Abstract
DNA methylation is also involved in the development and progression of cardiac diseases. Although studies have shown that DNA methylation and RNA m6A methylation play an important role in the development of myocardial hypertrophy, whether DNA methylation and RNA m6A methylation have a coordinated role in the development of myocardial hypertrophy and influence each other is still unknown. Here, we found that DNMT1 expression was downregulated in TAC mice and Ang II-treated NRCMs. Moreover, DNMT1 overexpression inhibited Ang II-induced apoptosis of NRCMs. Furthermore, we found that the expression of METTL3 was up-regulated after inhibiting the expression of DNMT1 by a DNMT1 inhibitor or small interfering RNA. In addition, ectopic expression DNMT1 inhibited METTL3 expression in NRCMs. Furthermore, METTL3 expression was elevated in NRCMs treated with Ang II, and suppression of METTL3 inhibited cell apoptosis induced by Ang II in NRCMs.In addition, this study revealed that the DNMT1/METTL3 pathway affected Ang II-induced apoptosis in NRCMs. Finally, this study found that DNMT1, but not METTL3, might directly regulated the ANP and BNP expression. Collectively, our findings revealed the role of the DNMT1/METTL3 pathway in cardiac hypertrophy and provided a novel molecular mechanism describing the physiological and pathological processes.
Keywords: Cardiac hypertrophy, DNA methylation, DNMT1, m6A RNA modification, METTL3
1. Introduction
More than 23 million people worldwide are affected by heart failure, which is the main cause of mortality and morbidity in cardiovascular diseases and a huge medical burden on society [1,2]. Hypertrophy is a physiological and pathological stimulus-induced adaptive cardiac remodeling that leads to increased cardiac wall stress and serves as a crucial predictor of cardiovascular mortality in the developmental and pathological progression of HF [3]. Cardiac hypertrophy undergoes from concentric to eccentric, resulting in heart fibrosis, enlargement, apoptotic loss of cardiomyocytes, eventually leading to HF which is often fatal [3,4]. Angiotensin II (Ang II), a neurohumoral factor, plays a crucial role in the developmental progression of heart hypertrophy through the activation of intracellular signaling pathways by interacting with membrane receptors, leading to the transcriptional modulation of hypertrophy-related genes [[5], [6], [7]]. Therefore, further investigation of the molecular mechanisms underlying the action of Ang II will provide a huge advantage in protecting cardiomyocytes from enlargement and cell death during cardiac hypertrophy.
DNA methylation, identified as the fifth base, is the procedure for the selective adjunction of methyl groups to cytosine at CpG islands of gene promoters [8,9]. There are two main classes of DNA methyltransferases in eukaryotic cells: DNMT3A and DNMT3B, which are responsible for de novo synthesis of methylation, and DNMT1, which is responsible for the maintenance of methylation synthesis [10]. DNA demethylation induces gene reactivation and promotes gene expression, whereas hypermethylation normally suppresses gene expression [11]. DNA methylation exerts important effects on a variety of cellular processes, including chromatin structure maintenance, genomic imprinting, inactivation of the X chromosome, embryonic development, and carcinogenesis by modulating gene expression [11,12]. DNA methylation is involved and further, enforced influence in the occurrence and development of cardiac hypertrophy [13,14], which was demonstrated by Tong-Tong Wu and his colleagues in the heart tissues of HF mice [15]. Meder et al. analyzed plasma-treated hiPSC-CMs and cardiac biopsies using array-based DNA methylation analysis and revealed prevalent alterations in cardiac DNA methylation; they further revealed that hypomethylation of the ATG promoter could serve as a diagnostic marker of HF [16]. Cameron et al. found large differences in DNA methylation at over 16,400 CpG methylation sites, particularly in the canonical signaling pathway of Cardiac Hypertrophy in neonates [17]. However, further studies are needed to elucidate the involvement and function of DNA methylation in cardiac hypertrophy.
N6-methyladenosine (m6A) methylation is defined as the adjunction of methyl to adenosine (A) in RNA transcripts [18]. The m6A process is reversible, and the main recognition site for most RNA methylases is RRACH [19,20]. The dynamic m6A RNA modification is maintained by m6a writers—adenosine methyltransferases, which consist of WTAP, METTL14, METTL3, METTL16 and; m6a erasers—demethylases, which include ALKBH5 and FTO; and m6a readers—binding proteins comprising HNRNPC, HNRNPA2B1, YTHDF2, YTHDF1, and eIF3 [21,22]. Methylation modification of mRNA 3′UTR region affects the RNA export, nuclear transport, translation initiation, and polyA binding protein maintain the structural stability of mRNA, and methylation modification in the 5′UTR region exerts a crucial role in RNA splicing, editing, polyadenylation, stability, degradation, etc. [23]. m6A RNA modification participates in the development and progression of cardiovascular diseases. Dorn et al. demonstrated that enhanced METTL3-mediated m6A methylation leads to cardiac hypertrophy [24]. However, further investigation is needed to determine whether mettl3 has a significant effect on cardiac hypertrophy.
Thus, the aim of our study was to verify the potential molecular mechanisms underlying the actions of DNA methylation in cardiomyocyte hypertrophy and to reveal the hypothesis that, during the occurrence and development of myocardial hypertrophy, the expression of DNA methyltransferase 1 was down-regulated, which leads to the decrease of DNA methylation level of the promoter of RNA methyltransferase METTL3, thus upregulating the expression of METTL3, causing the increase of apoptosis of cardiomyocytes, and finally promoting the pathological process. These findings improve our theoretical understanding and suggest that the DNMT1/METTL3 pathway is a potential diagnostic and therapeutic biomarker for cardiomyocyte hypertrophy.
2. Methods
2.1. Experimental animals
All animal studies were supervised by the Ethics Committee on Animal Experimentation at the First Affiliated Hospital of USTC and were conducted according to the guidelines for the care and use of laboratory animals issued by the China Animal Welfare Commission. Slack Laboratory Animal Co., Ltd. (Shanghai, China) supplied the six-week-old C57BL/6 male mice. Animals were reared at constant temperature (21 ± 1 °C) and maintained in a room with water and food during a 12 h dark/light cycle. After acclimatizing for a week, the animals were randomly assigned to two groups: model (TAC) and control (sham). In the TAC group, the aortic arch of the mice was constricted using a 27 G needle and 6.0 sutures. The Sham group was operated in the same way as the TAC group without ligation of the aorta. All mice were housed for four weeks for further research.
2.2. Echocardiography (ECG)
The mice were anesthetized after four weeks. Cardiac function was examined using high-frequency ultrasonic equipment (VEVO 2100) equipped with a high-frequency transducer (MS 400; VisualSonics, Fujifilm).
2.3. Hematoxylin-Eosin (HE) staining and Masson staining
The pathological condition of the hearts of mice was measured by HE and Masson staining. Briefly, after the anesthetization of all mice, the hearts were extracted and fixed in paraformaldehyde (PFA, 4 %). Hematoxylin and eosin (HE and Masson staining were performed and the results were recorded by a professional technology company (Servicebio, Wuhan, China).
2.4. NRCMs isolation and culture
Isolation of NRCMs from Sprague–Dawley rats (Male,1 to 3-day-old) was performed according to a previously described [24]. Ventricles from neonatal rats were shredded into ~1 mm3 pieces using surgical scissors. After digesting with an enzyme solution (Junxing, Suzhou, China) at 37 °C for 1.5 h, all cells were harvested by centrifugation. After pre-plating for 1 h, the NRCMs were transferred to a new plate coated with 0.1 % gelatin. NRCMs were growth in Dulbecco's modified Eagle's medium (Servicebio, Wuhan, China) and placed in a closed environment with 5 % CO2 at 37 °C. NRCMs cultures were supplemented with 1 % streptomycin/penicillin (Sangon) and 10 % fetal bovine serum (Sangon).
2.5. Identification of NRCMs
Immunofluorescence staining was performed to identify NRCMs. NRCMs were washed with PBS which was pre-cold in °C for 1.5 h, and the following steps were performed: set with PFA (4 %) for 35 min at room temperature; permeated use Triton X-100 (0.5 %) for 10 min at room temperature; incubated with BSA (3 %) at RT for 60 min; marked with α-Actinin antibodies (Abcam 50599, dilution of 1:100) about 16 h at 4 °C; marked with the Alexa-fluor-488 labeled secondary antibodies (Biolegend, USA, 1:2000 dilution). Hoechst staining was used to stain nuclei. The results were recorded using a fluorescence microscope (Olympus, Tokyo, Japan).
2.6. Plasmid construction, cell transfection, virus package and infections
Overexpression of DNMT1 (Oe-DNMT1) and METTL3 (Oe-METTL3) was achieved using the lenti-viral plasmid pLVX-Puro (Oe-NC). DNMT1 and METTL3 depletion (shDNMT1s and shMETTL3s) was achieved by inserting short hairpin RNA against DNMT1 and METTL3 into the lenti-viral plasmid pLKO.1-puro (shNC).
All plasmids were transfected into 293t cells using high-efficiency transfection reagents (Invitrogen, USA) for 48 h following the manufacturer's protocol. Media containing the virus particles were collected, used to infect NRCMs, and prepared for further experiments.
2.7. Reverse transcription-quantitative PCR (RT-qPCR)
An RNAs extraction kit (Junxin) was used to isolate RNAs from the neonatal rat cardiomyocytes and mouse hearts. A reverse transcription kit (CWbio, Beijing, China) was used for reverse transcription. A qPCR detection kit (Sangon, Shanghai, China) was used for real-time PCR according to the manufacturer's instructions. The relative gene expressions were calculated by normalizing to internal standard gene β-Actin according to equation 2−ΔΔCT. The primer sequences (Genewiz, Suzhou, China) are listed in Table 1.
Table 1.
The primers used in study.
| Genes | Sequence | |
|---|---|---|
| DNMT1 | Forward | 5′-GAGAGCCACTGTCCTGGTTC-3′ |
| Reverse | 5′-AGCTTATGGGCTATGACGCC-3′ | |
| METTL3 | Forward | 5′-TCATCTTGGCTCTATCCGGCG-3′ |
| Reverse | 5′-CGTGTCCGACATCCTAGCTC-3′ | |
| ANP | Forward | 5′-TCGGAGCCTACGAAGATCCA-3′ |
| Reverse | 5′-ACACACCACAAGGGCTTAGG-3′ | |
| BNP | Forward | 5′-GGAGAACACGGCATCATTGC-3′ |
| Reverse | 5′-GGCTAGGACTTCCCAGAGGA-3′ | |
| β-Actin | Forward | 5′-GCAGGAGTACGATGAGTCCG-3′ |
| Reverse | 5′-ACGCAGCTCAGTAACAGTCC-3′ | |
2.8. Western immunoblotting analysis
Neonatal rat cardiomyocytes were subjected to western blotting following standard procedures. First, proteins were prepared in RIPA buffer (Junxin, Suzhou, China). Secondly, 40 μg lysate samples were loaded by SDS-PAGE (10 %) gel and migrated via electrophoresis to nitrocellulose (NC) membranes (Solarb, Beijing, China). Then the blots were incubated with non-fat dry milk (5 %) at RT for 1 h. Subsequently, proteins were hybridized with different primary antibodies (anti-β-Actin, dilution of 1:1000; anti-DNMT1, dilution of 1:1000; anti-METTL3, dilution of 1:1000; anti-ANP, dilution of 1:1000; anti-BNP, dilution of 1:1000) from Abcam about 12 h at 4 °C. Next, the membrane was incubated with different secondary antibodies (Abcam) at room temperature for 1 h. Antigen-antibody binding was determined using an ECL reagent (Junxin) according to the manufacturer's instructions.
2.9. Assessment of cell viability
Cell Counting Kit 8 (CCK8) kits (Solarbio, Beijing, China) was used to assess NRCMs viability. Briefly, added 10 μL CCK-8 regents into the 96-plate well, plated the cells in a cell incubator for 2 h. A microplate reader (Shanpu, Shanghai, China) was used to measure the absorbance of the cells at 450 nm. Cell survival rates were computed using the following equation: cell survival rate (%) = (Atest group– Ablank)/(Acontrol group–Ablank) × 100 %.
2.10. Apoptosis assay
Apoptosis in NRCMs was observed using a TUNEL kit (Elabscience, Wuhan, China), according to the manufacturer's instructions. Briefly, cells were washed by pre-cold PBS, and the following steps were performed: set with PFA (4 %) at RT for 20 min; permeated use Triton X-100 (0.5 %) at RT for 20 min; incubated with 50 μl TUNEL detection regents at 37 °C for 60 min in dark, and stained with Hoechst solution at 37 °C for 15 min in dark. The results were recorded using a fluorescence microscope (Olympus, Tokyo, Japan).
2.11. Chromatin immunoprecipitation (Chip) and RNA immunoprecipitation (RIP) assay
Chip assay was operated using Chip Immunoprecipitation Kit (Millipore) based on the manufacturer's protocol. In short, the cells were collected and treated with formaldehyde so that the DNA-protein interaction was cross-linked and fixed. The whole cell lysate was obtained by RIPA lysate. Ultrasound treats the cells, breaking the genomic DNA to 100-500bp. Then, the lysates were incubated with protein A/B beads (Biolinkedin, Shanghai, China), anti-DNMT1 antibody (Abcam) or control IgG. The next day, elution is performed and the DNA is uncrosslinked. The DNA was isolated using an DNA extraction kit (Tiangen). The enrichment of promoters were measured using qPCR assay.
RIP assay was operated using RIP Immunoprecipitation Kit (Millipore) based on the manufacturer's protocol. In short, cells were lysed in RIPA buffer combined with RNase inhibitor and protease inhibitor. Then, the lysates were incubated with protein A/B beads (Biolinkedin, Shanghai, China), anti-METTL3 antibody (Abcam) or control IgG. Next day, the immunoprecipitated RNA were digested with proteinase K and isolated using an RNAs extraction kit (Junxin). The enrichment of RNA transcripts were measured using qPCR assay.
2.12. Statistical analysis
All data were expressed as means ± standard error of the mean (SEM). Statistical analysis was performed using one-way analysis of variance (ANOVA) or an unpaired two-tailed Student's t-test. The p-value of 0.05 and 0.01 was set to represent statistical significance level of two-tailed p-value. All the figures were drawn using Adobe Illustrator CS6 (Adobe Corporation, USA).
3. Results
3.1. DNMT1 was decreased in transverse aortic constriction mice
To determine whether DNMT1 participates in cardiac hypertrophy, we established a mouse model of TAC-induced cardiac hypertrophy. This was successfully verified by electrocardiography (ECG), hematoxylin and eosin (HE) staining, and Masson staining (Fig. 1A–C). Furthermore, qPCR results of qPCR discovered that both ANP and BNP expression was elevated in the cardiac hypertrophy mouse model (TAC group) compared to control mice (Sham group) (Fig. 1D and E). As shown by qPCR and western blotting assays, this study concluded that DNMT1 was downregulated in the cardiac hypertrophy mouse model (TAC group) compared to control mice (sham group) (Fig. 1F and G). Collectively, these results indicated that DNMT1 influences the progression of cardiac hypertrophy.
Fig. 1.
DNMT1 expression was elevated in Transverse aortic constriction (TAC) mice.
(A) Identification of the TAC mouse model by electrocardiography (ECG). FS, fractional shortening; EF, ejection fraction. (B and C) Identification of the TAC mouse model by Hematoxylin-Eosin (HE) staining and Masson analysis. (D and E) ANP and BNP expressions were discovered by RT-qPCR analysis. (F and G) DNMT1 expression was measured by RT-qPCR and western blots analysis. β-Actin was determined to normalize the gene expression. Statistically difference was marked as *P < 0.05, statistically significant difference was marked as **P < 0.01.
3.2. Ang II induced apoptosis of NRCMs
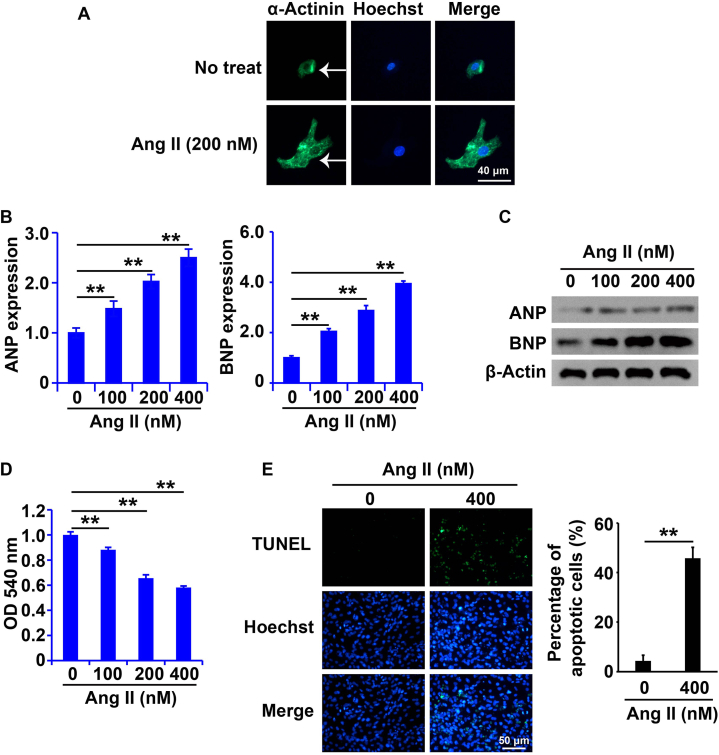
To determine whether Ang II-treated NRCMs could serve as cell models of cardiac hypertrophy, we isolated and verified primary NRCMs. As shown in Fig. 2A, we successfully isolated NRCMs and identified by α-Actinin immunofluorescence assay (Fig. 2A), which were then treated with Ang II (0, 100, 200, and 400 nM) for 48 h. Ang II increased the mRNA and protein levels of ANP and BNP in NRCMs (Fig. 2B and C). Following Ang II treatment, cell viability was reduced (Fig. 2D), and apoptosis was induced in NRCMs (Fig. 2E). These results clearly confirm that Ang II induces cardiomyocyte apoptosis.
Fig. 2.
Cell apoptosis was induced in NRCMs handled with Ang II.
(A) Identification of NRCMs by immunofluorescence staining analysis. (B and C) ANP and BNP expressions were measured by RT-qPCR and western blots analysis. (D) Cell viability was tested by CCK8 analysis. (E) Cell apoptosis was discovered by TUNEL analysis. β-Actin was determined to normalize the gene expression. Statistically difference was marked as *P < 0.05, statistically significant difference was marked as **P < 0.01.
3.3. Ang II inhibited DNMT1 expression in NRCMs
To determine whether DNMT1 expression was altered in NRCMs following Ang II treatment, RT-qPCR combined with Western blot assays were employed to examine DNMT1 expression in cardiac hypertrophic models. The expression of DNMT1 in NRCMs decreased with increasing concentrations of Ang II (Fig. 3A). The lenti-virus system was used to overexpress or silence DNMT1 expression in NRCMs. Compared with oe-NC, oe-DNMT1 markedly elevated DNMT1 expression in NRCMs (Fig. 3B). Meanwhile, shDNMT1 dramatically inhibited DNMT1 expression in NRCMs (Fig. 3C).
Fig. 3.
DNMT1 expression was decreased in NRCMs handled with Ang II.
(A and B) DNMT1 expression was disclosed by RT-qPCR and western blots assay. (C) DNMT1 expressions were disclosed by RT-qPCR and western blots analysis. (D and E) ANP and BNP expression was measured by RT-qPCR and western blots analysis. (F) CCK8 kit was introduced to analysis the cell viability. (G) TUNEL analysis was applied to test the cell apoptosis. β-Actin was determined to normalize the gene expression. Statistically difference was marked as *P < 0.05, statistically significant difference was marked as **P < 0.01.
To elucidate whether DNMT1 plays a crucial role in cardiac hypertrophic models, NRCMs were divided into four groups: DMSO + oe-NC, Ang II + oe-NC, DMSO + oe-DNMT1, and Ang II + oe-DNMT1. Interestingly, oe-DNMT1 inhibited the increased ANP and BNP mRNA and protein levels in NRCMs compared to control cells and repressed the elevation of genes in NRCMs with Ang II treatment (Fig. 3D and E). In addition, oe-DNMT1 enhanced cell viability and reduced apoptosis in NRCMs treated with or without Ang II (Fig. 3F and G). Therefore, these results demonstrated that decreased DNMT1 promotes apoptosis in NRCMs treated with Ang II.
3.4. DNMT1 inhibited METTL3 expression by influencing the DNA methylation of METTL3 promoters in NRCMs
RT-qPCR and western blotting were performed to determine whether DNMT1 regulates METTL3 expression in NRCMs. The results showed that oe-DNMT1 decreased METTL3 expression (Fig. 4A and B), and conversely, shDNMT1 increased METTL3 expression in NRCMs (Fig. 4C and D).
Fig. 4.
DNMT1 inhibited METTL3 expression by influencing DNA methylation of METTL3 promoter in neonatal rat cardiomyocytes (NRCMs).
(A–D) METTL3 expression was disclosed by RT-qPCR and western blots analysis. (E–F) DNMT1 and METTL3 expression was disclosed by RT-qPCR and western blots analysis. (G) The DNA methylation level in METTL3 promoter was disclosed by MSP analysis. β-Actin was determined to normalize the gene expression. Statistically difference was marked as *P < 0.05, statistically significant difference was marked as **P < 0.01.
To elucidate the molecular mechanism underlying METTL3 regulation by DNMT1, NRCMs were treated with a DNMT1 inhibitor, AZA (5-Aza-2′-deoxycytidine) (1 μM), for 24 h. Real-time PCR and Western blot analyses showed increased DNMT1 expression and decreased METTL3 expression in NRCMs (Fig. 4E and F). MSP analysis showed that oe-DNMT1 increased DNA methylation of the METTL3 promoter (Fig. 4G). Collectively, these results indicate that DNMT1 inhibits METTL3 expression in NRCMs.
3.5. Ang II up-regulated METTL3 expression in cardiac hypertrophy
RT-qPCR and Western blot analyses were used to determine METTL3 expression in cardiac hypertrophy models. First, METTL3 expression was elevated in the cardiac hypertrophic models (Fig. 5A). This study revealed that METTL3 expression in NRCMs was elevated with increasing concentrations of Ang II (Fig. 5B). Subsequently, the lenti-virus system was used to overexpress METTL3, and qPCR and western blotting assays revealed that shMETTL3 dramatically inhibited METTL3 expression in NRCMs (Fig. 5C).
Fig. 5.
Ang II elevated the METTL3 expression in NRCMs.
(A–C) METTL3 expression was disclosed by RT-qPCR and western blots analysis. (D and E) ANP and BNP expressions were measured by RT-qPCR and western blots analysis. (F) CCK8 kit was introduced to analysis the cell viability. (G) TUNEL analysis was applied to test the cell apoptosis. β-Actin was determined to normalize the gene expression. Statistically difference was marked as *P < 0.05, statistically significant difference was marked as **P < 0.01.
To determine whether METTL3 exerts an important influence on cardiac hypertrophy, NRCMs were divided into four different groups: DMSO + shNC, Ang II + shNC, DMSO + shMETTL3, and Ang II + shMETTL3. shMETTL3 inhibited the increases in ANP and BNP mRNA and protein levels in NRCMs treated with or without Ang II (Fig. 5D and E). In addition, hMETTL3 enhanced cell viability and reduced apoptosis in NRCMs treated with or without Ang II (Fig. 5F and G). Thus, this study demonstrated that increased METTL3 expression promotes apoptosis in NRCMs treated with Ang II.
3.6. DNMT1/METTL3 pathway affected the apoptosis of Ang II-treated NRCMs
Now that we know that DNMT1 can repress METTL3 expression in NRCMs, we determined whether DNMT1 can suppress the upregulation of METTL3 expression by Ang II. NRCMs were divided into three groups: DMSO + oe-NC, Ang II + oe-NC, and DMSO + oe-DNMT1. oe-METTL3 inhibited the increased expression of METTL3 in NRCMs following Ang II treatment (Fig. 6A and B). The lenti-virus system was used to overexpress METTL3, and oe-METTL3 markedly promoted the expression of METTL3 in NRCMs (Fig. 6C and D).
Fig. 6.
DNMT1/METTL3 pathway affected cardiomyocyte hypertrophy and apoptosis of NRCMs handled with Ang II.
(A–D) METTL3 expression was disclosed by RT-qPCR and western blots analysis. (E) ANP and BNP expressions were measured by RT-qPCR analysis. (F) CCK8 kit was introduced to analysis the cell viability. (G) TUNEL analysis was applied to test the cell apoptosis. β-Actin was determined to normalize the gene expression. Statistically difference was marked as *P < 0.05, statistically significant difference was marked as **P < 0.01.
To test the role of the DNMT1/METTL3 pathway in NRCMs treated with Ang II, NRCMs were divided into four groups: DMSO + oe-NC, Ang II + oe-NC, DMSO + oe-DNMT1, and Ang II + oe-DNMT1+oe-METTL3. Interestingly, oe-METTL3 alleviated the effects of oe-DNMT1 on hypertrophy and apoptosis of NRCMs handled with Ang II (Fig. 6E–G). In summary, this study showed that treatment with Ang II induces apoptosis of NRCMs via modulation of the DNMT1/METTL3 pathway.
3.7. DNMT1, but not METTL3, might directly regulated the ANP and BNP expression
This study demonstrated that DNMT1/METTL3 pathway affected the ANP and BNP expression. Thus, we wonder that weather DNMT1/METTL3 pathway could directly modulated the expression of ANP and BNP. Firstly, the results of Chip assay demonstrate that DNMT1 antibody could recruit the promoter of ANP and BNP (Fig. 7A), suggesting that DNMT1 might directly regulated the expression of ANP and BNP via binding the promoter of ANP and BNP. Then, the results of RIP demonstrated that METTL3 couldn't recruit the RNA transcripts of ANP and BNP (Fig. 7B), indicating that METTL3 couldn't directly regulated the expression of ANP and BNP. Collectively, this study showed that DNMT1, but not METTL3, might directly regulated the ANP and BNP expression.
Fig. 7.
DNMT1, but not METTL3, might directly regulated the ANP and BNP expression.
(A) The binding of DNMT1 and the promoter of ANP and BNP was disclosed by Chip analysis. (B) The binding of METTL3 and mRNA transcripts of ANP and BNP was disclosed by RIP analysis. Statistically difference was marked as *P < 0.05, statistically significant difference was marked as **P < 0.01.
4. Discussion
Cardiac hypertrophy is a pathological phenomenon of HF and the main cause of mortality and morbidity in cardiac diseases [25]. Cardiac hypertrophy causes changes in cardiac function, shape, and structure by modulating gene expression, resulting in altered cell growth, remodeling, and metabolism [26,27]. Thus, investigating the regulatory mechanisms of genes that exert an important influence on cardiac hypertrophy is important.
As the main class of DNA modification, DNA methylation is initiated by several DNMTs, including DNMT3a, DNMT3b, and DNMT1 [8]. DNA methylation represses gene transcription, resulting in expression at both the RNA and protein levels [28]. DNA methylation is involved in cellular progression, including tumorigenesis, cell development, metastasis, apoptosis, cell cycle, and growth [29]. Increasing evidence indicates that DNA methylation influences the occurrence and development of human diseases [30,31]. However, the effects and molecular mechanisms of DNA methylation in hypertrophy remain controversial. DaLiao Xiao and colleagues revealed that the inhibitor of DNA methylation, 5-aza-2′-deoxycytidine, attenuated norepinephrine-induced cardiac hypertrophy [32]. Fang et al. reported that DNA hypomethylation alters the expression profile of genes involved in cAMP-induced cardiac hypertrophy [33]. Several studies have focused on the effects and molecular mechanisms of DNA methylation in cardiac hypertrophy. Kao et al. demonstrated that DNA methylation is elevated in cardiomyocyte hypertrophy induced by Ang II [34]. Xiao et al. demonstrated that perinatal nicotine upregulates ATR type 1a expression by decreasing DNA methylation of CpG islands in the AT1aR promoter [35]. Consistent with these observations, this study discovered that DNMT1 was downregulated in primary NRCMs following Ang II treatment and that ectopic expression of DNMT1 attenuated Ang II-induced apoptosis, indicating its important role in the progression of cardiac hypertrophy.
Another main form of epigenetic modification, m6A RNA modification, mainly occurs with the addition of a methyl code to the adenosine (A) of RNA transcripts, leading to the modulation of RNA export splicing, editing, stability, degradation, polyadenylation, nuclear transport, and translation initiation [36,37]. An increasing number of reports have demonstrated that m6A RNA modification is involved in many cellular functions and pathological processes of human diseases, such as cardiovascular diseases, including coronary heart disease, hypertension, HF, stroke, and cardiac hypertrophy [38]. The first study focusing on the effects of m6A on cardiac hypertrophy demonstrated that hypertrophic stimuli promote METTL3-mediated m6A modification and repress m6A, resulting in eccentric cardiomyocyte remodeling and dysfunction [24]. Another study demonstrated that METTL3-mediated m6A modification is blocked by CHAPIR–PIWIL4 complexes and thus, which leads to the elevation of Parp10 expression and the pathological progression of hypertrophy [39]. Considering these results, the function of DNA methylation in cardiac hypertrophy remains controversial. Considering the observations of this study, METTL3 was upregulated in Ang II-induced cells and mouse models of cardiac hypertrophy, and reduced METTL3 expression inhibited Ang II-induced apoptosis in NRCMs. However, studies on the effects of m6A modifications and their molecular mechanisms on cardiac hypertrophy are scarce.
Both atrial NP (ANP) and brain (or B-type) NP (BNP) are main family members of natriuretic peptides which are secreted by heart myocardial cells (PMID: 36430893). Together with C-type natriuretic peptides (CNP), ANP and BNP controls the blood pressure through the functional antirenin-angiotensin system (PMID: 28378069). Thus, they could exert function on prevention of cardiomyopathy, arrhythmia, fibrosis and hypertrophy, and combating the occurrence and development of heart failure (PMID: 33852110). In this study, the expression changes of ANP and BNP were detected as indicators of myocardial hypertrophy, and it was found that DNMT1/METTL3 pathway affected the expression of ANP and BNP. Therefore, we believe that DNMT1 and METTL3 affect the occurrence and development of myocardial hypertrophy. However, we also wondered that whether DNMT1/METTL3 pathway can directly regulate the expression of ANP and BNP. It is well known that DNMT1 regulates the expression of downstream target genes by influencing the DNA methylation modification of downstream target genes by binding to the promoter region of genes, while METTL3 regulates the expression of downstream target genes by influencing the methylation modification of downstream target genes by binding to RNA transcripts [40,41]. The results of Chip assay demonstrated that DNMT1 might directly regulated the expression of ANP and BNP via binding the promoter of ANP and BNP and meanshile, METTL3 couldn't recruit the RNA transcripts of ANP and BNP. However, there needs further research to investigate the regulation of the expression of ANP and BNP by DNMT1.
Thus, this study was the first to identify the correlation between DNA methylation and RNA M6A methylation in the development and development of myocardial hypertrophy.
5. Conclusions
Our research first focused on the function of DNMT1 and then dissected the role of the DNMT1/METTL3 pathway in cardiac hypertrophy, providing a novel pathway that shows potential as a target for the diagnosis, therapy, and prognosis of cardiac hypertrophy.
Data availability statement
Data will be availability under request.
CRediT authorship contribution statement
Xidong Zhang: Writing – original draft, Validation, Project administration, Data curation, Conceptualization. Yanhua Nie: Visualization, Validation, Investigation, Formal analysis, Data curation. Rui Zhang: Resources, Methodology. Jiquan Yu: Software, Resources. Jianjun Ge: Writing – review & editing, Visualization, Supervision, Project administration.
Declaration of competing interest
The authors declare that there is no conflict of interests.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (grant number NSFC 81470530 to JG), the Natural Science Foundation of Anhui Province (2008085MH240), and the Major Science and Technology Project of Anhui Province (18030801132).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e24572.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
References
- 1.Savarese G., et al. Global burden of heart failure: a comprehensive and updated review of epidemiology. Cardiovasc. Res. 2023;118(17):3272–3287. doi: 10.1093/cvr/cvac013. [DOI] [PubMed] [Google Scholar]
- 2.Heidenreich P.A., et al. 2022 AHA/ACC/HFSA guideline for the Management of heart failure: Executive summary: a report of the American college of cardiology/American heart association joint committee on clinical practice guidelines. Circulation. 2022;145(18):e876–e894. doi: 10.1161/CIR.0000000000001062. [DOI] [PubMed] [Google Scholar]
- 3.Nakamura M., Sadoshima J. Mechanisms of physiological and pathological cardiac hypertrophy. Nat. Rev. Cardiol. 2018;15(7):387–407. doi: 10.1038/s41569-018-0007-y. [DOI] [PubMed] [Google Scholar]
- 4.Mann D.L., Barger P.M., Burkhoff D. Myocardial recovery and the failing heart: myth, magic, or molecular target? J. Am. Coll. Cardiol. 2012;60(24):2465–2472. doi: 10.1016/j.jacc.2012.06.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhullar S.K., Dhalla N.S. Angiotensin II-induced signal transduction mechanisms for cardiac hypertrophy. Cells. 2022;11(21) doi: 10.3390/cells11213336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding K., et al. Transient receptor potential channels, natriuretic peptides, and angiotensin receptor-neprilysin inhibitors in patients with heart failure. Front Cardiovasc Med. 2022;9 doi: 10.3389/fcvm.2022.904881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enzan N., et al. The use of angiotensin II receptor blocker is associated with greater recovery of cardiac function than angiotensin-converting enzyme inhibitor in dilated cardiomyopathy. ESC Heart Fail. 2022;9(2):1175–1185. doi: 10.1002/ehf2.13790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mattei A.L., Bailly N., Meissner A. DNA methylation: a historical perspective. Trends Genet. 2022;38(7):676–707. doi: 10.1016/j.tig.2022.03.010. [DOI] [PubMed] [Google Scholar]
- 9.Yousefi P.D., et al. DNA methylation-based predictors of health: applications and statistical considerations. Nat. Rev. Genet. 2022;23(6):369–383. doi: 10.1038/s41576-022-00465-w. [DOI] [PubMed] [Google Scholar]
- 10.Fitz-James M.H., Cavalli G. Molecular mechanisms of transgenerational epigenetic inheritance. Nat. Rev. Genet. 2022;23(6):325–341. doi: 10.1038/s41576-021-00438-5. [DOI] [PubMed] [Google Scholar]
- 11.Kan R.L., Chen J., Sallam T. Crosstalk between epitranscriptomic and epigenetic mechanisms in gene regulation. Trends Genet. 2022;38(2):182–193. doi: 10.1016/j.tig.2021.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li S., Peng Y., Panchenko A.R. DNA methylation: precise modulation of chromatin structure and dynamics. Curr. Opin. Struct. Biol. 2022;75 doi: 10.1016/j.sbi.2022.102430. [DOI] [PubMed] [Google Scholar]
- 13.Lei H., et al. The role and molecular mechanism of epigenetics in cardiac hypertrophy. Heart Fail. Rev. 2021;26(6):1505–1514. doi: 10.1007/s10741-020-09959-3. [DOI] [PubMed] [Google Scholar]
- 14.Russell-Hallinan A., et al. Epigenetic regulation of endothelial cell function by nucleic acid methylation in cardiac homeostasis and disease. Cardiovasc. Drugs Ther. 2021;35(5):1025–1044. doi: 10.1007/s10557-020-07019-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu T.T., et al. Myocardial tissue-specific Dnmt1 knockout in rats protects against pathological injury induced by Adriamycin. Lab. Invest. 2020;100(7):974–985. doi: 10.1038/s41374-020-0402-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oeing C.U., et al. Indirect epigenetic testing identifies a diagnostic signature of cardiomyocyte DNA methylation in heart failure. Basic Res. Cardiol. 2023;118(1):9. doi: 10.1007/s00395-022-00954-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cameron V.A., et al. DNA methylation patterns at birth predict health outcomes in young adults born very low birthweight. Clin Epigenetics. 2023;15(1):47. doi: 10.1186/s13148-023-01463-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y., et al. N6-methyladenosine-mediated gene regulation and therapeutic implications. Trends Mol. Med. 2023;29(6):454–467. doi: 10.1016/j.molmed.2023.03.005. [DOI] [PubMed] [Google Scholar]
- 19.Jiang X., et al. m6A modification on the fate of colorectal cancer: functions and mechanisms of cell proliferation and tumorigenesis. Front. Oncol. 2023;13 doi: 10.3389/fonc.2023.1162300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Y., et al. Analysis approaches for the identification and prediction of N(6)-methyladenosine sites. Epigenetics. 2023;18(1) doi: 10.1080/15592294.2022.2158284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li P., et al. N (6)-methyladenosine RNA methylation: from regulatory mechanisms to potential clinical applications. Front. Cell Dev. Biol. 2022;10 doi: 10.3389/fcell.2022.1055808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu B., et al. Readers, writers and erasers of N(6)-methylated adenosine modification. Curr. Opin. Struct. Biol. 2017;47:67–76. doi: 10.1016/j.sbi.2017.05.011. [DOI] [PubMed] [Google Scholar]
- 23.Roundtree I.A., et al. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169(7):1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dorn L.E., et al. The N(6)-methyladenosine mRNA methylase METTL3 controls cardiac homeostasis and hypertrophy. Circulation. 2019;139(4):533–545. doi: 10.1161/CIRCULATIONAHA.118.036146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giamouzis G., et al. Left ventricular hypertrophy and sudden cardiac death. Heart Fail. Rev. 2022;27(2):711–724. doi: 10.1007/s10741-021-10134-5. [DOI] [PubMed] [Google Scholar]
- 26.Martin T.G., Juarros M.A., Leinwand L.A. Regression of cardiac hypertrophy in health and disease: mechanisms and therapeutic potential. Nat. Rev. Cardiol. 2023;20(5):347–363. doi: 10.1038/s41569-022-00806-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winkle A.J., et al. Emerging therapeutic targets for cardiac hypertrophy. Expert Opin. Ther. Targets. 2022;26(1):29–40. doi: 10.1080/14728222.2022.2031974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 29.Moore L.D., Le T., Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38(1):23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krolevets M., et al. DNA methylation and cardiovascular disease in humans: a systematic review and database of known CpG methylation sites. Clin Epigenetics. 2023;15(1):56. doi: 10.1186/s13148-023-01468-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jurkowska R.Z., Jeltsch A. Mechanisms and biological roles of DNA methyltransferases and DNA methylation: from past achievements to future challenges. Adv. Exp. Med. Biol. 2022;1389:1–19. doi: 10.1007/978-3-031-11454-0_1. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y., et al. Inhibition of DNA methylation in newborns reprograms ischemia-sensitive biomarkers resulting in development of a heart ischemia-sensitive phenotype late in life. Reprod. Toxicol. 2021;105:198–210. doi: 10.1016/j.reprotox.2021.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fang X., et al. cAMP induces hypertrophy and alters DNA methylation in HL-1 cardiomyocytes. Am J Physiol Cell Physiol. 2015;309(6):C425–C436. doi: 10.1152/ajpcell.00058.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kao Y.H., et al. DNA methylation inhibition: a novel therapeutic strategy for heart failure. Int. J. Cardiol. 2014;176(1):232–233. doi: 10.1016/j.ijcard.2014.06.010. [DOI] [PubMed] [Google Scholar]
- 35.Ke J., et al. Role of DNA methylation in perinatal nicotine-induced development of heart ischemia-sensitive phenotype in rat offspring. Oncotarget. 2017;8(44):76865–76880. doi: 10.18632/oncotarget.20172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sendinc E., Shi Y. RNA m6A methylation across the transcriptome. Mol Cell. 2023;83(3):428–441. doi: 10.1016/j.molcel.2023.01.006. [DOI] [PubMed] [Google Scholar]
- 37.Zaccara S., Ries R.J., Jaffrey S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019;20(10):608–624. doi: 10.1038/s41580-019-0168-5. [DOI] [PubMed] [Google Scholar]
- 38.Yang Y., et al. Research progress on N(6)-adenosylate methylation RNA modification in heart failure remodeling. J Transl Int Med. 2022;10(4):340–348. doi: 10.2478/jtim-2022-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao X.Q., et al. The piRNA CHAPIR regulates cardiac hypertrophy by controlling METTL3-dependent N(6)-methyladenosine methylation of Parp10 mRNA. Nat. Cell Biol. 2020;22(11):1319–1331. doi: 10.1038/s41556-020-0576-y. [DOI] [PubMed] [Google Scholar]
- 40.Oerum S., et al. A comprehensive review of m6A/m6Am RNA methyltransferase structures. Nucleic Acids Res. 2021;49(13):7239–7255. doi: 10.1093/nar/gkab378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robertson K.D. DNA methylation and human disease. Nat. Rev. Genet. 2005;6(8):597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be availability under request.