Abstract
The novel loquat cultivar ‘Chunhua No.1′ (CH1) is a promising commercial cultivar. However, CH1 has texture characteristics different from those of common loquat, and its formation mechanism remains unclear. Here, we first identified the phenolic compounds of CH1 and its parent (‘Dawuxing’, DWX) and the effect on texture formation. The special presence of stone cells explained the flavor differences in CH1. Chlorogenic acid, neochlorogenic acid, and coniferyl alcohol were the main phenolic compounds in loquat, and the high content of coniferyl alcohol was a potential factor for the rough texture of CH1. Transcriptome reveals that phenylpropanoid metabolism was activated during CH1 fruit texture formation. Kyoto Encyclopedia of Genes and Genomes (KEGG) identified 51 structural genes involved in phenylpropanoid biosynthesis, and Weighted Gene Co-expression Network Analysis (WGCNA) identified four structural genes and 88 transcription factors. These findings provide new insights into the phenolic metabolism and flavor formation of loquat fruit.
Keywords: Loquat, Flavor, Phenolics, Fruit texture, Transcriptome, Phenylpropanoid biosynthesis
Introduction
Loquat (Eriobotrya japonica Lindl.), which originated in China and belongs to the Rosaceae family, contains many phenolics and triterpenes that exhibit anticancer, anti-inflammatory, and hypoglycemic effects (Chi et al., 2023, Xu et al., 2014). Unlike other temperate fruit crops, loquat trees bloom in autumn and early winter, their flowers and young fruits susceptible to damage from low temperatures and winter frost (Deng et al., 2023). The newly released loquat cultivar CH1 (E. japonica Lindl.), created in our loquat breeding project, blooms in spring, thereby avoiding chilling injuries during winter, and matures almost one month after the common loquat (Deng et al., 2023). CH1 is the first new spring-flowering cultivar in China, and the parent is DWX (E. japonica Lindl.). However, the flavor and taste of the common loquat and CH1 fruit differ, with those of CH1 being similar to pear.
Flavor is generally considered a combination of aroma, taste, and texture (An et al., 2023, Shukla et al., 2019). Texture is the tactile perception of a food product on the surface of the mouth. Phenolic compounds, critical secondary metabolites in plants, have strong antioxidant activity and provide flavor, color, and fruit texture to fruits (Chi et al., 2023, Ferreres et al., 2009). Phenolic compounds include phenolic acids, oligophenols, and polyphenols (Marchiosi et al., 2020). Phenolic acid has a sour, bitter, astringent, and phenolic-like flavor, which significantly affects the taste, nutritional attributes, and appearance of fruits (Liu et al., 2023). In plants, phenolic acids are classified as benzoic and hydroxycinnamic acids, mainly including chlorogenic, vanillic, caffeic, and p-coumaric acids (Marchiosi et al., 2020). Oligophenols are primarily associated with defense mechanisms and pharmacological properties and are predominantly composed of flavonoids (Xu et al., 2021). Flavanols and anthocyanins are among the widely distributed plant flavonoids of fruits and vegetables, which also provide color and flavor to fruits and vegetables (Soto-Vaca, Gutierrez, Losso, Xu, & Finley, 2012). Polyphenols produce astringent and bitter flavors, enhancing “the overall mouthfeel” of fresh fruit, which is characterized by a rough sensation throughout the mouth, especially on the tongue. When polyphenols react with proline in saliva, they precipitate onto the surface of the mouth, leading to a rough sensation (Luck et al., 1994). Lignin, under the category of polyphenols, is another critical factor that affects fruit texture because it hardens the pulp (Wang et al., 2021). For example, lignin deposition causes thin-walled cells to form stone cells (Huang, Zhu, Zhu, Wu, & Chen, 2019). Thus, research on loquat fruit quality has also considered pulp lignification, primarily focusing on lignification after harvest (Chi et al., 2023, Wang et al., 2021).
Studies on fruit flavor have often focused on the accumulation of soluble sugars and organic acids (Zhou et al., 2023, Zou et al., 2020). However, the phenolic compounds contributing to the specific textural properties of CH1 fruit and the underlying key metabolic mechanisms responsible for this difference remain unknown. Rough texture is an undesirable quality in fruit that is commonly observed in the flesh of pears and seldom observed in the flesh of loquats (Mamat, Tusong, Xu, Yan, Mei, & Wang, 2021). Furthermore, many factors, namely raw material, composition, and environment, are directly or indirectly related to the formation of flavor components. Notably, CH1 is a new cultivar for which the fruit flavor characteristics must be determined before commercialization.
To gain insights into the metabolic pathways involved in forming fruit texture in CH1, we selected CH1 and its parent DWX. Using high-performance liquid chromatography (HPLC) and transcriptomics methods, we have investigated the effects of phenolic composition on the unique taste properties and astringency formation of CH1 fruit. The objectives of this study were to (i) characterize the phenolic composition of CH1 and common loquat DWX fruits; (ii) analyze the variation and formation of CH1 fruit texture characteristics; and (iii) identify the key metabolic pathways and differentially expressed genes based on which contribute to fruit astringency and roughness. Our results demonstrate the regulatory mechanisms of phenolics in the formation of CH1 fruit taste characteristics and provide a basis for further germplasm improvements. Breeders can be provided with specific information about the phenolic characteristics of new cultivars, which can help pass on these superior traits to future generations.
Materials and methods
Plant materials growth conditions, and sample preparation
This study was conducted using 6-year-old CH1 and 9-year-old DWX fruit-bearing trees from the loquat germplasm resource nursery of Sichuan Agricultural University, Shimian County, Ya’an City, Sichuan Province, China (29°18′55″N–29°18′56″N, 102°32′16″E). CH1 was grafted onto a DWX rootstock with an interrow and interplant spacing of 4 m. All the plants were managed according to standard horticultural practices throughout the experiment.
Based on our observations and other reports (Chi et al., 2023), loquat fruits were collected at five developmental stages (Fig. 1A): preliminary development (S1), fruitlet (S2), swollen fruit (S3), breaker (S4), and maturation (S5). At least 30 fruits of the same size and color were collected from three plants of each cultivar, and three biological replicates were performed. Except for the samples for stone cell analysis, all the collected fruit samples were chopped and frozen in liquid nitrogen and stored at –80 ℃ until further use.
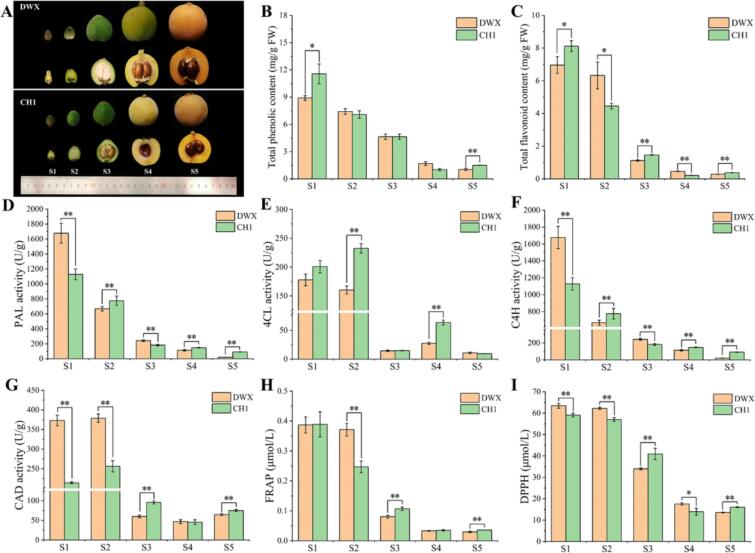
Fig. 1.
Phenotypic characterization and related indexes of antioxidant activity. (A) Fruit phenotypic. (B) Total phenolic content. (C) Total flavonoid content. (D) PAL activity. (E) 4CL activity. (F) C4H activity. (G) CAD activity. (H) Antioxidant capacities of FRAP. (I) Antioxidant capacities of DPPH. “*” and “**” indicate significant differences at p < 0.05 and p < 0.01, respectively.
Determination of total phenolic, total flavonoid, and antioxidant capacity
The total phenolic content and total flavonoid content were extracted using a previously described method with minor modifications (Wang, Shao, Madebo, Hou, Zheng, & Jin, 2020). Total phenolic content was determined using the Folin-Ciocalteu method, and the absorbance was recorded at 765 nm. The content was expressed as gallic acid equivalent (μgGAE/g·FW). Total flavonoids were extracted using solvent extraction. Two grams of loquat fruit were homogenized with 10 mL 80 % cold acetone and centrifuged at 12,000g for 20 min at 4 °C. The resulting extract (2 mL) was combined with 2 mL 3 % AlCl3 and 1 mL 70 % (v/v) ethanol in a reaction mixture, and then the absorbance was measured at 510 nm. The content is expressed as rutin equivalent (μg rutin/g·FW). DPPH and FRAP assays were determined according to a previous method (Rumpf, Burger, & Schulze, 2023). The results are expressed as μmol/L water-soluble vitamin E (Trolox) equivalent antioxidant capacity.
Determination of enzyme activities and lignin content
The activities of phenylalanine ammonia-lyase (PAL), cinnamate 4-hydroxylase (C4H), 4-coumarate: coenzyme A ligase (4CL), and cinnamyl alcohol dehydrogenase (CAD) were measured using a previously described method with slight modifications (Wang et al., 2019). Frozen fruit pulp powder (0.5 g) was extracted using 5 mL sodium phosphate buffer (50 mM, pH 7.8). One unit of PAL, C4H, 4CL, and CAD activity was defined as the amount of enzyme that caused an increase in absorbance of 0.01 at 290, 340, 333, and 340 nm/min, respectively. Lignin content was assessed using a method of lignin precipitation that used thioglycolic acid (Huang et al., 2019). The blank sample was prepared using 1 mL 2 M NaOH. Subsequently, 0.1 mL 7.5 M hydroxylamine hydrochloride was added, and the volume was adjusted to 10 mL using glacial acetic acid. The absorbance was measured at 280 nm, and the results are expressed as the percentage (%) of FW.
Determination of phenolic compounds and content
Phenolic compound extraction was performed using a previously described method with slight modifications (Wang et al., 2020). In brief, 1 g frozen tissue was extracted using 3 mL 80 % (v/v) cold methanol under light-free conditions. The mixture was extracted via sonification in a sonicator for 30 min and then centrifuged at 12,000g for 10 min at 4 °C. Subsequently, the supernatant was purified using a C18 Sep-Pak cartridge (Sep-Pak Vac 6 cc, Waters, Milford, MA, USA) and filtered through a 0.22 µm membrane. Phenolic compounds were analyzed using a HPLC system (Agilent, Santa Clara, CA, USA) equipped with UV photodiode array detection and a Comatex C18 column (4.6 × 250 mm, 5 μm; CoMetro, Princeton, NJ, USA). The operating temperature of the column was maintained at 30 °C, the injection volume was 20 μL, and the detection wavelengths of the UV photodiode array were 280 and 320 nm. The mobile phase comprised 100 % methanol (mobile phase A) and ultrapure water containing 0.1 % phosphoric acid (mobile phase B), with gradient elution at a flow rate of 1 mL·min−1. Gradient elution was programmed as follows: 5 %–71 % A (0–22 min), 71 %–5% A (22–30 min), and 5 % A (30–35 min). Individual phenolic compounds were quantified using the calibration curve, and the result is expressed as μg·g−1 FW.
Determination of stone cells and anatomical observation
The freeze separation method was used to determine the stone cell content (Lin et al., 2022). Approximately 30 g loquat fruit was weighed, frozen for 3 h, thawed completely, and frozen again for another 3 h. This freeze and thaw process was repeated thrice. The resulting homogenate was transferred to a beaker. Subsequently, 60 mL water was added, and the mixture was stirred with a glass rod for 1 min and left to stand for 30 s. Subsequently, the upper suspension was poured out. The rinsing process was performed and repeated four times. The rinsing solution collected from the previous rinses was used for the subsequent rinsing and collection of stone cells. The combined stone cells were filtered through coarse filter paper, spread out, and oven-dried at 60 °C until they reached a constant weight. Finally, dried stone cells were collected and weighed. The stone cell content (%) was calculated as (weight of extracted stone cells/weight of pulp) × 100 %.
Loquat fruits (0.5 × 0.5 × 0.5 mm) were fixed in FAA solution (i.e., formaldehyde, acetic acid, and ethanol) and embedded in paraffin for sectioning (Liu, Li, Feng, & Li, 2021). The sections were then stained with safranin-fast green, which stains the stone cells red. Subsequently, the slides were prepared for microscopic observation (Pannoramic MIDI, 3DHISTECH, Hungary) to capture images for determining the size and density of the stone cells.
RNA-seq analysis
Total RNA was extracted using the polysaccharide polyphenol kit (Tiangen, Beijing, China). RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA). The sequencing library construction and quality control were performed by Novogene Corporation (Beijing, China) and the Illumina NovaSeq6000 sequencing platform.
Reads containing adapters, poly-N, and low-quality reads were eliminated from the raw data to obtain clean reads. The clean reads were mapped to the reference genome of the yellow-fleshed E. japonica cultivar DWX using the HISAT2 software (Wang, 2021). Genes with an adjusted p value ≤ 0.05 and |log2(foldchange)| ≥ 1 identified determined using DESeq2 were designated as differentially expressed genes (DEGs) in the CH1 versus DWX comparison. Statistical enrichment of the DEGs in the KEGG pathway was performed using the R package “clusterProfiler” (http://www.genome.jp/kegg/). Gene trend analysis was performed using Short Time-series Expression Miner software. WGCNA was performed using R with the WGCNA package, and the networks were visualized using Cytoscape (version 3.10).
Validation by quantitative real-time PCR (qRT-PCR)
qRT-PCR primers for the selected genes were designed using Primer 5 (Premier Biosoft, Palo Alto, CA, USA.). Detailed primer sequences are shown in Supplementary Table 1. qRT-PCR was performed on a CFX96 Real-Time System C1000 Thermal Cycler (Bio-Rad, Hercules, CA, USA) using the ArtiCanCEO SYBR qPCR Mix (Tsingke, Beijing, China) according to the manufacturer’s instructions. Relative gene expression was normalized to that of the reference gene, Actin. Gene expression levels were calculated using the 2−ΔΔ CT method.
Statistical analysis
Student’s t-test was used to assess differences in means at the 0.05 or 0.01 significance level for all data except transcriptomics data. Data analysis was performed using SPSS 22.0 (IBM, Chicago, IL, USA). Data are expressed as mean ± standard deviation. Graphs were generated using OriginPro2021 (OriginLab, Northampton, MA, USA).
Results
Phenotypic characterization and related indicators for evaluating antioxidant activity
The phenotypic characterization of DWX and CH1 at different developmental stages is shown in Fig. 1A. The fruit morphology of CH1 was smaller than that of DWX. The total phenolic and flavonoid contents initially exhibited high values and subsequently a decreasing trend; the content of CH1 at S5 was significantly higher than that of DWX (Fig. 1B, C). The total phenolic and total flavonoid contents of CH1 at S5 were 43.81 % and 33.78 % higher than those of DWX, respectively. The PLA, C4H, and CAD activities of CH1 were higher than those of DWX in most periods, except for 4CL activity (Fig. 1D–G). Similar to the cumulative content of total phenolics, the antioxidant capacities of FRAP and DPPH were the highest at S1 and decreased according to the stage (Fig. 1H–I). At S5, the antioxidant capacity of CH1 was stronger than that of DWX, with 0.0359 (FRAP value) and 16.02 (DHHP value) μmol/mL, respectively.
Morphological observation of stone cells and lignin content
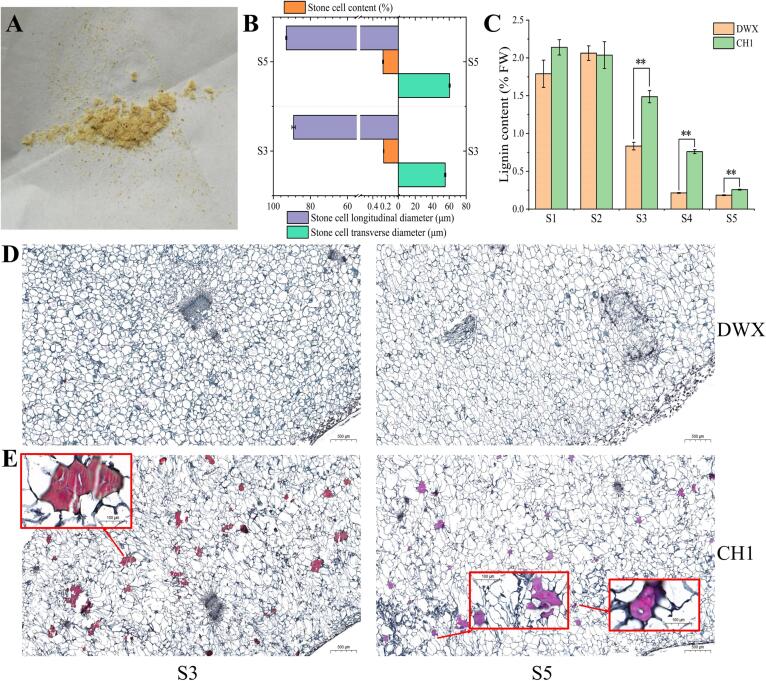
To investigate the granularity of CH1 pulp, we determined its stone cell content. The structure showed that stone cells were presented in CH1, with contents of 0.23 % (S3) and 0.24 % (S5), but not in DWX (Fig. 2A, B). The longitudinal and transverse diameters of the stone cells in CH1 underwent minimal changes during development. The lignin content decreased gradually with fruit development, and the lignin content of CH1 was significantly higher than that of DWX S3–S5 (Fig. 2C). At S5, the lignin content of DWX and CH1 reached 0.18 % and 0.26 % (103A280 kg−1), respectively. The results of paraffin sectioning showed that the stone cells of CH1 were dense and stabilized at S3 (Fig. 2E, F). Notably, newly formed stone cells were observed in CH1.
Fig. 2.
Morphological observation of stone cells and lignin content. (A) Stone cells appearance. (B) Stone cell size and content. (C) Lignin content. (D) Microscopic observation of fruit pulp collected from DWX at S3 and S5 (20× and 500 μm). (E) Microscopic observation of fruit pulp collected from CH1 at S3 and S5 (20× and 500 μm). Red boxes represent microscopic observations at 100× and 100 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Phenolic compounds and content
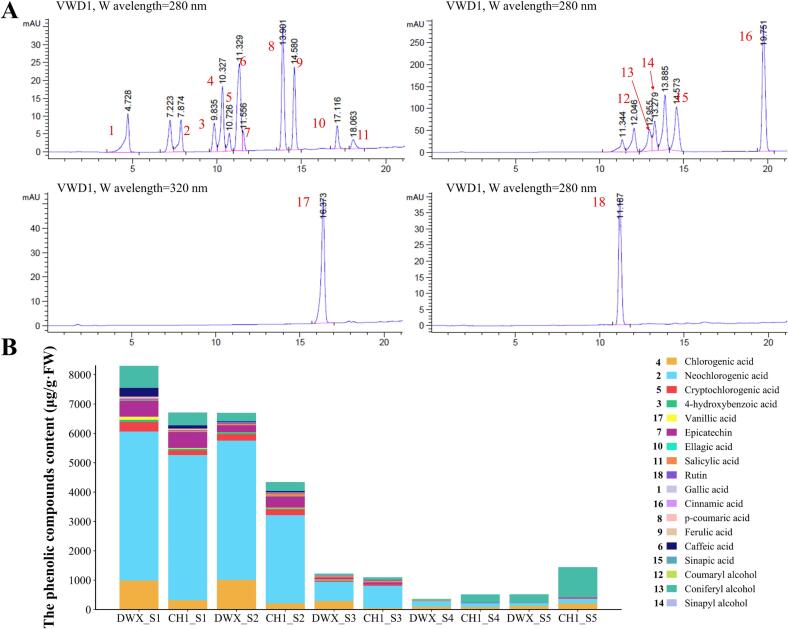
The following eighteen phenolic compounds were identified and quantified from the two cultivars using the HPLC method: 10 phenolic acids related to antioxidant capacity and 8 intermediates of lignin metabolism (Fig. 3A). Different detection wavelengths were used to effectively separate phenolic compounds. The standard curves and regression equations of the phenolic compounds are shown in Supplementary Table 2.
Fig. 3.
HPLC chromatogram traces, and dynamic changes of phenolic compounds. (A) HPLC phenolic peak profile. (B) Column stacked plot of the content of phenolic compounds. Numbers and colors represent different phenolic compounds.
The contents of phenolic compounds showed an overall gradually declining trend; the main phenolic compounds were chlorogenic acid, neochlorogenic acid, cryptochlorogenic acid, and coniferyl alcohol (Fig. 3B). Chlorogenic acid, a phenylpropanoid, showed high values at S1, reaching 987.51 and 316.58 μg/g FW in DWX and CH1, respectively, and then decreased to 124.97 and 206.65 μg/g FW at S5, respectively. At S1, neochlorogenic acid was the most abundant of all phenolic acids, reaching 5075.72 and 4946.68 μg/g FW in DWX and CH1, respectively. At S5, the cryptochlorogenic acid content of CH1 was significantly higher than that of DWX, reaching 27.00 and 5.43 μg/g FW, respectively. The coniferyl alcohol content showed a fluctuating trend, with low synthesis observed in the early stage of fruit development. The highest value was detected in CH1 at S5, reaching 1018.48 μg/g FW, which was significantly higher than that of DWX (281.49 μg/g FW). 4-hydroxybenzoic acid and salicylic acid were detected in CH1 at S5. In general, the content of other phenolic compounds was low.
Transcriptomic analysis
To further explore the molecular mechanisms through which phenolics influence flavor formation, we performed transcriptome analyses on fruit samples from five stages of DWX and CH1. Thirty libraries were obtained, each containing more than 6.27G clean reads, Q20 ≥ 97.71 %, Q30 ≥ 93.41 %, and GC content ≥ 44.97 % (Supplementary Table 3). Clean reads were mapped to the DWX genome, resulting in mapping percentages from 85.44 % to 93.33 % in the different libraries. Principal component analysis and correlation analysis of the transcriptome data showed high reproducibility (Supplementary Fig. 1A, B). Twenty-one genes involved in the phenylpropanoid biosynthesis pathway were selected to confirm the accuracy of the RNA-seq data. The correlation coefficients between RNA-seq and qRT-PCR data were calculated (R2 = 0.7684), which indicated that the expression trends of most genes were consistent with the RNA-seq results (Supplementary Fig. 2).
Venn diagram and KEGG annotation analysis of DEGs
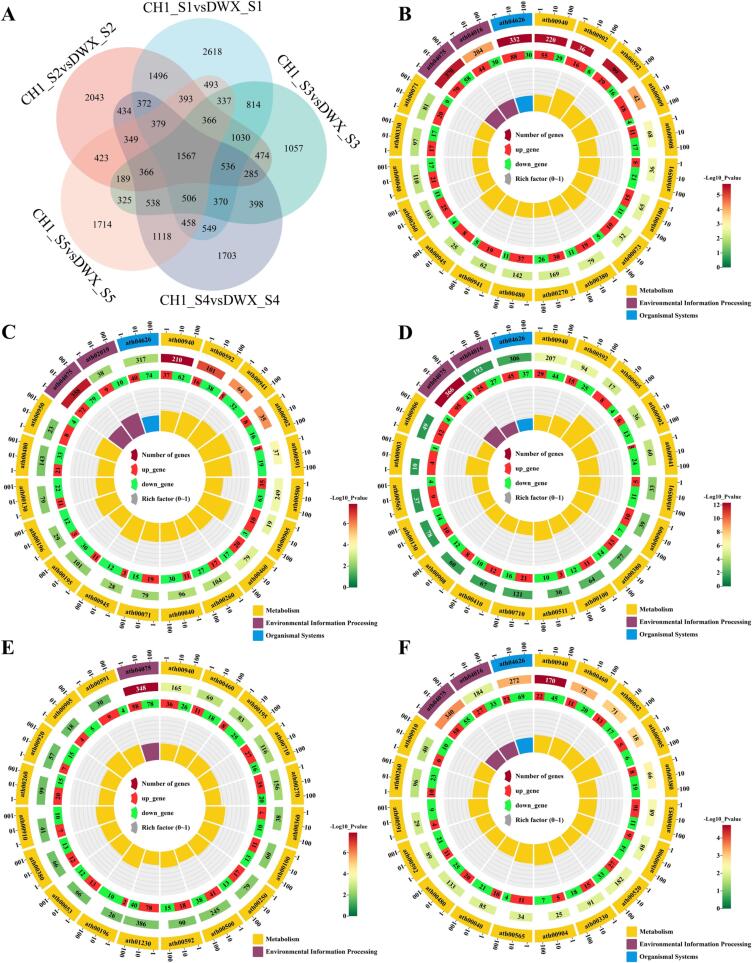
DEGs from different comparative combinations were analyzed using Venn diagrams (Fig. 4A). In total, 5795, 5142, 4868, 5162, and 4676 DEGs were up-regulated in the comparison of CH1 (experimental) versus DWX (control) at the five stages, respectively, whereas 4907, 7142, 4472, 4766, and 4845 DEGs were down-regulated, respectively, based on FPKM values. A total of 1567 DEGs were in the five comparative combinations. Subsequently, the differential genes were analyzed for metabolic enrichment (Fig. 4B–F). Five comparative combinations of DEGs were enriched for different metabolic pathways according to the KEGG database. Notably, the KEGG pathways “ath04075” for plant hormone signal transduction and “ath00940” for phenylpropanoid biosynthesis were significantly enriched in all five comparisons. Except for CH1_S4 versus DWX_S4, the KEGG databases showed three metabolic pathways: environmental information processing, metabolism, and organismal systems.
Fig. 4.
Venn diagram and KEGG enrichment circle maps based on DEGs. (A) Venn diagram. (B–F) KEGG enrichments of (B) CH1_S1 vs. DWX_S1, (C) CH1_S2 vs. DWX_S2, (D) CH1_S3 vs. DWX_S3, (E) CH1_S4 vs. DWX_S4, and (F) CH1_S5 vs. DWX_S5. From the outside to the inside of the circle, the expressions show the KEGG database ID, total gene number, p value, up- and downregulated genes, and rich factor.
DEG expression patterns of phenylpropanoid metabolism
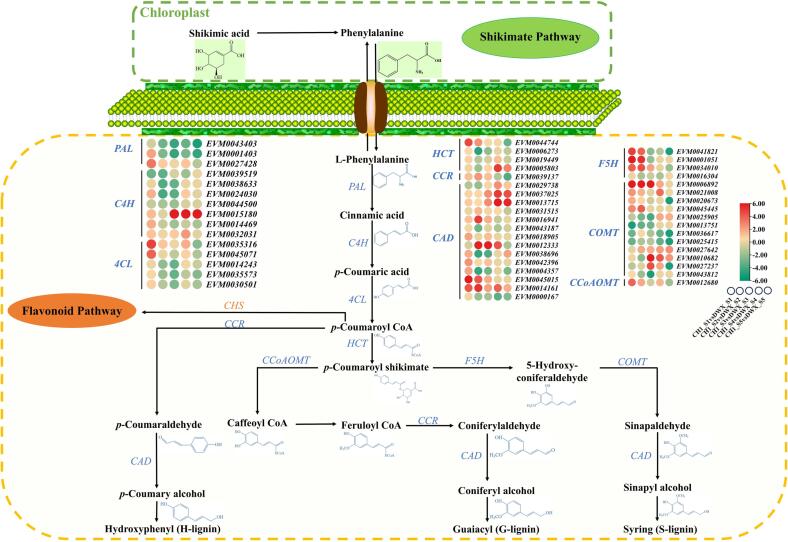
The synthesis of phenolic compounds primarily involved phenylpropanoid biosynthesis, of which 51 DEGs were identified in CH1 versus DWX, namely three PALs, seven C4Hs, five 4CLs, four HCTs, one CCR,14 CADs, four F5Hs, 12 COMTs, and one CCoAOMT (Fig. 5). During most developmental stages, the genes encoding C4H, HCT, CAD, F5H, and COMT were up-regulated in CH1 compared with those of DWX, and most of the genes encoding PAL and 4CL in CH1 were down-regulated from CH1_S2 to CH1_S5. By contrast, the relative expression of one C4H (EVM0015180), two CADs (EVM0037025 and EVM0013715), one F5H (EVM0016304), and two COMTs (EVM0013751 and EVM0027642) were significantly up-regulated in CH1 during fruit growth and development. Among the 4CL family genes, most of the 4CL expression was down-regulated in the late stages of fruit development. EVM0037025 expression in CH1 was significantly up-regulated compared with that in DWX, corresponding to the accumulation trend of p-coumaric acid (Fig. 3). The expression patterns of EVM0037025 and EVM0013715 in CH1 were consistent with the pattern of coniferyl alcohol accumulation during late fruit development (Fig. 3). These results suggest that these genes are responsible for the increase in the content of phenolic compounds and fruit flavor of CH1.
Fig. 5.
Heat map of DEGs involved in the phenylpropanoid metabolism. Colors indicate higher or lower levels based on the log2Foldchange value of the comparison between CH1 and DWX, respectively. DEGs involved in phenylpropanoid metabolism namely PAL; C4H; 4CL; HCT: hydroxycinnamoyl-CoA shikimate; CCR: cinnamoyl-CoA reductase; CAD, cinnamyl-alcohol dehydrogenase; F5H: ferulate-5-hydroxylase; COMT: Catechol-O-methyltransferase; and CCoAOMT: caffeoyl-CoA O-methyltransferase.
Analysis of WGCNA modules associated with phenolic biosynthesis
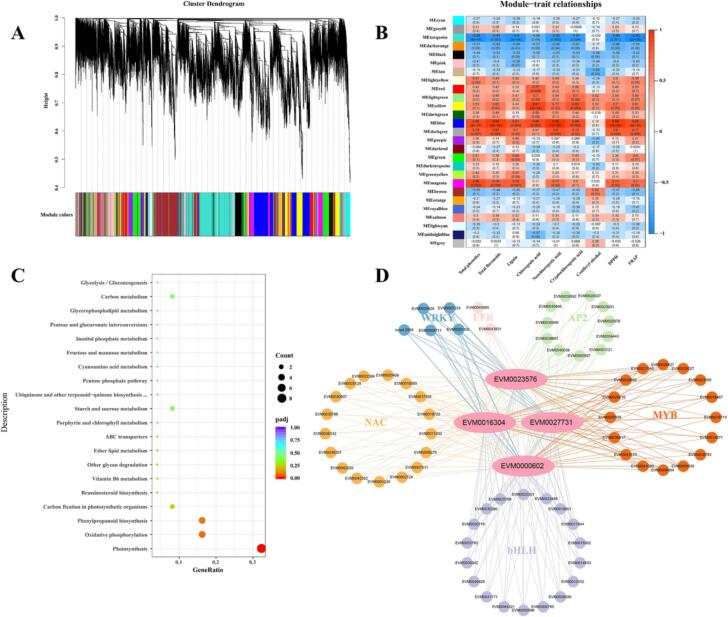
To obtain novel insights into the regulation mechanisms of phenolic changes during fruit ripening, we used WGCNA to select genes that conformed to a scale-free network distribution with a correlation coefficient greater than 0.8, and the soft threshold was set to 10 to construct a soft threshold map (Supplementary Fig. 3). We constructed a network based on a determined soft threshold, used the dissimilarity among genes for hierarchical clustering of genes in the network, and used dynamic shearing to divide the tree into different modules (Fig. 6A). A total of 28,964 genes were divided into 27 modules according to their expression patterns (Fig. 6B). In the heatmap of module-trait correlations, the blue module had the most genes (i.e., 4273 genes), which were highly and positively correlated with total phenolics, total flavonoids, lignin, chlorogenic acid, neochlorogenic acid, cryptochlorogenic acid, DPPH, FRAP, and had a low correlation with coniferyl alcohol.
Fig. 6.
Network analysis dendrogram by WGCNA. (A) Module hierarchical clustering tree diagram. (B) Heat map of key physiological indicators and inter-module correlations. (C) KEGG enrichment map of hub genes in the blue module. (D) Co-expression network of structural genes in phenylpropanoid biosynthesis pathway and transcription factors.
To identify key genes in the blue module, we constructed a gene correlation network using 4273 genes. Based on the degree of connectivity, the top 100 genes were considered hub genes. To investigate the pathways involved in the correlated hub genes in the blue module, we performed KEGG analysis and found that these genes were annotated to 22 metabolic pathways (Supplementary Table 4). The phenylpropanoid biosynthesis pathway was also involved (Fig. 6C). Among the 100 genes, four genes (EVM0000602, EVM0016304, EVM0027731, and EVM0023576) that might regulate the phenylpropanoid biosynthesis pathway in loquat were identified. Functional annotations of the four candidate genes are listed in Supplementary Table 5.
We constructed co-expression networks and demonstrated the connectivity between the four structural genes and transcription factors (Fig. 6D). The four candidate genes were regulated by the MYB, bHLH, NCA, AP2, WRKY, and EFR families. Heatmap analysis of clustering between structural genes and transcription factors based on normalized FPKM values is shown in Supplementary Fig. 4. EVM0023576 (4CL9), EVM0027731 (POD42), EVM0016304 (F5H), and EVM0000602 (BGH3B, beta-glucosidase) clustered with bHLH transcription factors. The expression patterns of EVM0016304 (F5H) and EVM0000602 (BGH3B) clustered with EVM0033763 (bHLH), suggesting that they are mutually regulated; thus, this finding requires further validation.
Discussion
Many secondary metabolic processes occur during loquat fruit development and ripening, among which phenolic-related metabolism plays a key role in food quality, namely, appearance, flavor, and nutrition (Ding, Chachin, Ueda, Imahori, & Wang, 2001). In this study, we described for the first time the dynamic changes in the phenolic composition of CH1 concerning the latest whole-genome sequencing of DWX and screened candidate genes for phenolic metabolism differences between CH1 and its parents.
Total phenolic, total flavonoid contents, and related enzyme activities
Flavor formation is associated with the presence of antioxidant compounds, such as polyphenols, and their interactions with other antioxidant compounds in foods (Soto-Vaca et al., 2012). Fruit growth and development are accompanied by the production of primary and secondary metabolites, among which phenolic compounds are the most widely distributed (Chi et al., 2023, Liang et al., 2020). Specifically, loquats are rich in phenolics and antioxidants that benefit human health (Ding et al., 2001). At maturity, the total phenolic and flavonoid contents of CH1 were significantly higher than those of DWX (Fig. 1B, C). Phenolics impart fruit flavor, which might explain the distinctive texture of CH1 (Zou et al., 2020). This study used the DPPH and FRAP methods, which are commonly used to evaluate the antioxidant capacity of loquat fruits. Different from that of DWX, the strong antioxidant capacity of CH1 was attributed to its richness in phenolics and stone cells (Fig. 1H, I, and Fig. 2) (Xu & Chen, 2011). The results were consistent with those of studies on loquat leaves and strawberry pulp, in which total phenolic content was positively correlated with antioxidant activity (Dhiman et al., 2022, Koraqi et al., 2023). Phenolics are primarily synthesized by the phenylpropane pathway, and PAL, C4H, and CAD are the essential rate-limiting enzymes (Qi et al., 2021, Zulhendri et al., 2021). In this study, the activities of PAL, 4CL, C4H, and CAD showed a decreasing trend, probably related to the decrease in phenolic content at the late stages of development (Zhang et al., 2023).
The phenolic characterization was thoroughly assessed during the breeding process. This provides breeders with specific information about the characteristics of new cultivars and helps them understand their genetic background. Additionally, the antioxidant capacity and phenolics of different cultivars were evaluated to produce fruits with health-promoting characteristics. Quality parents with flavorful antioxidant properties and rich phenolics were selected. This ensures that superior traits are passed on to the progeny.
Fruit texture of DWX and CH1
Fruit lignification is a crucial factor affecting fruit texture and flavor, and pulp granularity (stone cells) is a manifestation of fruit lignification (Chi et al., 2023, Lin et al., 2022). The pearly, grainy flavor is rarely observed in the fruit of loquat cultivars, and consumers are not concerned with the presence of stone cells in loquat fruit. The apparent pear-like granular texture of CH1 was attributed to stone cells, which were not detected in DWX (Fig. 2), similar to the results of a study on wild ‘Taiwanese’ loquats (Lin et al., 2022). CH1 is the first cultivar in which stone cells were found. The pulp containing stone cells is usually rough, and the flavor is astringent (Luck et al., 1994). Stone cells were mostly isolated, and even within a cluster of stone cells, the number of stone cells in the loquat fruit was typically less than four (Fig. 2D, E). Notably, the diameter of stone cells in pear fruits with a delicate texture and high juice yield was less than 150 μm (Huang et al., 2019). The longitudinal and transverse diameters of stone cells in CH1 were less than 100 μm. This finding explains why CH1 pulps with stone cells did not show significant lignification but had a satisfactory taste and texture. The complete morphology of stone cells in CH1 was observed at S3, and understanding the initial formation period of the stone cells requires further investigation. In addition, new stone cell generation was observed in S5. This finding is consistent with that of a study on the loquat cultivar ‘Jiefangzhong’ (Lin et al., 2022). The results suggest that the stone cell traits of CH1 were inherited from the parents of the progeny.
Fruit flavor formation is based on the flavor imparted by soluble metabolites (sugars, acids), chewing (texture) determined by lignin composition, and aroma determined by volatile metabolites (An et al., 2023). Stone cells are closely associated with lignin synthesis and deposition (Huang et al., 2019, Lin et al., 2022). Similarly, the lignin content in CH1 was significantly higher than that in DWX S3–S5 (Fig. 3C). Fruit lignification leads to reduced juice quality and flavor (Wang et al., 2021). In this study, the increased lignin, coniferyl alcohol, and p-coumaric acid contents might be responsible for the rough texture of CH1 fruit (Chi et al., 2023). However, stone cells and lignin did not lead to a decrease in CH1 fruit quality. This result might be attributed to the enrichment of CH1 fruits with phenolic compounds, as well as the lower acid content and higher sugar content than those of DWX (data related to sugar metabolism have not been published). In addition, the low amount of stone cells provided CH1 with a sandy texture and unique flavor.
Phenolic profiles accompanying fruit development and ripening
The extraction and identification of phenolics have been extensively studied. In this study, the extraction and detection methods were modified. Specifically, vanillic acid, coniferyl alcohol, and cinnamic acid were determined, which have not been detected in other loquat cultivars (Fig. 3A) (Ding et al., 2001, Xu et al., 2014). Chlorogenic and neochlorogenic acid contents were high between stages S1 and S2 (the fruitlet stage). Similar results have been obtained in other studies on phenolic compounds from different loquat fruits (Ding et al., 2001, Xu et al., 2014). Neochlorogenic acid content decreased gradually in the later developmental stages; a plausible explanation for this decreasing trend is its conversion to downstream forms (Liang et al., 2020). The taste of chlorogenic, neochlorogenic, and cryptochlorogenic acids is sour, bitter and astringent (Arai, Suzuki, Fujimaki, & Sakurai, 1966). The astringent taste of CH1 fruits resulted from the accumulation of these three acids, but the astringency was not strong and was mainly due to the rough texture.
In detail, coniferyl alcohol was also a main phenolic substance in the loquats (Fig. 3B). Notably, in this study, chlorogenic acid and coniferyl alcohol, suggested to be biochemical markers of loquat fruit maturity, increased after harvest (Ding et al., 2001). Lignin profile determines the chewing characteristics of the fruit. A study showed that the lignin in pomelo comprised the S/G (Syringyl/Gualacyl) type and that coniferyl alcohol was the main component affecting chewing (An et al., 2023). Coniferyl alcohol is derived from cinnamic acid and is regulated by 4CL, which is involved in chewing trait production and storability. However, information on loquat is limited. The coniferyl alcohol content of CH1 in this study was significantly higher than that of DWX, and the content of CH1 at maturity was approximately five times that of DWX. Therefore, the high content of coniferyl alcohol is a potential reason for the unique taste characteristics of CH1.
Moreover, lignin is crucial in maintaining cell wall structure and influencing fruit texture (Chi et al., 2023). The concentrations of several critical intermediates of the lignin synthesis pathway, namely, p-coumaric acid, sinapic acid, and coniferyl alcohol, were higher in CH1 than in DWX, particularly in S5. Thus, we hypothesized that the difference in phenolics between DWX and CH1 is a reason for the difference in flavor. The downregulation of sinapyl alcohol and coniferyl alcohol was the main factor responsible for the taste smoothness of triploid loquat (Chi et al., 2023). However, the up-regulation of these two phenolics in CH1 might explain the rough taste. These results provide novel insights into the dynamic changes in various phenolic substances in the newly released CH1 and their effects on fruit texture.
Molecular mechanisms underlying differences in flavor quality between DWX and CH1
To clarify the molecular mechanism of action underlying the phenolic compounds and fruit texture in CH1 and DWX, we sequenced the transcriptomes at five critical periods of fruit development. KEGG analysis showed that the DEGs were primarily enriched for phenylpropanoid biosynthesis and plant hormone signal transduction pathways (Fig. 4).
Phenolics, such as lignin, are the final metabolites of the phenylpropanoid synthesis pathway (Zulhendri et al., 2021). The special presence of stromal cells in CH1 was a major factor contributing to flavor, which was associated with lignin deposition (Fig. 2). PAL, C4H, and 4CL are the key genes involved in the initial stages of phenylpropanoid metabolism. PAL and 4CL exhibited low expression in CH1, and C4H exhibited high expression. The high expression of C4H may be a feedback inhibitory regulator of trans-cinnamate PAL activity and was associated with lignin accumulation (Lan, Liu, Liu, Li, Li, & Yuan, 2023). Notably, the EVM0015180 gene encoding C4H, which might be a key structural gene regulating the metabolism of the primary products of phenylpropanoid metabolism, was significantly and highly expressed (Fig. 5). CCR, CAD, F5H, COMT, and CCoAOMT are the key genes involved in lignin biosynthesis (Tang et al., 2019). In our study, most of the genes, namely, HCT, CCR, CAD, F5H, COMT, and CCoAOMT, were highly expressed at the early stages of CH1, suggesting that phenolics, such as lignin, are synthesized at the early stages of loquat development and accumulate at later developmental stages. A similar mechanism was revealed for phenolic metabolism in kiwifruit (Liang et al., 2020).
Lignin is polymerized from sinapyl, coniferyl, and p-coumaryl alcohols, in which CAD genes play a key regulatory role (Qi et al., 2021). In this study, 14 DEGs annotated to CAD were identified, among which EVM0037025 and EVM0013715 were up-regulated in CH1 during the late developmental stages, a phenomenon that we hypothesized was related to the accumulation of coniferyl alcohol during the late developmental stages of CH1 fruit (Fig. 2, Fig. 3). The low coniferyl alcohol content in ‘Shatian’ pomelo and the mutated CAD and COMT family genes suggest differences in the lignin biosynthesis pathway and are reflected in the soft chewing trait (texture) of the ‘Shatian’ (An et al., 2023). By contrast, the high expression of CAD in CH1 increased the accumulation of coniferyl alcohol, contributing to the rough taste characteristics. F5H is a key enzyme regulating the S/G lignin composition in plants (Liu, Luo, & Zheng, 2018). During fruit growth and development, the F5H in CH1 was down-regulated compared with that of DWX, and the content of coniferyl alcohols, key substrates for the formation of G lignin, was higher in CH1 than in DWX. This result is consistent with that of a study on rice, in which F5H down-regulation increased the G lignin content (Takeda et al., 2017). In addition, the DEGs were significantly enriched in the plant hormone signal transduction pathway (Fig. 4). On the one hand, we hypothesized that the phenylpropanoid biosynthesis pathway is associated with hormone-regulated genes related to fruit ripening (Hong et al., 2022). On the other hand, the single fruit weight of CH1 was significantly lower than that of DWX in our previous study, which may be another factor in the enrichment of the phytohormone signaling pathway (Deng et al., 2023). In conclusion, we used KEGG analysis and identified phenylpropanoid metabolism pathway DEGs, which might be key differential structural genes for phenolic compound formation and fruit texture in CH1.
Candidate genes co-expressed with EVM0023576 (4CL9), EVM0027731 (POD42), EVM0016304 (F5H), and EVM0000602 (β-glucosidase) were identified using WGCNA. These genes included members of the MYB, bHLH, NCA, AP2, EFR, and WRKY gene families, which may be involved in the regulation of loquat phenolics and fruit texture (Fig. 6). Notably, EVM0016304 was also differentially expressed in KEGG analysis. The deletion of F5H in the Arabidopsis fah1-2 mutant produced G-rich lignin (Fan et al., 2020). By contrast, F5H overexpression in the fah1 mutant resulted in fewer coniferyl alcohol-derived G-units but more sinapyl alcohol-derived S-units than those in the wild-type plants. However, F5H was not significantly correlated with the synthesis of coniferyl alcohol, but was significantly correlated with lignin content (Fig. 6B). This decrease is related to the high expression level of CAD (EVM0037025 and EVM0013715) in CH1, which leads to the accumulation of coniferyl alcohol in CH1 during late development, consequently synthesizing G-type lignin; the lignin content was significantly higher at S5 in CH1 than in DWX. CAD regulates lignin synthesis in pear fruit pulp (Qi et al., 2021). In this study, CAD was not identified by WGCNA, suggesting that CAD may not be involved in the synthesis of chlorogenic acid, cryptochlorogenic acid, and neochlorogenic acid. 4CL and POD are regulators of the phenylpropane pathway, suggesting that EVM0023576 (4CL9) and EVM0027731 (POD42) perform similar regulatory functions (Chen, Deng, Ruan, Yi, & Zeng, 2021). Several phenolic compounds, typically in the form of glycosides, can be hydrolyzed by β-glucosidase to release volatile phenolic components (Zhang, Zhang, Sirisena, Gan, & Fang, 2021). EVM0000602 encoding β-glucosidase is speculated to participate in the hydrolysis of phenolics. Up-regulation of β-glucosidase also plays an essential role in accelerating fruit lignification. Thus, further research should investigate volatile phenolics.
Transcription factors that affect the phenylpropanoid biosynthesis pathway by regulating structural genes are primarily found in the MYB, bHLH, and NAC transcription factor families. For example, the switchgrass PvMYB42/85 increased the lignin content, lignin S/G ratio, and expression of lignin biosynthesis genes through exons encoding F5H (Miyamoto, Tobimatsu, & Umezawa, 2020); bHLH regulated the phenylpropane pathway by forming a complex with MYB and WD40 (Liang et al., 2020); NAC acted as a master switch gene that was directly or indirectly regulated by other transcription factors implicated in the regulation of lignin biosynthesis, namely, the transcription factors MYB, WRKY, and bHLH (Miyamoto et al., 2020). In this study, the four hub genes were regulated by transcription factors, such as MYB, bHLH, and NCA, and the heat maps of the four structural genes and transcription factors were mapped (Fig. 6D and Supplementary Fig. 4). The expression patterns of EVM0000602, EVM0016304 with EVM0033763 (bHLH TF), and EVM0027731 with EVM0018467 (MYB TF) and EVM0038207 (NAC TF) were clustered; thus, we hypothesized that EVM0009881, EVM0033763, EVM0018467, and EVM0038207 were the key transcription factors regulating these structural genes. These results are similar to those of a study on kiwifruit, which identified the phenolic components regulated by the transcription factors MYB and bHLH (Liang et al., 2020). Thus, further research should verify the functions and mechanisms of action of these candidate genes in the phenolic acid metabolism of CH1.
Conclusions
The aim of this study was to investigate the effects and underlying molecular mechanisms of phenolics on fruit flavor in CH1 and DWX during growth and development. The results showed the main flavor differences between CH1 and DWX were in fruit texture. Stone cells were detected in CH1 but not in DWX. At maturity, the lignin content, antioxidant capacity, and phenolic content of CH1 were higher than that of DWX. HPLC analysis showed that CH1 contained high levels of coniferyl alcohol during the late stages of fruit development, a novel finding in loquat. Chlorogenic acid, neochlorogenic acid, and coniferyl alcohol are the main phenolic compounds in loquat, and the high content of coniferyl alcohol is a potential factor of the rough texture of CH1. Transcriptome analysis indicated that phenylpropanoid metabolism was activated in CH1 during the formation of phenolic compounds and fruit texture. KEGG analysis identified 51 candidate structural genes, and WGCNA identified four structural genes and 88 transcription factors. CAD and F5H were identified in KEGG analysis and WGCNA, respectively, which affected CH1 lignin synthesis and fruit texture, forming stone cells. The functions of these genes in CH1 remain unclear, but we plan to investigate them in further research.
CRediT authorship contribution statement
Kun Zhang: Conceptualization, Data curation, Visualization, Writing – original draft, Writing – review & editing. Qiaoli Ma: Data curation, Investigation. Yang Wang: Data curation, Methodology. Zhenchao Yuan: Methodology. Zhiwu Yang: Formal analysis. Xian Luo: Software, Validation. Huifen Zhang: Funding acquisition, Resources. Hui Xia: Validation, Visualization. Xiulan Lv: Writing – review & editing. Yongqing Wang: Supervision, Writing – review & editing. Qunxian Deng: Conceptualization, Funding acquisition, Project administration, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This study was funded by the program of Science and Technology Department of Sichuan Province (2021YFYZ0023-07), the Fruit Innovation Team of Sichuan Province within the National Modern Agricultural Industrial Technology System (sccxtd-2023-04), the Science and Technology Department of Sichuan Province program (2022NSFSC0092), the Agricultural Public Security and Resource Protection of Sichuan Province (035/2212129087), the Sichuan Tianfu New Area Rural Revitalization Research Institute “Unveiled the List of Marshals” project (XZY1-05), and the Project of Cooperation between Ya’an and Sichuan Agriculture University (063/063H2103).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fochx.2024.101145.
Contributor Information
Kun Zhang, Email: zhangkun@stu.sicau.edu.cn.
Qiaoli Ma, Email: 2020205001@stu.sicau.edu.cn.
Yang Wang, Email: 2020105004@stu.sicau.edu.cn.
Zhenchao Yuan, Email: 2023305054@stu.sicau.edu.cn.
Xian Luo, Email: ffeetalk@sicau.deu.cn.
Yongqing Wang, Email: yqw14@sicau.edu.cn.
Qunxian Deng, Email: dengqx@sicau.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- An Y., Zhu Q., Lv H., Zhang X., Huang F., Guo Y., Xu L. Genomic basis of metabolome-mediated cultivar-specific flavor formation in juice sacs of the pomelo (Citrus grandis (L.) Osbeck) cultivars Shatian and Guanxi honey. LWT. 2023;191 [Google Scholar]
- Arai S., Suzuki H., Fujimaki M., Sakurai Y. Studies on flavor components in soybean: Part II. Phenolic acids in defatted soybean flour. Agricultural and Biological Chemistry. 1966;30(4):364–369. [Google Scholar]
- Chen O., Deng L., Ruan C., Yi L., Zeng K. Pichia galeiformis induces resistance in postharvest citrus by activating the phenylpropanoid biosynthesis pathway. Journal of Agricultural and Food Chemistry. 2021;69(8):2619–2631. doi: 10.1021/acs.jafc.0c06283. [DOI] [PubMed] [Google Scholar]
- Chi Z., Liu X., Wen S., Wang Y., Lv W., Guo Q., Liang G. Integrated metabolomic profiling and transcriptome analysis of fruit quality and ripeness in early-maturing seedless triploid loquat. Scientia Horticulturae. 2023;316 [Google Scholar]
- Deng H., Li X., Wang Y., Ma Q., Zeng Y., Xiang Y., Liang D. Organic acid accumulation and associated dynamic changes in enzyme activity and gene expression during Fruit development and ripening of common loquat and its interspecific hybrid. Foods. 2023;12(5):911. doi: 10.3390/foods12050911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhiman A., Suhag R., Thakur D., Gupta V., Prabhakar P.K. Current status of Loquat (Eriobotrya japonica Lindl.): Bioactive functions, preservation approaches, and processed products. Food Reviews International. 2022;38(sup1):286–316. [Google Scholar]
- Ding C., Chachin K., Ueda Y., Imahori Y., Wang C.Y. Metabolism of phenolic compounds during loquat fruit development. Journal of Agricultural and Food Chemistry. 2001;49(6):2883–2888. doi: 10.1021/jf0101253. [DOI] [PubMed] [Google Scholar]
- Fan D., Li C., Fan C., Hu J., Li J., Yao S., Luo K. MicroRNA6443-mediated regulation of FERULATE 5-HYDROXYLASE gene alters lignin composition and enhances saccharification in Populus tomentosa. New Phytologist. 2020;226(2):410–425. doi: 10.1111/nph.16379. [DOI] [PubMed] [Google Scholar]
- Ferreres F., Gomes D., Valentão P., Gonçalves R., Pio R., Chagas E.A., Andrade P.B. Improved loquat (Eriobotrya japonica Lindl.) cultivars: Variation of phenolics and antioxidative potential. Food Chemistry. 2009;114(3):1019–1027. [Google Scholar]
- Hong C.P., Kim C.-K., Lee D.J., Jeong H.J., Lee Y., Park S.-G., Kwon S.-J. Long-read transcriptome sequencing provides insight into lignan biosynthesis during fruit development in Schisandra chinensis. BMC Genomics. 2022;23(1):1–14. doi: 10.1186/s12864-021-08253-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Zhu N., Zhu C., Wu D., Chen K. Morphology and cell wall composition changes in lignified cells from loquat fruit during postharvest storage. Postharvest Biology and Technology. 2019;157 [Google Scholar]
- Koraqi H., Petkoska A.T., Khalid W., Sehrish A., Ambreen S., Lorenzo J.M. Optimization of the extraction conditions of antioxidant phenolic compounds from strawberry fruits (Fragaria × ananassa Duch.) using response surface methodology. Food Analytical Methods. 2023;16:1030–1042. doi: 10.1007/s12161-023-02469-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan H., Liu R., Liu Z., Li X., Li B., Yuan Y. Biological valorization of lignin to flavonoids. Biotechnology Advances. 2023 doi: 10.1016/j.biotechadv.2023.108107. [DOI] [PubMed] [Google Scholar]
- Liang D., Deng H., Deng Q., Lin L., Lv X., Wang J., Xia H. Dynamic changes of phenolic compounds and their associated gene expression profiles occurring during fruit development and ripening of the Donghong kiwifruit. Journal of Agricultural and Food Chemistry. 2020;68(41):11421–11433. doi: 10.1021/acs.jafc.0c04438. [DOI] [PubMed] [Google Scholar]
- Lin S., Lin D., Wu B., Ma S., Sun S., Zhang T., Wu J. Morphological and developmental features of stone cells in Eriobotrya Fruits. Frontiers in Plant Science. 2022;13 doi: 10.3389/fpls.2022.823993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Luo L., Zheng L. Lignins: Biosynthesis and biological functions in plants. International Journal of Molecular Sciences. 2018;19(2):335. doi: 10.3390/ijms19020335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Li S., Feng X., Li L. Study on cell wall composition, fruit quality and tissue structure of hardened ‘Suli’ pears (Pyrus bretschneideri Rehd) Journal of Plant Growth Regulation. 2021;40:2007–2016. [Google Scholar]
- Liu X., Song L., Xue B., Chi Z., Wang Y., Wen S., Wang S. Organic acid and sugar components accumulation and flavor associated metabolites dynamic changes in yellow- and white-fleshed seedless loquats (Eriobotrya japonica) Food Chemistry. 2023;X doi: 10.1016/j.fochx.2023.101046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck G., Liao H., Murray N.J., Grimmer H.R., Warminski E.E., Williamson M.P., Haslam E. Polyphenols, astringency and proline-rich proteins. Phytochemistry. 1994;37(2):357–371. doi: 10.1016/0031-9422(94)85061-5. [DOI] [PubMed] [Google Scholar]
- Mamat A., Tusong K., Xu J., Yan P., Mei C., Wang J. Integrated transcriptomic and proteomic analysis reveals the complex molecular mechanisms underlying stone cell formation in Korla pear. Scientific Reports. 2021;11(1):7688. doi: 10.1038/s41598-021-87262-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchiosi R., dos Santos W.D., Constantin R.P., de Lima R.B., Soares A.R., Finger-Teixeira A., Abrahão J. Biosynthesis and metabolic actions of simple phenolic acids in plants. Phytochemistry Reviews. 2020;19:865–906. [Google Scholar]
- Miyamoto T., Tobimatsu Y., Umezawa T. MYB-mediated regulation of lignin biosynthesis in grasses. Current Plant Biology. 2020;24 [Google Scholar]
- Qi K., Song X., Yuan Y., Bao J., Gong X., Huang X., Tao S. CAD genes: Genome-wide identification, evolution, and their contribution to lignin biosynthesis in pear (Pyrus bretschneideri) Plants. 2021;10(7):1444. doi: 10.3390/plants10071444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumpf J., Burger R., Schulze M. Statistical evaluation of DPPH, ABTS, FRAP, and Folin-Ciocalteu assays to assess the antioxidant capacity of lignins. International Journal of Biological Macromolecules. 2023;233 doi: 10.1016/j.ijbiomac.2023.123470. [DOI] [PubMed] [Google Scholar]
- Shukla R., Chauhan N., Rajak C., Flora S. Flavors and fragrances. Flavor Development for Functional Foods and Nutraceuticals. 2019;141 [Google Scholar]
- Soto-Vaca A., Gutierrez A., Losso J.N., Xu Z., Finley J.W. Evolution of phenolic compounds from color and flavor problems to health benefits. Journal of Agricultural and Food Chemistry. 2012;60(27):6658–6677. doi: 10.1021/jf300861c. [DOI] [PubMed] [Google Scholar]
- Takeda Y., Koshiba T., Tobimatsu Y., Suzuki S., Murakami S., Yamamura M., Sakamoto M. Regulation of CONIFERALDEHYDE 5-HYDROXYLASE expression to modulate cell wall lignin structure in rice. Planta. 2017;246:337–349. doi: 10.1007/s00425-017-2692-x. [DOI] [PubMed] [Google Scholar]
- Tang Y., Liu F., Xing H., Mao K., Chen G., Guo Q., Chen J. Correlation analysis of lignin accumulation and expression of key genes involved in lignin biosynthesis of ramie (Boehmeria nivea) Genes. 2019;10(5):389. doi: 10.3390/genes10050389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Chen Q., Chen W., Guo Q., Xia Y., Wu D., Liang G. Melatonin treatment maintains quality and delays lignification in loquat fruit during cold storage. Scientia Horticulturae. 2021;284 [Google Scholar]
- Wang L., Shan T., Xie B., Ling C., Shao S., Jin P., Zheng Y. Glycine betaine reduces chilling injury in peach fruit by enhancing phenolic and sugar metabolisms. Food Chemistry. 2019;272:530–538. doi: 10.1016/j.foodchem.2018.08.085. [DOI] [PubMed] [Google Scholar]
- Wang L., Shao S., Madebo M.P., Hou Y., Zheng Y., Jin P. Effect of nano-SiO2 packing on postharvest quality and antioxidant capacity of loquat fruit under ambient temperature storage. Food Chemistry. 2020;315 doi: 10.1016/j.foodchem.2020.126295. [DOI] [PubMed] [Google Scholar]
- Wang Y. A draft genome, resequencing, and metabolomes reveal the genetic background and molecular basis of the nutritional and medicinal properties of loquat (Eriobotrya japonica (Thunb.) Lindl) Horticulture Research. 2021;8 doi: 10.1038/s41438-021-00657-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Chen J. Commercial quality, major bioactive compound content and antioxidant capacity of 12 cultivars of loquat (Eriobotrya japonica Lindl.) fruits. Journal of the Science of Food and Agriculture. 2011;91(6):1057–1063. doi: 10.1002/jsfa.4282. [DOI] [PubMed] [Google Scholar]
- Xu H., Li X., Chen J. Comparison of phenolic compound contents and antioxidant capacities of loquat (Eriobotrya japonica Lindl.) fruits. Food Science and Biotechnology. 2014;23:2013–2020. [Google Scholar]
- Xu S., Fang D., Tian X., Xu Y., Zhu X., Wang Y., Ma L. Subcritical water extraction of bioactive compounds from waste cotton (Gossypium hirsutum L.) flowers. Industrial Crops and Products. 2021;164 [Google Scholar]
- Zhang P., Zhang R., Sirisena S., Gan R., Fang Z. Beta-glucosidase activity of wine yeasts and its impacts on wine volatiles and phenolics: A mini-review. Food Microbiology. 2021;100 doi: 10.1016/j.fm.2021.103859. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Zhang W., Wang H., Shu C., Chen L., Cao J., Jiang W. The combination treatment of chlorogenic acid and sodium alginate coating could accelerate the wound healing of pear fruit by promoting the metabolic pathway of phenylpropane. Food Chemistry. 2023;414 doi: 10.1016/j.foodchem.2023.135689. [DOI] [PubMed] [Google Scholar]
- Zhou J., Yang S., Ma Y., Liu Z., Tu H., Wang H., Li M. Soluble sugar and organic acid composition and flavor evaluation of Chinese cherry fruits. Food Chemistry: X. 2023;20 doi: 10.1016/j.fochx.2023.100953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou S., Wu J., Shahid M.Q., He Y., Lin S., Liu Z., Yang X. Identification of key taste components in loquat using widely targeted metabolomics. Food Chemistry. 2020;323 doi: 10.1016/j.foodchem.2020.126822. [DOI] [PubMed] [Google Scholar]
- Zulhendri F., Chandrasekaran K., Kowacz M., Ravalia M., Kripal K., Fearnley J., Perera C.O. Antiviral, antibacterial, antifungal, and antiparasitic properties of propolis: A review. Foods. 2021;10(6):1360. doi: 10.3390/foods10061360. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.