Abstract
The Rel/NF-κB transcription factor Relish plays a key role in the humoral immune response in Drosophila. We now find that activation of this innate immune response is preceded by rapid proteolytic cleavage of Relish into two parts. An N-terminal fragment, containing the DNA-binding Rel homology domain, translocates to the nucleus where it binds to the promoter of the Cecropin A1 gene and probably to the promoters of other antimicrobial peptide genes. The C-terminal IκB-like fragment remains in the cytoplasm. This endoproteolytic cleavage does not involve the proteasome, requires the DREDD caspase, and is different from previously described mechanisms for Rel factor activation.
INTRODUCTION
Rel/NF-κB proteins are important regulators of innate immunity in both mammals and insects (Ghosh et al., 1998). These transcriptional activators reside in the cytoplasm as homo- or heterodimers, complexed with an inhibitory IκB molecule. Following an immune challenge, IκB is first phosphorylated and then degraded, via the ubiquitin/proteasome pathway, thus releasing active NF-κB dimers. The released dimers are translocated into the nucleus where they act as transcription factors. Two Rel proteins, p50 and p52, are produced from longer precursors, p105 and p100. The precursors have long IκB-like extensions at their C-terminal ends, which are degraded in a likewise proteasome-dependent step. However, the processing of p105 and p100 is largely constitutive and it is not clear whether this involves endoproteolytic cleavage.
We recently described a new member of the Rel/NF-κB family, Relish, which is crucial for the humoral immune response in Drosophila (Dushay et al., 1996). In Relish mutants the induction of the antimicrobial defense is severely reduced and the animals are extremely sensitive to bacterial and fungal infection (Hedengren et al., 1999). Much like human p100 and p105, Relish is a compound protein, comprising an N-terminal Rel homology domain (RHD) and a C-terminal IκB-like region. Unexpectedly, we have now found that the activation of Relish proceeds through an endoproteolytic step that is strictly signal dependent and that does not involve the proteasome.
RESULTS AND DISCUSSION
The full-length Relish protein
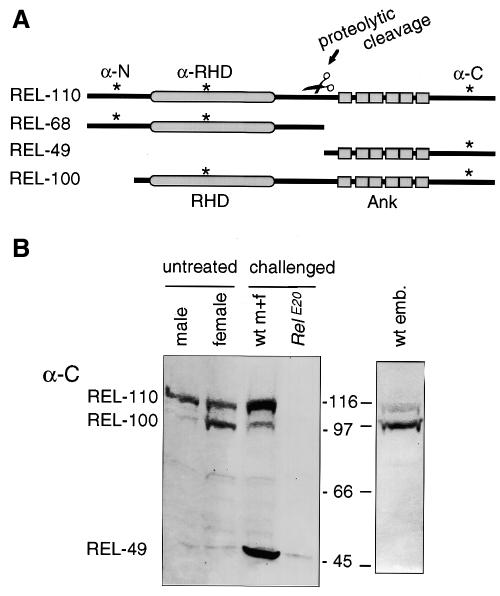
To follow the fate of different domains of the Relish protein we raised antibodies against a peptide from the Rel homology domain (α-RHD, see Figure 1A) that is not conserved between the known Rel proteins. We also made peptide antibodies against epitopes in the long N- and C-terminal extensions (α-N and α-C). These have no counterparts in other Rel proteins. Using these antibodies in immunoblotting we see that Relish appears as a major band of ∼110 kDa in male and female flies (REL-110, Figure 1B), in the Drosophila blood cell line mbn-2 (Figure 2A), and in third instar larvae (Figure 2B). This is in good agreement with the predicted molecular weight of 109 kDa for the open reading frame. The mobility of this band is identical to in vitro-translated Relish (not shown). Thus, under normal conditions Relish is mainly present as a full-length protein. Additionally, a 100 kDa band (REL-100, Figure 1B) is seen in female flies and embryos and is likely to correspond to the maternal Relish transcript (Dushay et al., 1996; Hedengren et al., 1999). This transcript encodes a protein with a shorter N-terminus, and with a calculated molecular weight of 98 kDa. A trace of REL-100 is also seen in males and it is possible that the maternal promoter is also active in some male tissues.
Fig. 1. Relish-derived proteins. (A) Map of proteins produced from the Relish gene, including the cleavage products REL-68 and REL-49. Positions of peptides used to raise specific antibodies are indicated by asterisks. (B) Western blot analysis of crude extracts from wild-type embryos, male and female flies and the Relish mutant RelE20, detected with α-C. Where indicated, flies have been injected with E. cloacae for 6 h. Molecular weight markers are given in kDa.
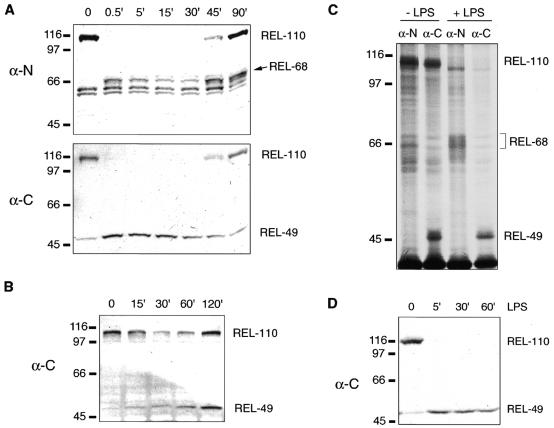
Fig. 2. Signal-induced processing of Relish. (A and B) Western blots show time courses (in minutes) of protein extracts from either induced mbn-2 cells [(A), 25 µg protein/lane] or infected wild-type larvae [(B), ∼0.5 animal equivalent/lane]. In (A) the membrane was stripped before the second detection. Antibodies used to detect different forms of Relish are indicated to the left of each membrane. (C) Pulse–chase of in vivo labeled Relish-derived proteins. [35S]methionine-labeled proteins were extracted from mbn-2 cells, either induced with LPS for 5 min or left untreated. Relish products were immunoprecipitated with the indicated rabbit antibody. The samples were separated by gel electrophoresis, the gel dried and exposed to X-ray film. (D) Immunoblot of extracts from mbn-2 cells treated with cycloheximide to inhibit protein biosynthesis prior to and during the challenge.
Figure 1B also shows that the mutant strain RelE20 is devoid of crossreacting material, confirming the specificity of the antiserum as well as the absence of gene product in this null mutant (Hedengren et al., 1999).
Signal-dependent cleavage of Relish
When the flies are challenged with Enterobacter cloacae, a 49 kDa C-terminal fragment (REL-49) appears in wild-type flies (Figure 1B). The same band is also seen in mbn-2 cells upon stimulation with lipopolysaccharide (LPS) (Figure 2A). The corresponding N-terminal part of Relish can be detected with α-N antiserum as a 68 kDa fragment (REL-68, Figure 2A). This antiserum also detects a few bands in the 60 kDa range. They are not seen with α-RHD (compare Figure 3A) and therefore are probably due to unspecific crossreactivity.
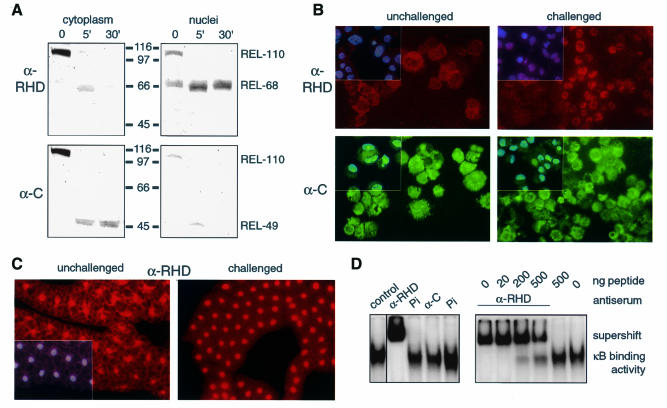
Fig. 3. Subcellular localization of Relish before and after an immune challenge. (A) Western blot with cytoplasmic and nuclear extracts from mbn-2 cells (50 µg protein/lane), developed with α-C, stripped and then developed with α-RHD. (B) Immunostaining of mbn-2 cells with α-RHD and α-C, untreated or 30 s after challenge. (C) Fatbody from wild-type larvae, untreated or taken 1 h after infection, immunostained with α-RHD. Insets in (B) and (C) show DAPI staining as overlays from the same sample area. (D) Gel shift assay for involvement of Relish in the κB binding activity. Mbn-2 cells were induced for 1 h and nuclear extracts were incubated with CecA1 κB oligonucleotide. Where indicated, an anti-Relish antiserum or the corresponding preimmune serum (Pi) was added (left panel). In a competition experiment we also added increasing amounts of the peptide, against which the serum was raised. Addition of 500 ng of peptide without antibody did not cause any effect.
The REL-68 and REL-49 fragments appear simultaneously, and at the same time the full-length REL-110 disappears completely in the mbn-2 cells (Figure 2A). This reaction is surprisingly rapid; in Figure 2A it occurs within 30 s. In other experiments we have seen complete Relish cleavage as soon as 13 s after LPS addition (not shown). The full-length protein is detectable again after a lag phase of 45 min, during which the Relish gene becomes transcriptionally upregulated (Dushay et al., 1996). The levels of REL-68 and REL-49 first decrease slowly and then increase again as more full-length Relish is formed (this increase is not obvious for REL-49 in Figure 2A, but was seen in several other experiments).
Infected larvae show kinetics of Relish cleavage and reappearance similar to the mbn-2 cells, although considerable levels of full-length REL-110 are always retained in the larvae (Figure 2B). Immunohistochemistry reveals the presence of Relish protein in most larval tissues, but not all respond to the bacterial challenge (not shown) and this may explain the remaining REL-110 in larval extracts. Conversely, low amounts of Relish cleavage products are sometimes detected in untreated animals or cells, indicating a low level of spontaneous activation of the system.
To test whether REL-68 and REL-49 are formed by cleavage from REL-110, rather than by de novo synthesis, we did a pulse–chase experiment. Figure 2C shows that radioactivity incorporated into REL-110 can be chased to REL-68, which appears as a radioactively labeled smear together with a number of unspecifically co-precipitated proteins. We could not draw any conclusions about REL-49 in this experiment, since this protein accumulated already in the unchallenged cells during the labeling period. We therefore tested whether REL-49 could be formed in the presence of cycloheximide, which blocks protein synthesis. Figure 2D shows that REL-49 is produced in the normal fashion in challenged cells, although the reappearance of the full-length protein is blocked. Thus, neither REL-68 nor REL-49 is formed by de novo synthesis, but by cleavage of Rel-110.
Nuclear translocation of REL-68
After subcellular fractionation of mbn-2 cells, most of the REL-110 protein is found in the cytoplasmic fraction (Figure 3A). However, after LPS stimulation the resulting REL-68 is localized in the nuclear fraction. In contrast, the IκB-like REL-49 is retained in the cytoplasm. The nuclear translocation of REL-68 can also be followed by immunohistochemistry. Figure 3B shows that Relish is mainly cytoplasmic in untreated mbn-2 cells. After LPS addition, the RHD-containing protein translocates to the nuclei, while the C-terminal fragment remains largely cytoplasmic. Nuclear translocation of the RHD is also evident in the fatbody (Figure 3C) and in lymph glands (not shown) of infected larvae.
REL-68 is part of the κB-binding activity
From the phenotype of Relish mutants, we know that Relish is required for the induction of antimicrobial peptide genes such as Cecropin A1 (CecA1; Hedengren et al., 1999). Here we have shown that Relish is cleaved into two parts within seconds of an immune challenge, and that the RHD-containing REL-68 fragment is translocated to the nucleus. To test whether the interaction between REL-68 and the CecA1 promoter is direct, we carried out gel shift assays on the conserved κB-like motif from the CecA1 promoter. Previous experiments demonstrated a complex between the κB oligonucleotide and an induced factor from nuclear extracts of challenged mbn-2 cells (Engström et al., 1993). Figure 3D demonstrates that α-RHD causes a complete supershift of the κB-binding complex. This effect is specific and is not observed with the preimmune serum. Similar results have been obtained with a κB-site from the Drosomycin promoter after overexpression of Relish (Han and Ip, 1999). Here we show that even at physiological levels of Relish, all protein complexes that bind to the κB site of the CecA1 gene contain the Relish REL-68 protein. No effect is seen with the α-C antibodies, indicating that neither REL-49 nor REL-110 interacts with the promoter.
The cleavage of Relish is the earliest observed consequence of a bacterial challenge, and is likely to be a key step in the activation of the humoral immune response in Drosophila. The resulting REL-68 fragment is translocated to the nucleus where it binds to the κB site in the promoter of the CecA1 gene and probably also to those of other genes that depend on Relish for their induction (Hedengren et al., 1999). That REL-68 is the active form of Relish is supported by earlier observations that overexpression of the N-terminal part of Relish is sufficient to activate transcription of the immune genes (Dushay et al., 1996; Han and Ip, 1999). Together with REL-68, other factors such as DIF may participate in the induction of CecA1 and other genes (Ip et al., 1993; Han and Ip, 1999; Rutschmann et al., 2000a). Surprisingly, Relish translocation is not inhibited by the continued presence of the IκB-like REL-49 part. The possible function of this fragment remains to be investigated.
Relish cleavage does not involve the proteasome
The rapid signal-dependent proteolysis of Relish and the persistence of both cleavage products is surprising and in strong contrast to what is known about the processing of mammalian p105. Whereas the proteasome is required for the processing of p105 (Palombella et al., 1994), we thought it unlikely that this protein complex is capable of endoproteolytic cleavage, considering current models for proteasome structure and function (Gottesman et al., 1997; Larsen and Finley, 1997). We therefore investigated the effect of well characterized proteasome inhibitors on Relish processing. Lactacystin and the peptide aldehyde MG132 have been shown to inhibit the processing of p105 to p50 and the degradation of IκBα (Palombella et al., 1994; Lin et al., 1998; Orian et al., 1999). MG132, lactacystin and calpain inhibitor I have also been used successfully to inhibit the proteasome-dependent degradation of the Timeless protein in Drosophila (Naidoo et al., 1999). Figure 4A shows that MG132 does not prevent the signal-dependent cleavage of REL-110, at concentrations that are known to block proteasome activity. The efficiency of proteasome inhibition was shown by the accumulation of multiubiquitylated proteins, as detected with a ubiquitin antibody. Similar results were obtained with lactacystin and calpain inhibitor I (not shown).
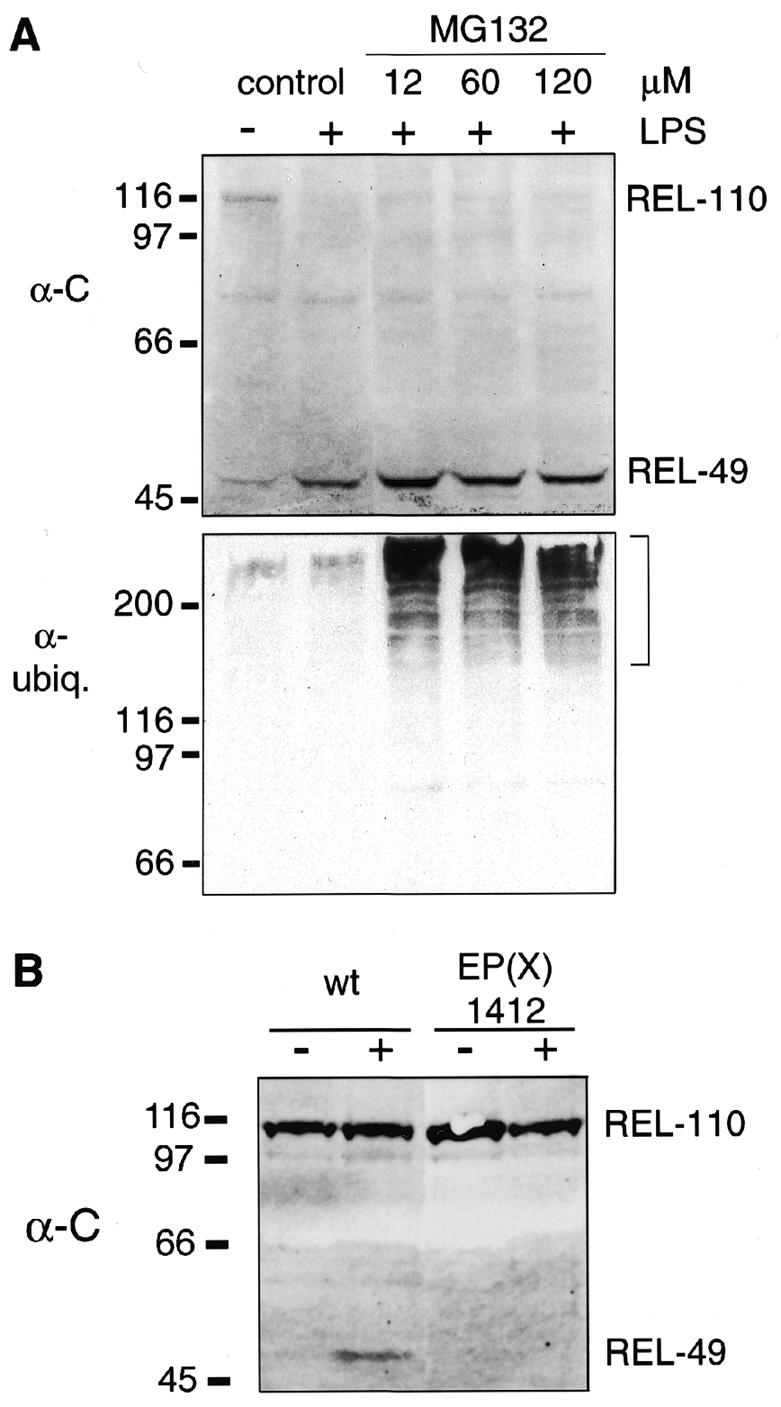
Fig. 4. Proteasome inhibition and Relish processing. (A) Immunoblotting of protein extracts from cell cultures, treated with different concentrations of MG132 prior to an immune challenge. The membrane was first developed with α-C and then with an anti-ubiquitin antibody to visualize the accumulation of polyubiquitylated proteins, indicated by the bracket. Addition of the solvent DMSO or of the inhibitors alone did not induce Relish processing. (B) Western blot with protein extracts from wild-type and EP(X)1412 third instar larvae. Where indicated (+) the animals have been infected 45 min before extract preparation. Different forms of Relish were detected with α-C.
A possible candidate for the Relish endoprotease is suggested by the recent observation that mutations in the Dredd gene, which codes for a caspase (Chen et al., 1998), block the induction of the antibacterial genes (Elrod-Erickson et al., 2000; Leulier et al., 2000). Figure 4B shows that no REL-49 is formed in infected larvae of the Dredd mutant EP(X)1412, while wild-type larvae are clearly induced. Thus, DREDD must act upstream of Relish in the signal transduction pathway. It may indeed be the enzyme that cleaves Relish, or may participate in a proteolytic cascade that leads to Relish activation.
It will be important to further dissect the machinery that controls the cleavage and activation of Relish, in order to understand the regulation of the humoral immune response in Drosophila. Recent experiments show that besides the protease an important role is also played by a kinase complex related to the human IKK. This kinase activity phosphorylates Relish in a signal-dependent manner and is required for Relish cleavage (Silverman et al., 2000). Mutations in the genes for two of the participating proteins show reduced levels of antimicrobial peptide expression upon infection (K. Anderson and D. Ferrandon, personal communication; Rutschmann et al., 2000)
On the basis of our results, we can now add a Rel protein to the growing family of factors such as SREBP, Notch and APP that are activated by endoproteolytic cleavage (Chan and Jan, 1998). The mechanism is clearly different from the constitutive processing of p105, which is strictly proteasome dependent and which leads to complete degradation of the IκB-like part (Palombella et al., 1994). However, p105 processing is complex and not fully understood, and may include an endoproteolytic step (Lin and Ghosh, 1996; Lin et al., 1998; Orian et al., 1999). Furthermore, signal-dependent processing of p105 has been described but rather leads to complete degradation of the entire molecule (Heissmeyer et al., 1999). Future experiments will show what relationships may exist between the mechanism of Relish activation and the activation of NF-κB in man.
METHODS
Cell and fly cultures. The Drosophila hemocyte-like cell line mbn-2 (malignant blood neoplasm; Gateff et al., 1980; Samakovlis et al., 1992) was grown in Schneider’s medium (Pansystems) with 5% fetal calf serum, 1× Glutamax I (Gibco-BRL), 50 U/ml penicillin, 50 µg/ml streptomycin and 50 µg/ml gentamicin. Flies were kept on agar media at 25°C and 60% humidity. As a wild-type strain we used Canton-S. The Relish-deficient strain RelE20 has been described (Hedengren et al., 1999). EP(X)1412 (Rorth et al., 1998) was provided by the Stock Centers in Bloomington and Szeged. Proteasome inhibitors lactacystin, MG132 (Calbiochem) and calpain inhibitor I (Sigma) or the diluent dimethylsulfoxide (DMSO) were added to mbn-2 cell cultures 1 h before LPS challenge. Cycloheximide (Sigma) was applied at 180 µM, 30 min before LPS addition.
Immune challenge. To stimulate mbn-2 cells we added 10 µg of LPS/ml (Escherichia coli O55:B5; Sigma) to the cultures. Mid to late third instar larvae were rolled in freshly grown bacteria (E. cloacae β12), pricked with a tungsten needle and put on apple juice plates for the indicated time. Adult flies were injected with E. cloacae (Hedengren et al., 1999).
Immunoreagents. Antibodies were raised in rabbits and in mice against keyhole limpet hemocyanin-conjugated peptides (Hancock and Evan, 1992) from the N-terminus (PGGNSPHQPPMANSP = α-N), the Rel homology domain (MNRRELSHKQLQEL = α-RHD) and the C-terminus (PLGHGSDPQDRKWM = α-C). The α-N rabbit serum was used at 1:10 000 in immunoblotting; the α-RHD antibody was affinity purified and used at 550 ng/ml. For detection of the Relish IκB part we also established mouse hybridoma cell lines. Supernatants were used at dilutions up to 1:100. All antibodies were tested for specificity by immunoprecipitation of in vitro-translated Relish and by western blotting on material from transiently Relish-overexpressing mbn-2 cells. Monoclonal anti-ubiquitin IgG was obtained from Santa Cruz (sc-8017). Secondary antibodies used in immunoblotting were goat anti-mouse and anti-rabbit horseradish peroxidase conjugates (Bio-Rad), pre-adsorbed to paraformaldehyde-fixed mbn-2 cells. For immunohistochemistry we used goat anti-mouse and anti-rabbit IgG conjugated to Cy2 and Cy3 fluorochromes (Jackson ImmunoResearch).
Protein extracts, immunoblotting and gel shift assay. Crude protein extracts were prepared from subconfluent mbn-2 cell cultures (Petersen et al., 1995), and cytoplasmic and nuclear proteins were enriched according to Grant et al. (1992). To inhibit protease and phosphatase activity we added protease inhibitor cocktail (Boehringer), 50 mM sodium fluoride and 1 mM sodium orthovanadate. Protein concentrations were determined by Bradford assay (Bio-Rad). For fly extracts, the animals were pulverized in liquid nitrogen (Petersen et al., 1995). Embryos were collected overnight, dechorionized, and lysed in SDS–PAGE sample buffer containing 4% SDS. Larvae were quickly ripped open and homogenized with an Eppendorff pestle in the same buffer. The material was boiled for 5 min and cleared by centrifugation. The samples were then separated on 7.5% SDS–polyacrylamide gels and blotted (Hybond-C extra; Amersham). Membranes were incubated with primary antibody overnight at 4°C in blocking buffer (50 mM Tris pH 8, 150 mM NaCl, 0.05% Tween 20, 2% casein) and washed for 4× 15 min in phosphate-buffered saline (PBS; Sambrook et al., 1989) with 0.1% Tween 20. Secondary antibody conjugates, diluted in blocking buffer, were applied for 1 h, followed by washing as above. Immunocomplexes were visualized with the ECLplus system and could also be stripped off the membranes (Amersham). Protein molecular weight standards were from Bio-Rad.
For the gel shift assay the deoxyoligonucleotide 5′-tcgagacacGGGGATTTTTgcac (κB site in uppercase) was used. The DNA-binding reaction was carried out according to Petersen et al. (1999), except that 0.5 ng of 32P-labeled probe and 2 µg of poly(dI-dC) were added. Nuclear extracts were incubated with 2 µl of antibody on ice for 30 min before adding the oligonucleotide probe whenever applicable. For competition with peptide antigens, the peptides were first incubated with the respective antibody for 10 min. The protein–DNA complex was separated on a 5% non-denaturing polyacrylamide gel and visualized by autoradiography.
In vivo labelling and immunoprecipitation. Subconfluent cell cultures were incubated in methionine-free Grace’s insect medium (Gibco-BRL) with 50 µCi of [35S]methionine/ml for 4.5 h. The cells were chased with normal medium for 30 min. We then induced one culture with LPS for 5 min and harvested the cells. Proteins were extracted in 10 mM Tris pH 8, 140 mM NaCl, 1% Triton X-100, 0.6% bovine serum albumin (BSA), inhibitors as above. Immunoprecipitation was carried out according to Edwards et al. (1997). We used ∼300 µg of total protein, adjusted to 1 µg/µl in lysis buffer, and 3 µl of rabbit antibody (either α-C antiserum, or a 1:1 mixture of α-N IgG and affinity-purified α-RHD).
Immunohistochemistry. Third instar larvae were dissected inside-out in PBS with 4% paraformaldehyde and fixed for 20 min. The specimens were washed for 6× 10 min with 0.1% Triton X-100, 1% BSA in PBS and then incubated with the first antibody for 2 h. After washing for 6× 10 min with 0.1% Triton X-100, 0.1% BSA in PBS the secondary antibody was applied for 1 h. All antibodies were diluted in the respective washing buffer. mbn-2 cells were spun onto glass slides at 1000 r.p.m. for 5 min in the cold. Slides were allowed to dry, then the cells were fixed with acetone and air dried. After blocking for 30 min in cell culture medium and two short washes with 0.1% BSA in 10 mM Tris pH 8, 150 mM NaCl, first and secondary antibodies were applied as above. In all tissue and cell samples, nuclei were stained with diamidinophenylindole (DAPI) and mounted with 80% glycerol in PBS.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Solveig Sundberg and Ann Sjölund for help with antibody preparation, and Raul Bettencourt, Anne Uv and Eva Kurucz for technical advice. We also thank Neal Silverman for helpful discussion of our results, and Tom Maniatis, Christos Samakovlis and Michael Williams for comments on the manuscript. This research was supported by grants from the Göran Gustafsson Foundation for Scientific Research, the Swedish Natural Science Research Council, the Swedish Cancer Society and the Swedish Medical Research Council.
REFERENCES
- Chan Y.-M. and Jan, Y.N. (1998) Roles for proteolysis and trafficking in Notch maturation and signal transduction. Cell, 94, 423–426. [DOI] [PubMed] [Google Scholar]
- Chen P., Rodriguez, A., Erskine, R., Thach, T. and Abrams, J.M. (1998) Dredd, a novel effector of the apoptosis activators Reaper, Grim and Hid in Drosophila. Dev. Biol., 201, 202–216. [DOI] [PubMed] [Google Scholar]
- Dushay M.S., Åsling, B. and Hultmark, D. (1996) Origins of immunity: Relish, a compound Rel-like gene in the antibacterial defense of Drosophila. Proc. Natl Acad. Sci. USA, 93, 10343–10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards D.N., Towb, P. and Wasserman, S.A. (1997) An activity-dependent network of interactions links the Rel protein Dorsal with its cytoplasmic regulators. Development, 124, 3855–3864. [DOI] [PubMed] [Google Scholar]
- Elrod-Erickson M., Mishra, S. and Schneider, D. (2000) Interactions between the cellular and humoral immune responses in Drosophila. Curr. Biol., 10, 781–784. [DOI] [PubMed] [Google Scholar]
- Engström Y., Kadalayil, L., Sun, S.-C., Samakovlis, C., Hultmark, D. and Faye, I. (1993) κB-like motifs regulate the induction of immune genes in Drosophila. J. Mol. Biol., 232, 327–333. [DOI] [PubMed] [Google Scholar]
- Gateff E., Gissmann, L., Shrestha, R., Plus, N., Pfister, H., Schröder, J. and zur Hausen, H. (1980) Characterization of two tumorous blood cell lines of Drosophila melanogaster and the viruses they contain. In Kurstak, E., Maramorosch, K. and Dübendorfer, A. (eds), Invertebrate Systems In Vitro. Elsevier/North-Holland Biomedical Press, Amsterdam, The Netherlands, pp. 517–533.
- Ghosh S., May, M.J. and Kopp, E.B. (1998) NF-κB and Rel proteins: evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol., 16, 225–260. [DOI] [PubMed] [Google Scholar]
- Gottesman S., Maurizi, M.R. and Wickner, S. (1997) Regulatory subunits of energy-dependent proteases. Cell, 91, 435–438. [DOI] [PubMed] [Google Scholar]
- Grant P.A., Arulampalam, V., Ahrlund-Richter, L. and Pettersson, S. (1992) Identification of Ets-like lymphoid specific elements within the immunoglobulin heavy chain 3′ enhancer. Nucleic Acids Res., 20, 4401–4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z.S. and Ip, Y.T. (1999) Interaction and specificity of Rel-related proteins in regulating Drosophila immunity gene expression. J. Biol. Chem., 274, 21355–21361. [DOI] [PubMed] [Google Scholar]
- Hancock D.C. and Evan, G.I. (1992) Synthesis of peptides for use as immunogens. In Manson, M.M. (ed.), Immunochemical Protocols, Vol. 10. Humana Press, Totowa, NJ, pp. 23–32.
- Hedengren M., Åsling, B., Dushay, M.S., Ando, I., Ekengren, S., Wihlborg, M. and Hultmark, D. (1999) Relish, a central factor in the control of humoral, but not cellular immunity in Drosophila. Mol. Cell, 4, 827–837. [DOI] [PubMed] [Google Scholar]
- Heissmeyer V., Krappmann, D., Wulczyn, F.G. and Scheidereit, C. (1999) NF-κB p105 is a target of IκB kinases and controls signal induction of Bcl-3–p50 complexes. EMBO J., 18, 4766–4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip Y.T., Reach, M., Engström, Y., Kadalayil, L., Cai, H., Gonzales-Crespo, S., Tatei, K. and Levine, M. (1993) Dif, a dorsal-related gene that mediates an immune response in Drosophila. Cell, 75, 753–763. [DOI] [PubMed] [Google Scholar]
- Larsen C.N. and Finley, D. (1997) Protein translocation channels in the proteasome and other proteases. Cell, 91, 431–434. [DOI] [PubMed] [Google Scholar]
- Leulier F., Rodriguez, A., Khush, R.S., Abrams, J.M. and Lemaitre, B. (2000) The Drosophila caspase Dredd is required to resist Gram-negative bacterial infection. EMBO Rep., 1, 353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L. and Ghosh, S. (1996) A glycine-rich region in NF-κB p105 functions as a processing signal for the generation of the p50 subunit. Mol. Cell. Biol., 16, 2248–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., DeMartino, G.N. and Greene, W.C. (1998) Cotranslational biogenesis of NF-κB p50 by the 26S proteasome. Cell, 92, 819–828. [DOI] [PubMed] [Google Scholar]
- Naidoo N., Song, W., Hunter-Ensor, M. and Sehgal, A. (1999) A role for the proteasome in the light response of the timeless clock protein. Science, 285, 1737–1741. [DOI] [PubMed] [Google Scholar]
- Orian A., Schwartz, A.L., Israël, A., Whiteside, S., Kahana, C. and Ciechanover, A. (1999) Structural motifs involved in ubiquitin-mediated processing of the NF-κB precursor p105: roles of the glycine-rich region and a downstream ubiquitination domain. Mol. Cell. Biol., 19, 3664–3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palombella V.J., Rando, O.J., Goldberg, A.L. and Maniatis, T. (1994) The ubiquitin–proteasome pathway is required for processing the NF-κB1 precursor protein and the activation of NF-κB. Cell, 78, 773–785. [DOI] [PubMed] [Google Scholar]
- Petersen U.-M., Björklund, G., Ip, Y.T. and Engström, Y. (1995) The dorsal-related immunity factor, Dif, is a sequence-specific trans-activator of Drosophila Cecropin gene expression. EMBO J., 14, 3146–3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen U.-M., Kadalayil, L., Rehorn, K.-P., Hoshizaki, D.K., Reuter, R. and Engström, Y. (1999) Serpent regulates Drosophila immunity genes in the larval fat body through an essential GATA motif. EMBO J., 18, 4013–4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorth P. et al. (1998) Systematic gain-of-function genetics in Drosophila. Development, 125, 1049–1057. [DOI] [PubMed] [Google Scholar]
- Rutschmann S., Jung, A.C., Hetru, C., Reichhart, J.-M., Hoffmann, J.A. and Ferrandon, D. (2000a) The Rel protein DIF mediates the antifungal, but not the antibacterial, host defense in Drosophila. Immunity, 12, 569–580. [DOI] [PubMed] [Google Scholar]
- Rutschmann S., Jung, A.C., Zhou, R., Silverman, N., Hoffmann, J.A. and Ferrandon, D. (2000b) Role of Drosophila IKKγ in a Toll-independent antibacterial immune response. Nature Immunol., in press. [DOI] [PubMed] [Google Scholar]
- Samakovlis C., Åsling, B., Boman, H.G., Gateff, E. and Hultmark, D. (1992) In vitro induction of cecropin genes—an immune response in a Drosophila blood cell line. Biochem. Biophys. Res. Commun., 188, 1169–1175. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch, E.F. and Maniatis, T. (1989) Molecular Cloning. A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Silverman N., Zhou, R., Stöven, S., Pandey, N., Hultmark, D. and Maniatis, T. (2000) A Drosophilia IκB kinase complex required for Relish endoproteolysis and antibacterial immunity. Genes Dev., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]