Abstract
Mycoplasma synoviae has two major membrane antigens, MSPA and MSPB, both of which are phase variable and which may be coordinately involved in adhesion of the organism to erythrocytes. A single gene (vlhA) from M. synoviae was characterized, and polypeptides were expressed from nonoverlapping 5′ and 3′ regions in Escherichia coli. The expression product of the vlhA 5′ region reacted with specific reagents against MSPB, while that of the 3′ region reacted with specific reagents against MSPA. Analysis of the predicted amino acid sequence showed a characteristic signal peptidase II cleavage site, and the presence of the acylation site was confirmed by identification of a lipid-associated membrane protein, similar in molecular mass to MSPB, in [3H]palmitate-labelled membrane proteins. Further sequence analysis of the vlhA gene revealed a high identity with the Mycoplasma gallisepticum pMGA1.7 gene, a member of a large translated family. The vlhA gene was shown to hybridize to multiple restriction fragments of the M. synoviae genome, suggesting that it was also a member of a multigene family. These findings indicate that coordinate phase variation of the two major surface antigens of M. synoviae WVU may be due to their expression from the same gene and that homologous gene families encode the major hemagglutinins of two phylogenetically distinct mycoplasmas. The presence of homologous multigene families in such phylogenetically distinct species, but not in the genomes of more closely related species, suggests that the families may have been transferred horizontally.
The use of large multigene families to encode phase-variable surface antigens is an emerging theme in mycoplasma pathogenesis. The identification of such families in phylogenetically diverse mycoplasma species, including Mycoplasma bovis (3), Mycoplasma gallisepticum (20), Mycoplasma hyorhinis (29), and Mycoplasma pulmonis (4), has suggested that they may be a common feature in mycoplasma genomes. However, none of the families identified thus far has had detectable similarity at the level of their primary sequence with families in other phylogenetically distinct species, and the mechanisms used for the control of their expression appear to be distinct in each species. Indeed, the largest of these multigene families, the pMGA family of M. gallisepticum, has no identifiable homolog in either Mycoplasma pneumoniae or Mycoplasma genitalium, both of which have had their complete genomic sequences determined (10, 13). In spite of the absence of this gene family, six distinct gene families of unknown function have been identified for M. pneumoniae, all of which, like the families characterized for other mycoplasma species, are predicted to encode lipoproteins (13).
Both Mycoplasma synoviae and M. gallisepticum are major poultry pathogens, producing respiratory diseases in chickens and turkeys (14). Recent work in our laboratory has identified two major phase-variable surface proteins of M. synoviae, MSPA and MSPB (22), and has shown that MSPA is a hemagglutinin. The sizes of these proteins are predicted to be 50 and 45 kDa, respectively, although multiple forms of each are seen in a single clone of M. synoviae. Expression of these proteins may be coordinate, as a clone which had lost the capacity to hemadsorb was shown to have ceased expressing both surface proteins.
The aim of this study was to identify the genes encoding the two phase-variable immunodominant surface proteins of M. synoviae and to establish any similarities that might exist between these genes and those of other phase-variable surface proteins found in other mycoplasma species.
Bacterial strains and growth media.
In this study, all experiments on M. synoviae were conducted with strain WVU-1853. The origins of M. synoviae WVU-1853 and M. gallisepticum S6 have been described previously (19, 21). The identity of each species was confirmed by immunofluorescence, restriction fragment length polymorphism, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and colony immunoblotting techniques, as described previously (28). The organisms were grown in mycoplasma broth (MB) to late logarithmic phase and harvested at approximately pH 6.8 (28). Escherichia coli DH5α and JM109, electrotransformed with recombinant plasmids, were grown at 37°C in SOC broth (containing, per liter, 20 g of Bactotryptone, 5 g of Bacto-yeast extract, 0.5 g of NaCl, and 20 mM glucose) or on Luria agar, both containing 50 μg of ampicillin/ml (25).
Genomic libraries.
Isolation of M. synoviae genomic DNA was performed as described previously (20). The initial expression library was prepared by ligating M. synoviae DNA partially digested with Sau3AI into plasmid pGEX-4T-1 (Pharmacia Biotech) and using this to transform E. coli DH5α as described previously (8). A pool of monospecific rabbit antisera to each antigen (22) was used to screen for recombinant clones expressing regions of the MSPA and MSPB gene(s) in the expression library. The selected clones were further examined by probing a Western blot of their whole-cell proteins with monospecific antiserum to MSPB or MSPA. Additional genomic libraries were prepared by ligating M. synoviae DNA digested to completion with EcoRI or BglII (Boehringer Mannheim) into compatibly digested pUC18 (Pharmacia Biotech). Competent E. coli DH5α cells were transformed with the recombinant plasmids (25) and grown on Luria agar, and the resultant colonies were screened by DNA hybridization as described by Sambrook et al. (25).
PCR.
Table 1 describes the oligonucleotide primers used for PCR amplification and their target sites. The PCR protocol was adapted from the procedure previously described by Sambrook et al. (25). Briefly, a 50-μl reaction mixture containing 300 μM each dATP, dCTP, dGTP, and dTTP, 1.75 mM MgCl2, 250 μM each primer, 1.25 U of Taq DNA polymerase (Promega), 5 μl of 10× Taq DNA polymerase buffer, and 1 μl of template DNA (plasmid containing the complete gene [or fragments of it] or diluted M. synoviae genomic DNA) was prepared. The reaction mixture was incubated at 94°C for 60 s and then subjected to 30 cycles of 51°C for 20 s, 72°C for 90 s, and 94°C for 10 s. For cloning, the PCR product was purified by using Bio-Spin 6 chromatography columns (Bio-Rad). The purified PCR product of the entire length of the gene was treated with the Klenow fragment of E. coli DNA polymerase and bacteriophage T4 polynucleotide kinase (Boehringer Mannheim) to fill and phosphorylate the recessed 3′ termini created by Taq DNA polymerase (25). The resultant product was purified by using the Wizard DNA clean-up system kit (Promega) and ligated into pUC18 (Pharmacia Biotech) digested with SmaI. The ligated plasmid was used to transform E. coli DH5α cells, and transformants lacking β-galactosidase activity were examined by agarose gel electrophoresis to confirm the presence of an inserted fragment of the expected size (25). For expression of polypeptides from regions I and II of the gene, the relevant PCR product was ligated into the plasmid vector PinPoint Xa1-T (Promega) as instructed by the manufacturer. Electrotransformation of the E. coli JM109 cells was performed as described above, and the resultant colonies were screened for expression of corresponding polypeptides by SDS-PAGE analysis of their whole-cell proteins.
TABLE 1.
Sequences, binding sites, and orientations of the PCR oligonucleotide primers
| Name | Orientationa | Binding siteb | Sequence |
|---|---|---|---|
| PCRF | Forward | 1–23 | GATGCGTAAAATAAAAGGATTT |
| PCRR | Reverse | 2405–2382 | ATGTTTTTGGTTTTATTATTATTA |
| B1 | Forward | 104–129 | TGGATCCCAAACTCCAGCACCTGAAC |
| B2 | Reverse | 1071–1052 | GTTTGAATTCTGATTTGTCT |
| A1 | Forward | 1598–1625 | TGGATCCTCTAAAGGAAATGTTACTAAA |
Forward and reverse refer to orientations of the coding and complementary strands, respectively.
Positions of bases correspond to the nucleotide sequence presented in Fig. 1.
Southern blot hybridization, DNA sequencing, and sequence analysis.
For Southern blot hybridization, genomic DNAs from M. synoviae and M. gallisepticum were isolated and digested with restriction endonuclease BglII, EcoRI, or HindIII. The resultant fragments were separated by electrophoresis through a 0.8% agarose gel and Southern transferred to a nylon membrane (Amersham Hybond-N+) as described previously (25). The PCR product, amplified with 5′ and 3′ sequencing primers for the recombinant plasmid, was radiolabelled with [α-32P]dCTP by using a random-primed-labelling kit (Boehringer Mannheim). The radiolabelled probe was purified by using Bio-Spin 6 chromatography columns and incubated with the nylon membrane at 57°C in 6× SSC (1× SSC is 0.15 M NaCl plus 15 mM Na3 citrate) overnight. The membrane was washed three times, each time for 10 min at 57°C, in 2× SSC–0.1% SDS for washes of low stringency and 0.1× SSC–0.1% SDS for washes of high stringency. The membrane was autoradiographed as described previously (25).
The dideoxy-chain termination method was performed with T7 DNA polymerase (Promega) as instructed by the manufacturer to determine the nucleotide sequences of the recombinant plasmids. Both strands of the DNA were completely sequenced with synthetic oligonucleotide primers designed based on the previously determined DNA sequence. The DNA sequence was analyzed by using computer programs provided by the Australian National Genomic Information Service.
A gene which codes for a protein with a molecular mass higher than that of the native protein.
Twenty-four colonies were selected from an expression library of M. synoviae DNA partially digested with Sau3AI and were found to be immunoreactive with reagents directed to the 45-kDa MSPB. The plasmid from these clones was purified, and the M. synoviae genomic fragments contained by the plasmid were partially or completely sequenced. All of these plasmids shared a 332-bp Sau3AI fragment of M. synoviae (results not shown). By using pGEX 3′- and 5′-sequencing primers, the 332-bp Sau3AI fragment from one clone was amplified by PCR, and the resultant product was radiolabelled and used to probe a Southern blot of M. synoviae genomic DNA digested with EcoRI. The autoradiograph of the blot, washed at high stringency, showed that the probe hybridized to an EcoRI fragment of 2.6 kb (results not shown). A library of M. synoviae EcoRI fragments in the plasmid vector pUC18 was screened by colony blot hybridization with the 332-bp Sau3AI fragment as a probe. Analysis of the sequence data derived from a clone containing a recombinant plasmid with a 2.6-kb EcoRI fragment revealed a putative 5′ end of an open reading frame (ORF). In the same manner, the 2.6-kb EcoRI fragment was used to identify a 2.8-kb BglII fragment of M. synoviae genomic DNA, which was cloned, sequenced, and found to contain the 3′ end of the ORF. As there were sequence differences between the corresponding regions of the EcoRI and BglII fragments, two PCR primers, PCRF and PCRR (Table 1), were designed, and the entire ORF of the gene was amplified directly from M. synoviae genomic DNA. The resultant 2.4-kb PCR product was purified and ligated into pUC18, and the recombinant plasmid was used to transform E. coli DH5α cells.
The nucleotide sequence of the cloned PCR product and its corresponding predicted amino acid sequence were determined (Fig. 1). The ORF is 2,364 bp long, starting from base 27 with an ATG start codon and finishing at base 2382 with three consecutive TAA stop codons. The overall G+C content of the putative protein-coding region is 36%, which is in the approximate range predicted previously for M. synoviae genomic DNA (12). The ORF was predicted to encode a protein with a molecular mass of 84.5 kDa containing six Trp residues (positions 317, 338, 444, 485, 521, and 740), all encoded by TGA codons. The protein sequence possessed a consensus sequence of the signal peptidase II cleavage (Fig. 1), suggesting that it encoded a lipoprotein. Adjacent to the amino-terminal Cys predicted for the mature protein were tandem repeats of a Pro- and Asn-rich region of 19 amino acids (Fig. 1). A Kyte-Doolittle hydrophobicity plot (16) of the predicted amino acid sequence showed a relatively hydrophilic protein with two exceptionally hydrophobic amino- and carboxyl-terminal regions (results not shown).
FIG. 1.
Truncated nucleotide and deduced amino acid sequences of the vlhA gene (uppercase letters). Two nearly identical tandemly repeated 19-amino-acid regions at the amino-terminal end of the sequence are underlined. The location of the putative signal peptidase II recognition sequence is underlined with asterisks. Lowercase letters indicate the target site of the oligonucleotide primers (PCRF and PCRR) used to amplify the gene by PCR. Numbers to the left show the positions of the nucleotides, relative to the first 5′ nucleotide in the target site of the PCRF oligonucleotide primer.
Like many other membrane proteins of mycoplasmas (6, 20, 23, 26), MSPB contains a proline-rich region. Recent studies of M. pneumoniae have established an essential role for proline-rich regions in cytadherence (2, 7, 18). It is notable that this region is tandemly repeated, and several mycoplasma proteins are capable of size variation due to expansion or contraction of such tandemly repeated regions (4, 29, 30).
Sequence analysis of the vlhA gene showed a major continuous ORF of 2,364 bp and three shorter ORFs in the complementary strand of the sequence (results not shown). Multiple complementary-strand ORFs have also been observed in variable lipoprotein genes (vlpA to vlpF) of M. hyorhinis and have been suggested to provide a reservoir of potential coding capacity (29). However, it is possible that they occur simply as a result of codon usage bias in these genes.
Two ends of the gene encode two antigenically distinct proteins, MSPA and MSPB.
As the predicted product of the identified gene was almost twice the molecular mass of MSPB, the reactivities of different regions of the gene product were examined with different reagents against MSPA and MSPB. Two pairs of oligonucleotide primers (B1-B2 and A1-PCRR [Table 1]) were used to amplify two nonoverlapping regions of the gene which did not contain TGA Trp codons at their 5′ ends. The resultant PCR products were ligated into the expression vector PinPoint Xa1-T, which was used to transform E. coli JM109 cells. Region I, containing bases 104 to 1071, was predicted to encode a polypeptide of 290 amino acids, and region II, containing bases 1598 to 2405, was predicted to encode a polypeptide of 215 amino acids (Fig. 2A) in E. coli cells. SDS-PAGE of the whole-cell proteins of E. coli cells containing the recombinant plasmids (results not shown) demonstrated the expression of fusion proteins of 50 kDa from the plasmid containing region I and of 40 kDa from the plasmid containing region II. Immunostaining of Western blots of the whole-cell proteins of the clone expressing the polypeptide encoded by region I showed that the fusion protein reacted with a pool of monoclonal antibodies (MAbs) to MSPB (MAbs 50, 97, and 334, as described previously [11]) and also with rabbit monospecific antiserum to MSPB, but not with that to MSPA (Fig. 2B, panel I, lanes 1, 3, and 2, respectively). Conversely, immunostaining of Western blots of the whole-cell proteins of the clone expressing polypeptide from region II showed that only rabbit monospecific antiserum to MSPA bound to the fusion protein (Fig. 2B, panel II, lane 2), while a pool of MAbs to MSPB (MAbs 50, 97, and 334) or rabbit monospecific antiserum to MSPB (lanes 1 and 3, respectively) did not bind to it. Thus, the coordinate expression of MSPA and MSPB can be explained by their translation from a single gene, vlhA (for variably expressed lipoprotein and hemagglutinin), with posttranslational cleavage generating the two antigenically distinct membrane proteins. Such posttranslational cleavage of an adhesion-related gene product has been suggested for M. pneumoniae (17). The cleavage site of the vlhA gene product remains to be accurately located, as several attempts in our laboratory to determine the amino-terminal sequence of MSPA have been unsuccessful. Previous studies on the expression of MSPA and MSPB in different strains of M. synoviae identified one strain (K1723) expressing a protein of 80 kDa which was immunoreactive with monospecific antisera to both MSPA and MSPB (22). Based on the findings presented here, this 80-kDa protein probably results from a failure of posttranslational cleavage of the vlhA gene product. The combined molecular mass of MSPA and MSPB as predicted from SDS-polyacrylamide gels (50 and 45 kDa, respectively) exceeds that predicted for the entire product of the vlhA gene. However, the fusion protein expressed from the recombinant plasmid containing the 5′ region of the vlhA gene had a predicted molecular mass of 43 kDa but had an apparent molecular mass of 50 kDa on SDS-PAGE. This difference, possibly due to the high proline content of the protein (23), would appear to partially account for the discrepancy between apparent and predicted molecular masses.
FIG. 2.
(A) The vlhA ORF. The region encoding the putative signal peptide (filled) and regions used to express fusion proteins in E. coli (hatched) are shown on the ORF. The scale on the top indicates amino acid residues encoded by the ORF. (B) Immunostaining of whole-cell proteins of E. coli cells expressing fusion proteins from recombinant plasmid PinPoint Xa1-T containing region I (panel I) or II (panel II) of the vlhA gene. In each panel, immunoblots were probed with a pool of MSPB-specific MAbs (lanes 1) and with rabbit monospecific antisera to MSPA (lanes 2) and MSPB (lanes 3).
The ability of the truncated MSPA, expressed in E. coli, to adhere to chicken erythrocytes was not examined in this study. However, as the full length of the protein could not be expressed in E. coli (due to the presence of TGA codons), and because more than one type of membrane component is usually involved in the process of mycoplasma adhesion (15), it is unlikely that this fragment would exhibit function as a hemagglutinin.
MSPB, a lipoprotein.
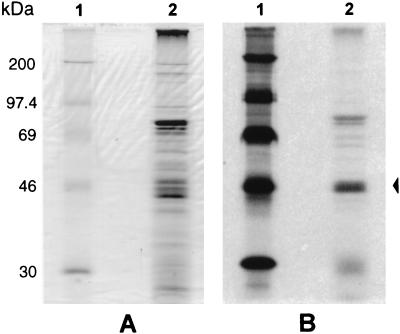
M. synoviae membrane proteins were metabolically labelled with [3H]palmitate (NEN, DuPont) by the method previously described by Bricker et al. (5) with some modifications. Briefly, M. synoviae cells grown in 20 ml of MB were harvested by centrifugation at 20,000 × g at 4°C and resuspended in 2 ml of fresh MB containing 20 μCi of [3H]palmitate per ml. The culture was incubated at 37°C for approximately 2 h and harvested by centrifugation before the pH reached 6.8. The integral membrane proteins of the radiolabelled culture were purified by Triton X-114 (TX-114) fractionation (22) and separated by SDS-PAGE, and the resultant gel was subjected to autoradiography. SDS-PAGE and autoradiographic analysis of [3H]palmitate-radiolabelled integral membrane proteins of M. synoviae (Fig. 3 A and B, respectively) showed that a strong band of approximately 40 to 45 kDa (similar in molecular mass to MSPB), a diffuse band of 25 to 30 kDa, and two bands of high molecular mass are bound by lipid moieties.
FIG. 3.
SDS-PAGE analysis of [3H]palmitate-labelled M. synoviae integral membrane proteins. (A) Lane 1, whole-cell proteins of M. synoviae were fractionated by TX-114 phase partitioning, and proteins in the detergent phase were separated by SDS-PAGE and stained with Coomassie brilliant blue. Lane 2, radiolabelled protein molecular mass markers. (B) The gel was autoradiographed as described previously (5). The arrow to the right indicates a lipoprotein similar in molecular mass to MSPB.
Our previous studies, using reversed-phase high-pressure liquid chromatography to separate the M. synoviae proteins in the TX-114 phase and immunoblotting to further characterize the relationships between these proteins, have established that the proteins in this fraction with molecular masses of 45 to 50 kDa were variants of either MSPA or MSPB (22). Those studies also demonstrated that a more diffuse band of proteins of 25 to 30 kDa (MSPC) in the TX-114 phase were antigenically related to MSPB. These findings, examined in the light of the [3H]palmitate-labelling experiments, and the consensus signal peptidase II cleavage site reported for the predicted amino acid sequence of the vlhA gene indicate that MSPB is a lipoprotein.
Previous studies have shown that both MSPA and MSPB are membrane-associated proteins (11, 22). Also, the findings of the current study suggest that MSPB may be anchored in the membrane by a covalently bound lipid moiety. Membrane localization of the carboxyl-terminal protein MSPA appears likely to be mediated by the carboxyl-terminal hydrophobic region of the predicted protein. The possibility of any covalent linkage between MSPA and MSPB was excluded by the absence of any Cys residue in the predicted sequence other than that found at the signal peptidase II cleavage site.
Identity of vlhA with pMGA1.7, a gene from the phylogenically distant species M. gallisepticum.
Comparison of the nucleotide and predicted amino acid sequences of vlhA with sequences in the databases revealed a high level of both nucleotide and amino acid sequence identity with two M. synoviae putative pcl42-56 membrane protein gene sequences, MS2/12 and MS2/28 (accession no. MSU66314 and MSU66315), and an M. gallisepticum S6 hemagglutinin precursor, pMGA1.7 (accession no. MGU90714). Comparison of nucleotide sequences by using the Genetics Computer Group program GAP found identities of 84.6% with MS2/12, 73.2% with pMGA1.7, and 67.5% with MS2/28. Similar comparisons with predicted amino acid sequences revealed an identity of 74.9% in 410 overlapping amino acids with MS2/12 (results not shown), 63.1% in 745 overlapping amino acids with pMGA1.7 (Fig. 4), 58.4% in 495 overlapping amino acids with ORF G2149017 of MS2/28, and 54.1% in 244 overlapping amino acids with ORF G2149016 of MS2/28 (results not shown).
FIG. 4.
Comparison of the amino acid sequence predicted from vlhA with that for the pMGA1.7 gene from M. gallisepticum. Numbers to the right of the sequences refer to the positions of adjacent residues relative to the first encoded amino acid. Vertical lines indicate identical residues, while colons (:) indicate conservative amino acid substitutions.
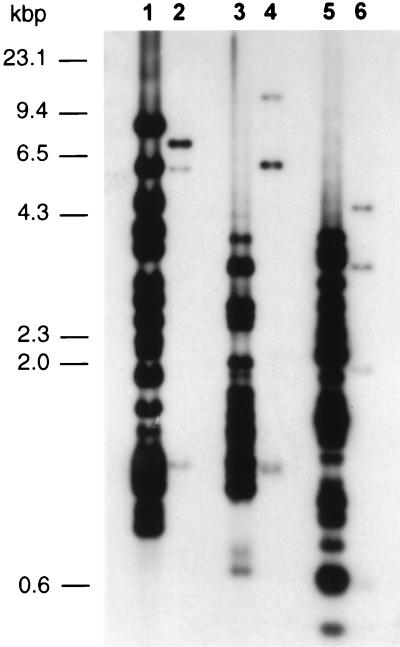
To confirm the similarity between the vlhA and pMGA1.7 genes, M. synoviae and M. gallisepticum genomic DNAs were digested with three separate restriction endonucleases, EcoRI, BglII, and HindIII, and the resultant fragments were separated in an agarose gel, Southern transferred to a nylon membrane, and probed with the radiolabelled vlhA gene. Autoradiography of the blot washed at low stringency (Fig. 5) showed that in each digestion, 3 fragments of M. gallisepticum DNA (lanes 2, 4, and 6) and about 20 fragments of M. synoviae DNA (lanes 1, 3, and 5) hybridized to the vlhA gene probe. High-stringency washing removed the vlhA gene probe bound to M. gallisepticum DNA, while at least 10 bands of M. synoviae DNA in each digestion remained hybridized to the probe, of which three BglII fragments (4, 1.2, and 1.1 kb), two EcoRI fragments (2.7 and 1.1 kb), and two HindIII fragments (2.1 and 1.5 kb) had the greatest intensity (results not shown).
FIG. 5.
Southern blot of genomic DNAs from M. synoviae WVU (lanes 1, 3, and 5) and M. gallisepticum S6 DNA (lanes 2, 4, and 6) digested with restriction enzymes BglII (lanes 1 and 2), EcoRI (lanes 3 and 4), and HindIII (lanes 5 and 6) and probed with the 32P-labelled vlhA gene. The blot was washed under low-stringency conditions and autoradiographed. Three fragments of M. gallisepticum digested with each restriction endonuclease (BglII, 7.4, 5.8, and 1.2 kb; EcoRI, 11.2, 6, and 1.1 kb; and HindIII, 4.5, 3.2, and 1.5 kb) hybridized to the vlhA gene probe. Numbers on the left indicate the sizes of the nucleic acid molecular size markers (HindIII-digested λ phage).
Hybridization of the vlhA gene probe to Southern blots of M. synoviae and M. gallisepticum genomic DNAs established that the vlhA gene of M. synoviae had some similarity to several regions of the M. gallisepticum genome but had highest identity with numerous regions of the M. synoviae genome. These data, along with the fractionation of MSPBs or MSPAs with different hydrophobicities by high-pressure liquid chromatography (22), suggest that the vlhA gene is a member of a gene family in M. synoviae. These results also explain the sequence differences observed between two apparently overlapping EcoRI and BglII fragments of M. synoviae DNA, as they were probably derived from two closely related members of the vlhA gene family.
Sequence data obtained from the vlhA gene revealed high levels of identity with a member of the pMGA family of M. gallisepticum. This gene family, the largest known translated gene family in procaryotes, occupies 5 to 10% of the M. gallisepticum genome and contains up to 70 members (1). It is particularly notable that this is the first identification of homologous multigene families encoding lipoproteins in phylogenetically distinct mycoplasmas. Although M. synoviae and M. gallisepticum share the same host species, M. gallisepticum lies within the M. pneumoniae phyletic group, while M. synoviae is within the M. hominis group. As there are no detectable homologs of these gene families in either M. pneumoniae or M. genitalium, for which genomic sequences have been fully determined (10, 13), these observations suggest that one or both families have arisen by relatively recent horizontal transfer, possibly as a result of a shared habitat. Intraspecies gene transfer, by an unknown mechanism, has already been demonstrated for Spiroplasma and Acholeplasma spp. (9). Also, a number of in vitro studies have used gram-positive bacteria as donors to transfer plasmids carrying antibiotic resistance genes into mycoplasmas (9, 24, 27); however, intraspecies transfer of a gene family has not been described for mycoplasmas to date.
Further studies will be necessary to examine whether multiple members of the family could have been transferred or whether there has been expansion of the family after transfer of a single member. It is remarkable that a mechanism which appears to have evolved to facilitate evasion of the immune response in one species should be used by a second species in the same host.
Nucleotide sequence accession number.
The vlhA sequence has been submitted to GenBank under accession no. AF035624.
REFERENCES
- 1.Baseggio N, Glew M D, Markham P F, Whithear K G, Browning G F. Size and genomic location of the pMGA multigene family of Mycoplasma gallisepticum. Microbiology. 1996;142:1429–1435. doi: 10.1099/13500872-142-6-1429. [DOI] [PubMed] [Google Scholar]
- 2.Baseman J B, Reddy S P, Dallo S F. Interplay between mycoplasma surface proteins, airway cells, and the protean manifestations of mycoplasma-mediated human infections. Am J Respir Crit Care Med. 1996;154:S137–S144. doi: 10.1164/ajrccm/154.4_Pt_2.S137. [DOI] [PubMed] [Google Scholar]
- 3.Behrens A, Heller M, Kirchhoff H, Yogev D, Rosengarten R. A family of phase- and size-variant membrane surface lipoprotein antigens (Vsps) of Mycoplasma bovis. Infect Immun. 1994;62:5075–5084. doi: 10.1128/iai.62.11.5075-5084.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhugra B, Voelker L L, Zou N X, Yu H L, Dybvig K. Mechanism of antigenic variation in Mycoplasma pulmonis—interwoven, site-specific DNA inversions. Mol Microbiol. 1995;18:703–714. doi: 10.1111/j.1365-2958.1995.mmi_18040703.x. [DOI] [PubMed] [Google Scholar]
- 5.Bricker T M, Boyer M J, Keith J, Watson M R, Wise K S. Association of lipids with integral membrane surface proteins of Mycoplasma hyorhinis. Infect Immun. 1988;56:295–301. doi: 10.1128/iai.56.2.295-301.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dallo S F, Chavoya A, Baseman J B. Characterization of the gene for a 30-kilodalton adhesion-related protein of Mycoplasma pneumoniae. Infect Immun. 1990;58:4163–4165. doi: 10.1128/iai.58.12.4163-4165.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dallo S F, Lazzell A L, Chavoya A, Reddy S P, Baseman J B. Biofunctional domains of the Mycoplasma pneumoniae P30 adhesin. Infect Immun. 1996;64:2595–2601. doi: 10.1128/iai.64.7.2595-2601.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duffy M F, Walker I D, Browning G F. The immunoreactive 116 kDa surface protein of Mycoplasma pneumoniae is encoded in an operon. Microbiology. 1997;143:3391–3402. doi: 10.1099/00221287-143-10-3391. [DOI] [PubMed] [Google Scholar]
- 9.Dybvig K, Voelker L L. Molecular biology of mycoplasmas. Annu Rev Microbiol. 1996;50:25–57. doi: 10.1146/annurev.micro.50.1.25. [DOI] [PubMed] [Google Scholar]
- 10.Fraser C M, Gocayne J D, White O, Adams M D, Clayton R A, Fleischmann R D, Bult C J, Kerlavage A R, Sutton G, Kelley J M, Fritchman J L, Weidman J F, Small K V, Sandusky M, Fuhrmann J, Nguyen D, Utterback T R, Saudek D M, Phillips C A, Merrick J M, Tomb J, Dougherty B A, Bott K F, Hu P, Lucier T S, Peterson S N, Smith H O, Hutchinson III C A, Venter J C. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 11.Gurevich V A, Ley D H, Markham J F, Whithear K G, Walker I D. Identification of Mycoplasma synoviae immunogenic surface proteins and their potential use as antigens in the enzyme-linked immunosorbent assay. Avian Dis. 1995;39:465–574. [PubMed] [Google Scholar]
- 12.Herrmann R. Genome structure and organization. In: Maniloff J, editor. Mycoplasmas: molecular biology and pathogenesis. Washington, D.C: American Society for Microbiology; 1992. pp. 157–169. [Google Scholar]
- 13.Himmelreich R, Hilbert H, Plagens H, Pirkl E, Li B C, Herrmann R. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 1996;24:4420–4449. doi: 10.1093/nar/24.22.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jordan F T W. Avian mycoplasmosis. In: Jordan F T W, Pattison M, editors. Poultry diseases. W. B. London, United Kingdom: Saunders; 1996. pp. 81–93. [Google Scholar]
- 15.Kahane I, Horowitz S. Adherence of mycoplasma to cell surfaces. In: Rottem S, Kahane I, editors. Mycoplasma cell membranes. New York, N.Y: Plenum; 1993. pp. 255–243. [DOI] [PubMed] [Google Scholar]
- 16.Kyte H, Doolittle R F. A simple method for displaying the hydrophobic character of a protein. J Mol Biol. 1982;156:2906–2913. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 17.Layhschmitt G, Herrmann R. Localization and biochemical characterization of the ORF6 gene product of the Mycoplasma pneumoniae P1 operon. Infect Immun. 1992;60:2906–2913. doi: 10.1128/iai.60.7.2906-2913.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Layhschmitt G, Himmelreich R, Leibfried U. The adhesin related 30-kDa protein of Mycoplasma pneumoniae exhibits size and antigen variability. FEMS Microbiol Lett. 1997;152:101–108. doi: 10.1111/j.1574-6968.1997.tb10415.x. [DOI] [PubMed] [Google Scholar]
- 19.Markham P F, Glew M, Brandon M R, Walker I D, Whithear K G. Characterization of a major hemagglutinin protein from Mycoplasma gallisepticum. Infect Immun. 1992;60:3885–3891. doi: 10.1128/iai.60.9.3885-3891.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Markham P F, Glew M D, Whithear K G, Walker I D. Molecular cloning of a member of the gene family that encodes pMGA, a hemagglutinin of Mycoplasma gallisepticum. Infect Immun. 1993;61:903–909. doi: 10.1128/iai.61.3.903-909.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morrow C J, Whithear K G, Kleven S H. Restriction endonuclease analysis of Mycoplasma synoviae strains. Avian Dis. 1990;34:611–616. [PubMed] [Google Scholar]
- 22.Noormohammadi A H, Markham P F, Whithear K G, Walker I D, Ley D H, Browning G F. Mycoplasma synoviae has two distinct phase-variable major membrane antigens, one of which is a putative hemagglutinin. Infect Immun. 1997;65:2542–2547. doi: 10.1128/iai.65.7.2542-2547.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogle K F, Lee K K, Krause D C. Nucleotide sequence analysis reveals novel features of the phase-variable cytadherence accessory protein HMW3 of Mycoplasma pneumoniae. Infect Immun. 1992;60:1633–1641. doi: 10.1128/iai.60.4.1633-1641.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roberts M C, Kenny G E. Conjugal transfer of transposon Tn916 from Streptococcus faecalis to Mycoplasma hominis. J Bacteriol. 1987;169:3836–3839. doi: 10.1128/jb.169.8.3836-3839.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 26.Su C J, Tryon V V, Baseman J B. Cloning and sequence analysis of cytadhesin P1 gene from Mycoplasma pneumoniae. Infect Immun. 1987;55:3023–3029. doi: 10.1128/iai.55.12.3023-3029.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Voelker L L, Dybvig K. Gene transfer in Mycoplasma arthritidis: transformation, conjugal transfer of Tn916, and evidence for a restriction system recognizing AGCT. J Bacteriol. 1996;178:6078–6081. doi: 10.1128/jb.178.20.6078-6081.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whithear K G. Avian mycoplasmosis. In: Corner L A, Bagust T J, editors. Australian standard diagnostic techniques for animal diseases. East Melbourne, Australia: CSIRO for the Standing Commitee on Agriculture and Resource Management; 1993. pp. 1–12. [Google Scholar]
- 29.Wise K S. Adaptive surface variation in mycoplasmas. Trends Microbiol. 1993;1:59–63. doi: 10.1016/0966-842X(93)90034-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Q J, Wise K S. Molecular basis of size and antigenic variation of a Mycoplasma hominis adhesin encoded by divergent vaa genes. Infect Immun. 1996;64:2737–2744. doi: 10.1128/iai.64.7.2737-2744.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]