data, and inferring possible function to cDNA products without any identified homologies. Such a GFP fusion localization approach in live cells is advantageous since it is rapid, generates reagents of value for further analysis of interesting candidates, and is extremely amenable to automation and greater high-throughput capacity. In addition, it allows the identification of groups of proteins localizing to the same subcellular compartment. This should help to concentrate subsequent work only on those cDNAs where the localization, as has been determined here, correlates with specific study areas [for example, gene transcription (nucleus) or secretory membrane traffic (ER, Golgi, plasma membrane)]. Furthermore, this approach does not require the subsequent identification and cloning of the cDNA after an interesting localization has been identified, which has always been a time-consuming factor in other localization-based strategies (reviewed by Gonzalez and Bejarano, 2000). Clearly the challenge of human genomics is to assign relevant functional data to the wealth of cDNAs now being sequenced, and the approach and data presented here is surely one step towards this goal.
METHODS
Sources of ORFs used in this study. The aim of the German cDNA Consortium is to generate and analyse cDNAs of as yet unknown human transcripts. In order to identify novel cDNAs, arrayed cDNA libraries from fetal brain, fetal kidney, testis, amygdala, melanoma cell line (MeWo) and several other tissues have been generated. The libraries are systematically sequenced and >30 000 ESTs have been generated thus far. After bioinformatic analysis of the 5′ ESTs, targets for full-length sequencing are selected. The resulting sequences are finally characterized with bioinformatic tools and analysed for the presence of novel ORFs. In the project described here, identified ORFs are amplified on a sequential basis with the eventual aim being to screen all proteins encoded by the full coding cDNAs identified by the consortium.
Amplification of ORFs to generate products compatible with the Gateway™ cloning system. ORFs were amplified from cDNA (either clone pools of 384 clones, single clones or primary cDNA) omitting the 5′ and 3′ UTRs. PCR primer pairs were selected using the PRIDE program (Haas et al., 1998). The ORF-specific part of the 5′ amplification primer was fixed to include the initiator ATG. The 3′ amplification primer was designed to contain the codon encoding the C-terminal amino acid residue but leaving out the terminator triplet, in order to allow for the expression of C-terminal fusion proteins. PCR primers were purchased from commercial vendors (Life Technologies). Amplification of ORFs was done using the high fidelity amplification systems (Roche, Mannheim) in order to minimize the number of PCR errors. PCR products were purified with the help of Qiaquick spin columns (Qiagen, Hilden).
Modification of expression vectors. Original EGFP vectors (pEGFP-N3 and pEGFP-C1) and their colour variants (pEYFP-N1 and pECFP-C1) were obtained from Clontech (Heidelberg). The kanamycin resistance genes in the pEYFP and pECFP vectors were replaced with gentamycin and ampicillin resistance genes, respectively, which had been amplified from standard vectors (pLacUV5-gen, Life Technologies and pBluescript, Stratagene). Then, Gateway™ rf cassettes were cloned into the blunted XhoI and AgeI sites of the pEYFP and the blunted BglII and BamHI sites of the pECFP vectors to generate Gateway™-compatible ‘destination vectors’. These vectors were propagated in the host B462 to compensate for the expression of the ccdB gene present on the cassette (Bernard et al., 1994). The correct orientation and reading frames of the constructs were verified by sequencing.
Cloning protocol. Cloning of ORFs with the help of the Gateway™ system takes advantage of the phage Lambda recombination system, overcoming the need for restriction digests and ligation reactions. Cloning of ORFs with the Gateway™ system was carried out in two steps. First, the amplified ORF was cloned into an ‘entry vector’ via the BP reaction. The resulting ‘entry clones’ then formed a universal source of material for the subsequent generation of any expression constructs. Two independent entry clones were picked and analysed for every ORF. The integrity of the ORFs was verified by sequencing to exclude errors introduced in the amplification step. The cloning into the EYFP and ECFP expression vectors was done in the LR reaction. Both the pdEYFP and pdECFP vectors were combined together with the entry clone in the same tubes to perform simultaneous transfer of the ORF from the entry clone into the destination vectors. After transformation into DH10B (Life Technologies), the respective destination clones were selected for by plating on suitable selective agar plates (ampicillin for the pdECFP-ORF destination clones and gentamycin for the pdEYFP-ORF destination clones).
Purification of plasmid DNA. Plasmid DNA was prepared in 96-well format using the Macherey and Nagel Nucleobond preps (Macherey, Nagel, Dueren) with the help of a Qiagen Biorobot 9600 (Qiagen, Hilden). Alternatively, DNA was prepared with the Qiawell Ultra kit (Qiagen, Hilden). The DNA prepared with the help of the Nucleobond preps was of sufficient quality for the transfections and was thus predominantly used to prepare the expression plasmid DNAs.
Transfections. Vero cells (ATCC CCL81) were routinely cultured in MEM supplemented with 10% fetal bovine serum, 100 IU penicillin and 100 µg/ml streptomycin at 37°C in a humidified 5% CO2 incubator. The day prior to transfection cells were plated into 35 mm glass-bottomed dishes (MatTek Corp., MA) at a density of 20%. On the day of transfection, 1 µg of each DNA was used with 3 µl of FuGENE6 (Roche, Mannheim) to transfect the cells, according to the manufacturer’s instructions.
Data collection and image analysis. All data acquisition and image analysis was carried out at the Advanced Light Microscopy Facility at EMBL, Heidelberg (http://www.EMBL-Heidelberg.DE/ExternalInfo/almf/index.html).
Cells were imaged at 16, 24 and 40 h after transfection in carbonate-free culture medium equilibrated with 10 mM HEPES pH 7.4 (Shima et al., 1999) on a Leica DM/IRBE microscope with a 63× NA 1.4PL Apo objective using custom designed CFP or YFP filters (Stephens et al., 2000). Images were captured with a Hamamatsu charge-coupled device camera (ORCA 1) using the Openlab 2.0 software (Improvision, Coventry, UK). Images were analysed using Adobe Photoshop 5.0.
Immunofluorescence and determination of the localization of GFP fusion proteins. Identification of certain structures was relatively clear: for example, the cytoplasm, nucleus, nucleolus, nuclear envelope, mitochondria, ER and plasma membrane. For other structures seen, we also considered the bioinformatic predictions before assigning a localization category: for example, peroxisomes and focal adhesion sites. In the cases where N- and C-terminal fusion localizations were not identical (one fusion order giving distinct structures, the other giving nucleus and cytoplasm), the bioinformatic predictions were also taken into account before regarding the nucleus and cytosol localization as being aberrant, due to GFP masking appropriate targeting signals. For other structures, the cells were fixed in methanol at –20°C for 4 min, then washed with phosphate-buffered saline. Immunofluorescence was carried out using primary antibodies against known proteins, for example, microtubules (anti-α-tubulin; Amersham) and the Golgi complex (anti-giantin and anti-GM130 from David Shima, ICRF, UK) (anti-beta COP; Pepperkok et al., 1993).
Bioinformatic analyses. Every cDNA sequence was compared with the sequences in EMBL and EMBL-EST databases using BLASTN. Putative protein sequences were identified by searches for the longest ORF (with a minimum length of 90 codons) in each of the three forward frames. The deduced protein sequences were compared against a non-redundant protein database comprising PIR, SWISSPROT and TREMBL using the BLASTX program. Hits were screened for significance by analysing for functional domains (PFAM), and finally the most significant hit was selected as reference.
Acknowledgments
ACKNOWLEDGEMENTS
We thank David Shima for antibodies, Philippe Bastiaens, Jan Ellenberg, Wilhelm Just and David Stephens for known DNA clones, and Cayetano Gonzalez and Martin Lowe for critical reading of this manuscript. We thank Daniel Bongartz, Oliver Heil and Lars Ebert for help with the web presentation of the data. We thank the German cDNA Consortium for providing cDNA clones and sequences. We thank Life Technologies for advanced access to the Gateway technology. The Advanced Light Microscopy Facility (ALMF) at EMBL is supported by Bitplane (Switzerland), Eppendorf (Germany), Improvision (UK), Zeiss (Germany), Leica (Germany), Olympus (Europe), Omicron (Germany), T.I.L.L. Photonics (Germany) and Perkin Elmer (UK). J.C.S. is supported by an EMBO Long Term Fellowship.
REFERENCES
- Bernard P., Gabant, P., Bahassi, E.M. and Couturier, M. (1994) Positive-selection vectors using the F plasmid ccdB killer gene. Gene, 148, 71–74. [DOI] [PubMed] [Google Scholar]
- Dunham I. et al. (1999) The DNA sequence of human chromosome 22. Nature, 402, 489–495. [DOI] [PubMed] [Google Scholar]
- Eisenhaber F. and Bork, P. (1998) Wanted: subcellular localization of proteins based on sequence. Trends Cell Biol., 8, 169–170. [DOI] [PubMed] [Google Scholar]
- Gonzalez C. and Bejarano, L.A. (2000) Protein traps: using intracellular localization for cloning. Trends Cell Biol., 10, 162–165. [DOI] [PubMed] [Google Scholar]
- Haas S., Vingron, M., Poustka, A. and Wiemann, S. (1998) Primer design for large scale sequencing. Nucleic Acids Res., 26, 3006–3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuno R., Nagase, T., Suyama, M., Waki, M., Hirosawa, M. and Ohara, O. (2000) HUGE: a database for human large proteins identified in the Kazusa cDNA sequencing project. Nucleic Acids Res., 28, 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepperkok R., Scheel, J., Horstmann, H., Hauri, H.P., Griffiths, G. and Kreis, T.E. (1993) Beta-COP is essential for biosynthetic membrane transport from the endoplasmic reticulum to the Golgi complex in vivo. Cell, 74, 71–82. [DOI] [PubMed] [Google Scholar]
- Scales S.J., Pepperkok, R. and Kreis, T.E. (1997) Visualization of ER-to-Golgi transport in living cells reveals a sequential mode of action for COPII and COPI. Cell, 90, 1137–1148. [DOI] [PubMed] [Google Scholar]
- Shima D.T., Scales, S.J., Kreis, T.E. and Pepperkok, R. (1999) Segregation of COPI-rich and anterograde-cargo-rich domains in endoplasmic-reticulum-to-Golgi transport complexes. Curr. Biol., 9, 821–824. [DOI] [PubMed] [Google Scholar]
- Stephens D.J., Lin-Marq, N., Pagano, A., Pepperkok, R. and Paccaud, J.-P. (2000) COPI coated ER-to-Golgi transport complexes segregate from COPII at ER exit sites. J. Cell Sci., 113, 2177–2185. [DOI] [PubMed] [Google Scholar]
- Walhout A.J.M., Sordella, R., Lu, X., Hartley, J.L., Temple, G.F., Brasch, M.A., Thierry-Mieg, N. and Vidal, M. (2000) Protein interaction mapping in C.elegans using proteins involved in vulval development. Science, 287, 116–122. [DOI] [PubMed] [Google Scholar]