Abstract
Pre-mRNA splicing has to be coordinated with other processes occurring in the nucleus including transcription, mRNA 3′ end formation and mRNA export. To analyze the relationship between transcription and splicing, we constructed a network of nested introns. Introns were inserted in the 5′ splice site and/or branchpoint of a synthetic yeast intron interrupting a reporter gene. The inserted introns mask the recipient intron from the cellular machinery until they are removed by splicing. Production of functional mRNA from these constructs therefore requires recognition of a spliced RNA as a splicing substrate. We show that recurrent splicing occurs in a sequential and ordered fashion in vivo. Thus, in Saccharomyces cerevisiae, intron recognition and pre-spliceosome assembly is not tightly coupled to transcription.
INTRODUCTION
While nuclear pre-mRNA splicing is understood at the biochemical level, our knowledge of how this process is integrated with other RNA metabolism events in eukaryotic cell nuclei remains limited. Pre-mRNA splicing has indeed to be coordinated with other processes such as transcription.
Co-localization experiments indicate that, in mammalian nuclei, splicing occurs close to transcription sites (Sleeman and Lamond, 1999). Several studies indicated that transcription and pre-mRNA splicing can occur concomitantly, by showing that pre-mRNA splicing and spliceosome assembly can take place while the downstream part of the transcript is still being transcribed (LeMaire and Thummel, 1990; Baurén and Wieslander, 1994; Tennyson et al., 1995). However, not all splicing events are co-transcriptional and, for mammalian polycistronic genes, the order of intron removal depends on intron-specific efficiency of excision, cross-talk between introns and on the proximity to the cap or poly(A) tail structures (Lewis et al., 1996). The small nuclear size and the paucity of the introns have limited similar studies in Saccharomyces cerevisiae (Elliott et al., 1992; Lopez and Séraphin, 1999; Spingola et al., 1999). Moreover, characterization of one of the few yeast pre-mRNA containing two introns did not show a well-defined intron excision order (Miller, 1984; Howe and Ares, 1997). Nevertheless, cotranscriptional splicing probably occurs, at least in a sub-population of yeast pre-mRNAs (Elliott and Rosbash, 1996). The observation that engineered RNAs synthesized in eukaryotic cells by pol I, pol III or T7 polymerases are often defective in some steps of mRNA processing (capping, polyadenylation and/or splicing) suggested a requirement for pol II for efficient and accurate mRNA processing (Kohrer et al., 1990; Gunnery and Mathews, 1995; McCracken et al., 1997; Lo et al., 1998). A model involving the C-terminal domain (CTD) of the large subunit of pol II as a landing pad for proteins involved in RNA maturation was proposed to explain the role of pol II in pre-mRNA processing (Greenleaf, 1993; Neugebauer and Roth, 1997). Consistent with this model, the CTD of the large subunit of pol II has been shown to associate with proteins implicated in pre-mRNA processing (for review see Bentley, 1999; Hirose and Manley, 2000) and affected pre-mRNA splicing in various assays (e.g. Du and Warren, 1997; McCracken et al., 1997; Hirose et al., 1999). However, both mRNA capping and cleavage/polyadenylation are also CTD dependent (reviewed in Minvielle-Sebastia and Keller, 1999) and also influence pre-mRNA splicing (Talerico and Berget, 1990; Izaurralde et al., 1994). Therefore, it still remains unclear whether the requirement of the pol II CTD for pre-mRNA splicing is direct or indirect through the involvement of both pre-mRNA ends in the splicing process, and whether this link between transcription and splicing extends to yeast.
To address the coordination between transcription and splicing in S. cerevisiae, we constructed a network of nested introns. Introns were inserted in the 5′ splice site and/or in the branchpoint of a synthetic intron. Splicing of the newly inserted intron(s) regenerates the recognition site(s) of the synthetic intron. We show that, in vivo, the splicing machinery still efficiently recognizes pre-mRNA segments that have already undergone splicing, indicating that splicing does not need to be tightly coupled to transcription.
RESULTS
Construction of a nested-intron network
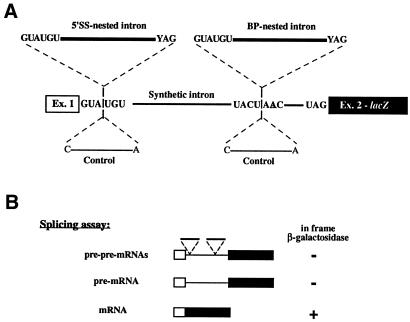
In the present study, an efficiently spliced pre-mRNA reporter harboring an artificial intron (Luukkonen and Séraphin, 1999) was modified to remove putative cryptic splice sites. Then, the yeast RPS16B intron was inserted in the 5′ splice site (5′SS), in the branchpoint sequence (BP) or combined insertions in 5′SS and BP of the synthetic intron (Figure 1A). The RPS16B intron was chosen for this analysis because it contains consensus recognition sequences and a polypyrimidine tract. In all constructs, RPS16B intron insertion interrupted the original splicing signals in such a way that they were no longer recognizable. The precursor RNAs encoded by the single insertion constructs were named pre-pre-mRNA as their splicing would form a pre-mRNA while the precursor encoded by the double insertion construct was named pre2-pre-mRNA to indicate that two independent splicing events were required to form the pre-mRNA. As a control, we inserted at the same locations a non-spliceable 15 mer sequence that maintains the pre-mRNA frame (Figure 1A). In our constructs the mRNA was in-frame while the pre-mRNA was out-of-frame; these allow the measure of the splicing efficiency of the synthetic intron (Figure 1B). All pre-pre-mRNAs and pre2-pre-mRNAs containing the RPS16B insertion do not code for β-galactosidase because of the presence of premature stop codons.
Fig. 1. Design of reporter constructs and assays. (A) Schematic representation of the nested-introns networks. A synthetic intron was inserted within the coding sequence of the lacZ gene. The sequences of the 5′SS, branchpoint, and 3′SS are indicated (middle line, note that they correspond to the consensus). The sites of insertion of nested introns (upper line) or control insertions (lower line) interrupting the 5′SS and the branchpoint sequences are indicated. (B) Reporter constructs used to test recursive splicing. Only the fully spliced mRNA species leads to synthesis of β-galactosidase (+) the pre-pre-mRNAs and pre-mRNAs contain in-frame stop codons (–).
Efficient recursive splicing
These various constructs were introduced into yeast cells and pre-mRNA splicing was assessed by measuring β-galactosidase activity after 2 h of induction of the reporter promoter. The synthetic intron was efficiently spliced since it gave one fourth of the activity obtained with the very efficiently natural RP51A intron (data not shown). In contrast, insertion of the control oligonucleotide into the 5′SS and/or BP abolished splicing, reducing β-galactosidase activity to background levels (Table I). These results indicate that insertion into either the 5′SS or the BP sequence masks the synthetic intron from the splicing machinery. The observation that control insertion in the 5′SS and/or BP abolishes pre-mRNA splicing, and re-directs the corresponding RNA to the cytoplasm (data not shown), confirms that intron recognition depends on the presence of these intact sequences.
Table I. Assaying recursive splicing by monitoring β-galactosidase production.
| Nested-RPS16B |
Nested-control |
||||||
|---|---|---|---|---|---|---|---|
| Uninduced | No | 5′SS | BP | 5′SS & BP | 5′SS | BP | 5′SS & BP |
| 0.06 | 209 | 200 | 135 | 88 | 0.28 | 0.36 | 0.42 |
| ± 0.04 | ± 3.0 | ± 16 | ± 12 | ± 8.0 | ± 0.04 | ± 0.15 | ± 0.03 |
For the different reporters the nature and the position of the insertion within the synthetic intron are indicated (5′SS, BP or 5′SS & BP). The column ‘No’ relates to the synthetic intron without insertion and the column ‘Uninduced’ to the same reporter but from a culture that had not been induced. The values correspond to an average of three to seven independent determinations done in duplicates. Standard deviations are indicated as a second row.
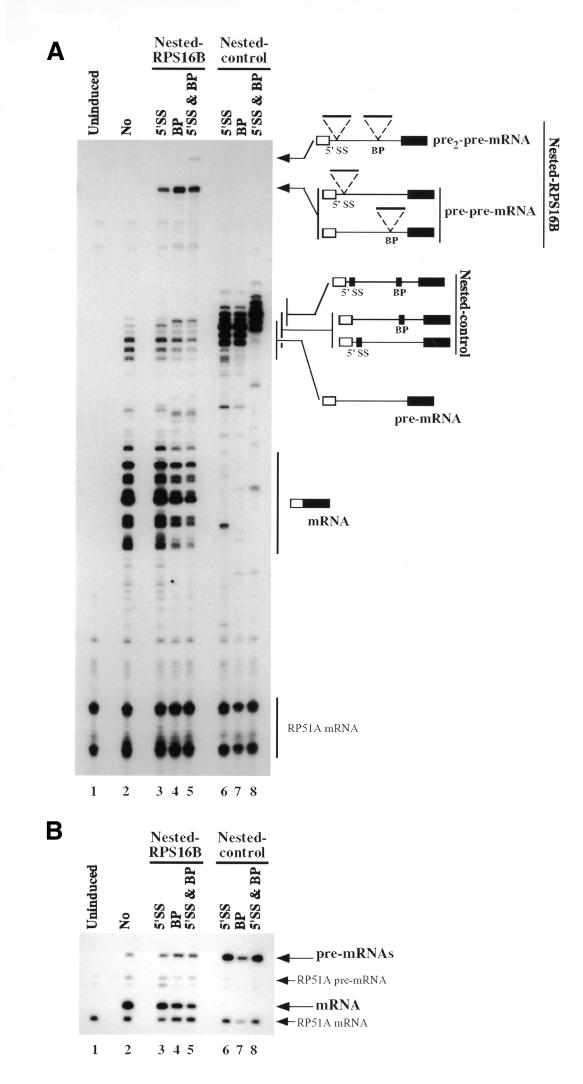
We then tested whether splicing of the nested RPS16B intron(s) would generate a pre-mRNA that would be efficiently spliced, i.e. would recursive splicing occur? As the splicing signals of the pre-mRNA are masked during transcription, loading of splicing factors on these sequences could consequently not have been tightly coupled to their synthesis. The β-galactosidase levels produced by mRNAs derived from the nested constructs were slightly lower than those observed for the synthetic intron alone (Table I; see also below). This result suggested that recursive splicing occurred. To confirm that β-galactosidase production resulted from recursive splicing rather than aberrant RNA processing events, total RNA was extracted and used in a primer extension analysis with an exon 2-specific oligonucleotide. Two groups of multiple bands corresponding to the pre-mRNA and mRNA were detected for the synthetic intron construct (Figure 2A, lane 2). These multiple bands result from the heterogeneous transcription start sites of the reporter promoter used (Guarente et al., 1982). Signals corresponding to the endogenous RP51A mRNA served as internal controls. These were the only signals detected in RNA extracted from uninduced cultures (Figure 2A, lane 1). Reverse-transcription products, slightly larger than the pre-mRNA, were detected in RNA extracted from cells expressing the control constructs with 15 mer insertions (Figure 2A, lanes 6–8). The size of these products correlated with the sizes of the sequence insertions confirming that they represent unspliced RNA. Consistent with the β-galactosidase analysis, no mRNA species was apparent (Figure 2A, lanes 6–8). Analysis of the construct containing the inserted RPS16B intron revealed the expected pre2-pre-mRNA precursors for the construct carrying insertion at both the 5′SS and the BP and pre-pre-mRNA for constructs carrying insertions at either the 5′SS or the BP (Figure 2A, lanes 3–5). In addition all spliced products derived from these precursors were detected, including a significant amount of mRNA (Figure 2A, lanes 3–5). Consistent with the β-galactosidase assay (Table I), reduced levels of mRNA were detected for the recursively spliced RNA. Measurement of the ratio of mRNA to pre-mRNA (the most sensitive indicator of splicing efficiency; Pikielny and Rosbash, 1985) shows that the splicing efficiency of the synthetic intron varies within 2-fold between the different constructs (data not shown).
Fig. 2. Recursive splicing. (A) Total RNA was extracted from a wild-type strain harboring the different reporters and splicing was analyzed by primer extension using an exon 2 primer. Extension products were resolved on a 6% acrylamide-denaturing gel. For each species the multiple bands correspond to the multiple initiation sites characteristic of the GAL–CYC1 promoter used (Guarente et al., 1982). 5′SS, BP or 5′SS & BP indicates the position of the insertions. The uninduced control corresponds to RNA derived from the construct without insertion (No) extracted before induction of the GAL–CYC1-promoter. The different extension products are schematically depicted on the right of the figure. Signals derived from the endogenous RP51A RNA (bottom) are used as internal control. (B) Primer extension was performed with the same RNAs used in (A) but in the presence of ddG. Extension products were analyzed on 8% acrylamide-denaturing gel. Extension products corresponding to fully spliced mRNA and the various pre-mRNAs (pre-mRNA, pre-pre-mRNA, pre2-pre-mRNA) are schematically depicted on the right of the figure together with signals derived from the endogenous RP51A species.
Due to the multiple transcriptions start sites, it was not possible from this primer extension analysis to demonstrate that recursive splicing produced accurately processed mRNAs. To rule out aberrant events or low level of spliced RNA, we repeated the primer extension analysis using the oligonucleotide complementary to exon 2 in the presence of dideoxyguanosine (see Methods). In these conditions a single band corresponds to the various pre-mRNAs and a second band to the mRNA (Figure 2B, lanes 2–5). The sizes of these products differ from those resulting from extension of the endogenous RP51A precursor (Figure 2B, lane 1). This experiment confirmed that insertion of the control sequence prevented splicing and that the mRNAs produced through recursive splicing events were accurately processed. They also corroborated the quantitative results described above even though the level of individual pre-mRNA species could not be determined.
Overall, these experiments demonstrated that recursive pre-mRNA splicing occurred efficiently and accurately.
Recursive splicing occurs with various inserted introns
To confirm that the results reported above for the RPS16B intron could be a general property, we repeated these experiments using a different intron. We chose the KIN28 intron for this analysis because it is much smaller than the RPS16B intron (81 instead of 432 nucleotides). To avoid analysis problems associated with the presence of multiple transcription initiation sites, RNA products were assayed using a quantitative RT–PCR assay (see Methods). Analysis of the products of this reaction indicated that recursive splicing occurred with similar efficiency for all constructs (Figure 3A) even though some RNA products resulting from alternative splicing events were also visible (asterisk in Figure 3A). Those were products from alternative 5′SS or BP selection and accumulated mainly when the KIN28 intron was present (P.J. Lopez and B. Séraphin, unpublished). Their presence did not affect the main conclusion, i.e. that recursive splicing is not specific for the RPS16B insertion. Interestingly, the size difference between the KIN28 and RPS16B introns allowed us to determine whether introns were removed in a preferential order when they were present in both the 5′SS and the BP (Figure 3A, lanes 2 and 3). This demonstrated that the 5′ proximal intron was quantitatively spliced first. A similar conclusion was obtained by analyzing RNA from the double RPS16B insertion construct by RT–PCR followed by restriction digestion (data not shown). This is likely to result from the similar splicing efficiencies of the inserted introns: RPS16B and/or KIN28. Indeed, with other combinations of introns inserted in the 5′SS and in the BP, we observed that the BP-nested intron could be spliced prior to the 5′SS-nested one (e.g. with the combinations RPS16B or MRPL44 introns in the 5′ SSs and ACT1 intron in the BP; Figure 3B). Because of the lower splicing efficiency of the MRPL44 intron, higher levels of pre-pre-mRNA precursors harboring this intron in the 5′SS accumulated with this construct (compare lane 5, which shows two kinds of pre-pre-mRNAs, with lane 4, where only one accumulated). Nevertheless, recursive splicing still occurred efficiently (Figure 3B). This demonstrates that recursive splicing is not dependent upon a defined order of removal of the inserted introns that would mimic the emergence of the recipient intron from pol II. Overall, these results demonstrate that recursive splicing is an efficient and versatile process.
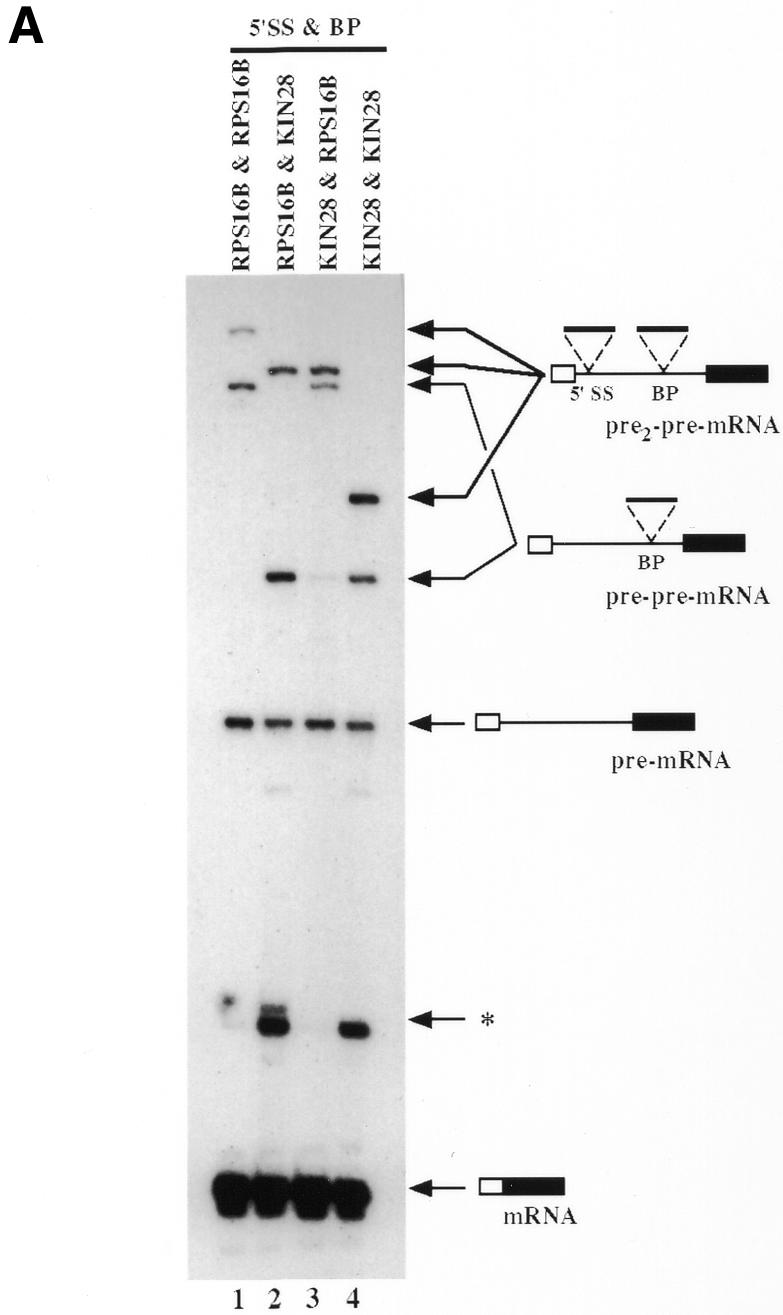
Fig. 3. Ordered splicing events. (A) RT–PCR analyses of constructs carrying all combinations of insertion of the KIN28 and RPS16B nested introns using exon 1 and exon 2 primers. PCR products were analyzed on 8% denaturing gel. The structure of the different PCR products is schematically depicted on the right of the figure. The asterisk corresponds to alternative splicing RNA species. For the different constructs, the intron name and position of insertion are indicated at the top of each lane. β-galactosidase activities measured, from four different experiments, for the constructs in lanes 1, 2, 3 and 4 were 88 ± 8.0, 120 ± 15, 95 ± 12 and 108 ± 8.1, respectively. (B) Primer extension analysis of two different 5′SS-nested intron combinations with the ACT1 BP-nested intron. The structure of the different extension products is schematically depicted on the sides of the figure. For the different constructs, the intron name and position of insertion are indicated at the top of each lane. β-galactosidase activities measured, from two different experiments, for the constructs in lanes 1, 2, 3, 4 and 5 were 154 ± 28, 117 ± 45, 183 ± 14, 129 ± 11 and 139 ± 8.0, respectively.
DISCUSSION
To address the coordination between transcription and splicing, we have constructed a network of nested introns by inserting genuine S. cerevisiae introns in the 5′SS and/or the BP of a synthetic intron and used these constructs to test for the ability of yeast recursively to recognize and splice pre-mRNAs. We show that the synthetic intron is efficiently spliced after removal of the newly inserted intron(s). Control insertion analysis confirms that the split splice sites of the synthetic intron are masked from the splicing machinery until splicing reveals them. To our knowledge, this situation has not been reported for natural spliceosomal introns. However, related situations have been observed. In Euglena chloroplasts an essential domain of a group II intron interrupting the cytochrome b559 coding sequence is itself interrupted by another group II intron that needs to be removed to allow splicing of the recipient intron (Copertino and Hallick, 1991). In Didymium iridis a spliceosomal intron has been found inserted in a group I intron, but the recipient group I intron was active in the presence of the insertion (Vader et al., 1999). A resplicing process has recently been described for the formation of ultrabithorax isoforms in Drosophila (Hatton et al., 1998). In this case the first splicing event brings consensus nucleotides in front of the downstream exon that contains a splice donor site at its 5′ end, thereby generating a perfect 5′SS. This 5′SS is then sometimes used in subsequent alternative splicing events. In the Drosophila system, before the first splicing event occurs the re-spliced 5′SS is only weak rather than masked from the splicing machinery. Furthermore, recognition of the BP and 3′SS region of the intron is still likely to occur co-transcriptionally and commit this sequence to splicing, even though the final step of spliceosome assembly would only occur after generation of an optimal 5′SS. This situation differs significantly from our yeast reporter, since in our case the two sequences that are critical for intron recognition, namely the 5′SS and BP of the recipient intron, are unrecognizable before splicing of the nested introns. Nevertheless, our results demonstrate that a network of interlocked introns can lead to efficient recursive splicing in S. cerevisiae. Even if this situation has not been described so far, it remains possible that it occurs in natural introns especially in organisms with large genomes and intricate alternative splicing patterns.
Our analysis of constructs containing two different introns inserted in the 5′SS and BP of the recipient intron revealed ordered splicing of the two nested introns. When two identical introns were present, the one located in the 5′SS was spliced first. While ordered splicing may have been interpreted as preferential loading of splicing factors on the first intron by the transcribing pol II, it may reflect the kinetic of availability of the two introns to the splicing machinery and/or the proximity to the cap structure, which is known to affect splicing (Izaurralde et al., 1994; Lewis et al., 1996). As the great majority of the yeast split genes contain a single intron, the order of intron removal had not been analyzed in detail in yeast (Miller, 1984; Howe and Ares, 1997). Overall, our results indicate that the order of intron removal is, like in metazoan systems, affected by the intron-specific splicing efficiency and its location relative to other pre-mRNA features in S. cerevisiae. This is consistent with the fact that two spliceosomes can assemble and process independently two neighboring introns (Christofori et al., 1987). In the future, it will be interesting to test whether the resplicing process is performed by the same or different spliceosomes.
We have used recursive splicing to probe the organization of the in vivo splicing pathway in yeast. Our demonstration that recursive splicing occurs efficiently even though the splice sites of the recipient intron were masked to the transcription machinery indicates that loading of splicing factors onto splice sites is not necessarily linked to their synthesis in vivo. Indeed, intron recognition occurs nearly as efficiently when the pre-mRNA is synthesized by transcription or when it is the product of the splicing of a pre-pre-mRNA. Moreover, we checked whether the highest number of CTD heptapetide deletions still allowing growth (Nonet and Young, 1989) would affect recursive splicing. We observed that this process is not sensitive to partial truncation of the CTD of pol II (our unpublished results). This strengthens the conclusion that the period window during which the pre-mRNA is available for inspection by the intron recognition machinery in the nucleus is not highly restricted after transcription (see Daneholt, 1999). Furthermore, efficient recursive splicing reveals that the RNA is not efficiently targeted to RNA export or localized in a different compartment incompetent for intron recognition after the splicing reaction. Overall, our results suggest that intron recognition can efficiently occur uncoupled from transcription in yeast. However, this conclusion might not always apply in mammalian systems. Indeed, in this system some SR-related proteins appear to associate with the pol II CTD and/or transcription factors thereby affecting alternative splicing (Cramer et al., 1999; Lai et al., 1999).
METHODS
Yeast strains and plasmids. The wild-type yeast stain MGD453-13D was used in this work (Séraphin et al., 1988). The starting plasmid, pBS983, has been described (Luukkonen and Séraphin, 1999) and contains a synthetic intron inserted upstream of the lacZ coding sequence. This sequence was designed to include unique restriction sites in the synthetic intron and flanking exons. For the control insertion, the 15 mer sequence (5′-CCACCGCGGACCTGA-3′) was introduced by replacing small DNA fragments at the 5′SS and/or BP by appropriate synthetic DNA fragments. For the intron insertions, fragments encompassing the full-length intron along with the flanking 5′SS and/or the BP sequences were obtained by PCR from genomic MGD453-13D DNA. A table describing the plasmids used in this study is available at http://www.embl-heidelberg.de/ExternalInfo/seraphin/recursive_splicing.html
Growth conditions and β-galactosidase assay. Cells were grown at 30°C in minimal medium containing 2% lactate, 2% glycerol, 0.05% glucose and 50 mM phosphate buffer pH 6.3 to an OD600 of 0.4 to 0.6. The cultures were then induced with 2% galactose for 2 h before processing cells for the determination of β-galactosidase activity and RNA analysis (Luukkonen and Séraphin, 1999).
RNA analysis. Primer extension analysis, using or not a chain termination nucleotide, was performed using the exon 2-specific EM38 oligonucleotide (5′-CACGCTTGACGGTCTTGGT-3′) as described previously (Pikielny and Rosbash, 1985). RT–PCR was performed by an initial cDNA synthesis step using primer EM38, and a subsequent amplification of the cDNA using EM38 (1:20 32P-labeled:unlabeled) and an exon 1-specific EM84 primer (5′-CACACTAAATTAATAATGACC-3′) (16 cycles of 42 s at 95°C, 1 min at 57°C and 1 min at 72°C) using AmpliTaq DNA polymerase. We checked that the signals obtained were proportional to the quantity of total RNA used for the primer extension, and of cDNA used for the PCR (not shown). Primer extensions and RT–PCRs were fractionated on denaturing polyacrylamide gels and quantified using a PhosphorImager.
Acknowledgments
ACKNOWLEDGEMENTS
We thank E. Bouveret, F. Caspary, E. Izaurralde, I. Mattaj, O. Puig, R. Ramirez-Morales, G. Rigaut and B. Rutz for careful reading of the manuscript and constructive criticisms. We gratefully acknowledge Lotti Frisen for constructive discussions and constant encouragement. P.J.L. is a recipient of an EMBO long-term fellowship. B.S. is on leave from the CNRS.
REFERENCES
- Baurén G. and Wieslander, L. (1994) Splicing of Babiani ring 1 gene pre-mRNA occurs simultaneaously with transcription. Cell, 76, 183–192. [DOI] [PubMed] [Google Scholar]
- Bentley D. (1999) Coupling RNA polymerase II transcription with pre-mRNA processing. Curr. Opin. Cell Biol., 11, 347–351. [DOI] [PubMed] [Google Scholar]
- Christofori G., Frendewey, D. and Keller, W. (1987) Two spliceosomes can form simultaneously and independently on synthetic double-intron messenger RNA precursors. EMBO J., 6, 1747–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copertino D.W. and Hallick, R.B. (1991) Group II twintron: an intron within an intron in a chloroplast cytochrome b559 gene. EMBO J., 10, 433–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer P., Caceres, J.F., Cazalla, D., Kadener, S., Muro, A.F., Baralle, F.E. and Kornblihtt, A.R. (1999) Coupling of transcription with alternative splicing: RNA pol II promoters modulate SF2/ASF and 9G8 effects on an exonic splicing enhancer. Mol. Cell, 4, 251–258. [DOI] [PubMed] [Google Scholar]
- Daneholt B. (1999) Pre-mRNP particles: From gene to nuclear pore. Curr. Biol., 9, R412–R415. [DOI] [PubMed] [Google Scholar]
- Du L. and Warren, S.L. (1997) A functional interaction between the carboxy-terminal domain of RNA polymerase II and pre-mRNA splicing. J. Cell Biol., 136, 5–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott D.J. and Rosbash, M. (1996) Yeast pre-mRNA is composed of two populations with distinct kinetic properties. Exp. Cell Res., 229, 181–188. [DOI] [PubMed] [Google Scholar]
- Elliott D.J., Bowman, D.S., Abovich, N., Fay, F.S. and Rosbash, M. (1992) A yeast splicing factor is localized in discrete subnuclear domains. EMBO J., 11, 3731–3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenleaf A.L. (1993) Positive patches and negative noodles: linking RNA processing to transcription? Trends Biochem. Sci., 18, 117–119. [DOI] [PubMed] [Google Scholar]
- Guarente L., Yocum, R.R. and Gifford, P. (1982) A GAL10-CYC1 hybrid yeast promoter identifies the GAL4 regulatory region as an upstream site. Proc. Natl Acad. Sci. USA, 79, 7410–7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunnery S. and Mathews,M.B. (1995) Functional mRNA can be generated by RNA polymerase III. Mol. Cell. Biol., 15, 3597–3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatton A.R., Subramaniam, V. and Lopez, A.J. (1998) Generation of alternative Ultrabithorax isoforms and stepwise removal of a large intron by resplicing at exon–exon junctions. Mol. Cell, 2, 787–796. [DOI] [PubMed] [Google Scholar]
- Hirose Y. and Manley, J.L. (2000) RNA polymerase II and the integration of nuclear events. Genes Dev., 14, 1415–1429. [PubMed] [Google Scholar]
- Hirose Y., Tacke, R. and Manley, J.L. (1999) Phosphorylated RNA polymerase II stimulates pre-mRNA splicing. Genes Dev., 13, 1234–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K.J. and Ares, M., Jr (1997) Intron self-complementarity enforces exon inclusion in a yeast pre-mRNA. Proc. Natl Acad. Sci. USA, 94, 12467–12472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E., Lewis, J., McGuigan, C., Jankowska, M., Darzynkiewicz, E. and Mattaj, I.W. (1994) A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell, 78, 657–668. [DOI] [PubMed] [Google Scholar]
- Kohrer K., Vogel, K. and Domdey, H. (1990) A yeast tRNA precursor containing a pre-mRNA intron is spliced via the pre-mRNA splicing mechanism. EMBO J., 9, 705–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.C., Teh, B.H. and Tarn, W.Y. (1999) A human papillomavirus E2 transcriptional activator. The interactions with cellular splicing factors and potential function in pre-mRNA processing. J. Biol. Chem., 274, 11832–11841. [DOI] [PubMed] [Google Scholar]
- LeMaire M.F. and Thummel, C.S. (1990) Splicing precedes polyadenylation during Drosophila E74A transcription. Mol. Cell. Biol., 10, 6059–6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J.D., Izaurralde, E., Jarmolowski, A., McGuigan, C. and Mattaj, I.W. (1996) A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5′ splice site. Genes Dev., 10, 1683–1698. [DOI] [PubMed] [Google Scholar]
- Lo H.J., Huang, H.K. and Donahue, T.F. (1998) RNA polymerase I-promoted HIS4 expression yields uncapped, polyadenylated mRNA that is unstable and inefficiently translated in Saccharomyces cerevisiae. Mol. Cell. Biol., 18, 665–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez P.J. and Séraphin, B. (1999) Genomic-scale quantitative analysis of yeast pre-mRNA splicing: implications for splice site recognition. RNA, 5, 1135–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luukkonen B.G. and Séraphin, B. (1999) A conditional U5 snRNA mutation affecting pre-mRNA splicing and nuclear pre-mRNA retention identifies SSD1/SRK1 as a general splicing mutant suppressor. Nucleic Acids Res., 27, 3455–3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S., Fong, N., Yankulov, K., Ballantyne, S., Pan, G., Greenblatt, J., Patterson, S.D., Wickens, M. and Bentley, D.L. (1997) The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature, 385, 357–360. [DOI] [PubMed] [Google Scholar]
- Miller A.M. (1984) The yeast MATa1 gene contains two introns. EMBO J., 3, 1061–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minvielle-Sebastia L. and Keller, W. (1999) mRNA polyadenylation and its coupling to other RNA processing reactions and to transcription. Curr. Opin. Cell Biol., 11, 352–357. [DOI] [PubMed] [Google Scholar]
- Neugebauer K.M. and Roth, M.B. (1997) Transcription units as RNA processing units. Genes Dev., 11, 3279–3285. [DOI] [PubMed] [Google Scholar]
- Nonet M.L. and Young, R.A. (1989) Intragenic and extragenic suppressors of mutations in the heptapeptide repeat domain of Saccharomyces cerevisiae RNA polymerase II. Genetics, 123, 715–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikielny C.W. and Rosbash, M. (1985) mRNA splicing efficiency in yeast and the contribution of nonconserved sequences. Cell, 41, 119–126. [DOI] [PubMed] [Google Scholar]
- Séraphin B., Kretzner, L. and Rosbash, M. (1988) A U1 snRNA:pre-mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5′ cleavage site. EMBO J., 7, 2533–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeman J.E. and Lamond, A.I. (1999) Nuclear organization of pre-mRNA splicing factors. Curr. Opin. Cell Biol., 11, 372–377. [DOI] [PubMed] [Google Scholar]
- Spingola M., Grate, L., Haussler, D. and Ares, J.M. (1999) Genome-wide bioinformatic and molecular analysis of introns in Saccharomyces cerevisiae. RNA, 5, 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talerico M. and Berget, S.M. (1990) Effect of 5′ splice site mutations on splicing of the preceding intron. Mol. Cell. Biol., 10, 6299–6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennyson C.N., Klamut, H.J. and Worton, R.G. (1995) The human dystrophin gene requires 16 hours to be transcribed and is cotranscriptionally spliced. Nature Genet., 9, 184–190. [DOI] [PubMed] [Google Scholar]
- Vader A., Nielsen, H. and Johansen, S. (1999) In vivo expression of the nucleolar group I intron-encoded I-dirI homing endonuclease involves the removal of a spliceosomal intron. EMBO J., 18, 1003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]