Abstract
Cervical squamous cell carcinoma (CESC) ranks among the primary contributors to global cancer-associated mortality. However, the role mediated by synaptotagmin 7 (SYT7) in CESC remains unclear. Our study employed immunohistochemistry to assess the level of SYT7 expression in the tissue microarray. Furthermore, lentiviral shRNA transduction was utilized to establish SYT7 knockdown cell line models based on HeLa and SiHa cell lines. The functional impacts of silencing SYT7 expression in vitro were evaluated. A subcutaneous xenograft model was employed to examine the tumorigenic potential of cells with or without SYT7. The content of SYT7 in CESC tissues was significantly elevated compared to adjacent normal tissues. Functionally, silencing SYT7 in HeLa and SiHa cells suppressed cell proliferation, colony formation ability, and apoptosis enhancement. Additionally, cells with suppressed SYT7 also exhibited inhibited cell migration and invasion. In vivo experiments demonstrated the loss of tumorigenic ability in SYT7 knockdown cells and suppressed tumor growth. Quantitative PCR PrimeView PathArray and apoptosis antibody array analyses revealed that upon elimination of SYT7, there was a significant upregulation observed in Caspase 8, TNF-R1 (TNF receptor superfamily member 1A), and HSPA5 (heat shock protein family A [Hsp70] member 5), while TGFBI (transforming growth factor beta-induced), RPL31 (ribosomal protein L31), LUM (lumican), HSDL2 (hydroxysteroid dehydrogenase-like 2), ITGB5 (integrin subunit beta 5), and Smad2 (SMAD family member2) were downregulated. Overall, we have demonstrated the tumor-promoting functions of SYT7 in CESC.
Keywords: Cervical squamous cell carcinoma, SYT7, Proliferation, Apoptosis, Migration
1. Introduction
Cervical cancer (CC) is a major contributor to global cancer-associated mortality. In 2018, around 570,000 cases were newly reported, and 311,000 deaths were observed due to CC [1]. The popularization of cervical screening programs and improved therapeutic strategies have contributed to the treatment of CC in most developed countries. However, it is still among the major contributors to cancer-associated mortality in developing countries. Almost 90 % of CC cases develop in low- and middle-income countries owing to insufficient detection and vaccination [2]. Therapeutic strategies, including surgery and chemoradiation, can achieve curative treatment for 80 % of female stage I-II patients and 60 % of stage III patients [3]. However, metastatic CC has a 5-year relative survival rate of 16.5 %. However, to date, the treatment of CC has been hampered by an unclear understanding of its underlying mechanism, which needs urgent investigation.
Research examining the significance of SYT7 in cancer development is in its infancy. Emerging clinical findings have indicated that high levels of SYT7 expression are correlated with the progression and worsened outcomes of patients [[4], [5], [6]]. Laboratory studies have shown that suppressing SYT7 may reduce the malignancy of lymphocytic leukemia and colorectal cancer [7,8], suggesting its role as a cancer-promoting factor. Nonetheless, the SYT7-mediated regulation of CC has not been elucidated.
In a preliminary investigation, we queried the GDC TCGA-CESC dataset (Ver. 18) using the Xena browser [9]. Transcriptomic sequencing revealed a high level of SYT7 mRNA in tissues of cervical squamous cell carcinoma (CESC), which might correlate with the shortened overall survival (OS) of patients. Therefore, we hypothesized that the increased SYT7 expression could contribute to the development of CESC. Herein, we, for the first time, investigate the function of SYT7 in CESC. Our findings might suggest that targeting SYT7 is regarded as a viable therapeutic alternative.
2. Materials and methods
2.1. Immunohistochemical analysis (IHC) of the tissue microarray
The tissue microarray of the CESC & adjacent normal tissue chip was used for IHC staining analysis, including 24 adjacent normal tissues and 546 CESC tissues. This work received ethical approval from the First Affiliated Hospital of Guangxi Medical University (approval number: 2021, KY-E−347). Participant age was between 25 and 76 in the cancer group, and 18 to 72 in the normal group, and their median age was 47 and 48.5, respectively. IHC was conducted via a 1:200 dilution of the SYT7 antibody (Tianjin Saierbio, Cat. SRP09489). The samples were collected and ethically processed, as approved by the First Affiliated Hospital of Guangxi Medical University and after obtaining informed consent from the patients.
2.2. Bioinformatics analysis
The CC patient RNA sequence (level 3) and associated clinical data were retrieved from the TCGA dataset (https://portal.gdc.com). The difference in survival rates between various cohorts was assessed via the log-rank test. The ROC (v0.4) analysis was utilized to evaluate the SYT7 mRNA's predictive potency. Then, we computed p-values and hazard ratios (HR) with 95 % confidence intervals (CI) from log-rank tests and univariate cox proportional hazards regression for Kaplan-Meier curve generation. All data analyses were conducted using the R package (2020) version 4.0.3, and a p-value ≤0.05 was set as the significance threshold.
2.3. Cell culture and lentiviral-based SYT7 knockdown
The CESC cell lines, HeLa (CCL-2), and SiHa (HTB-35), were obtained from ATCC. DMEM medium with 10 % FBS and 1 % Antibiotic-Antimycotic solution (ThermoFisher, USA, Cat. 15240062) were used to maintain cell culture in a 5 % CO2 incubator at 37 °C.
Shanghai Biosciences Co. Ltd. (Shanghai, China) constructed Lentiviral shRNA vectors targeting SYT7 (shSYT7). The SYT7 targeting sequences were synthesized and cloned into the backbone vector BR-V108 (Shanghai Biosciences Co. Ltd., Shanghai, China). 293T cells (Supplied by the Chinese Academy of Science) seeded in a 10-cm dish were used for cotransfection with the third-generation packaging system to produce the lentivirus. Lentiviral vectors containing control-shRNA were also provided by Sigma, USA.
In brief, BR-V108-RNAi (10 μg), pHelper X2 (7.5 μg), and pHelper 2.G (5 μg) were added and resuspended in 500 μL Opti-MEM R1 medium and kept for 5 min at room temperature (RT). Meanwhile, 60 μL of the transfection reagent PEI (PEI 25K™, CAS No. 23966–1) was added to another 500 μL Opti-MEM R1 medium before a 5-min RT incubation. Then the two combinations were mixed and left at RT for an additional 20 min before being added to 293T cells for virus packaging under 37 °C incubation, lasting for 48–72 h. The virus was harvested by ultracentrifugation. The negative control was treated with the shCtrl lentivirus, while the test cells (shSYT7) were treated with shSYT7. Fresh media with the virus were used to replace the previous media and incubated at 37 °C for 18–20 h for lentiviral infection. Then, the medium was replaced with normal media without the virus. After 72 h of lentiviral infection, the green fluorescent protein (GFP) levels were examined in SiHa and HeLa cells under a fluorescence microscope to assess infection proficiency. The MOI of transfection is 10 for HeLa or SiHa.Human shSYT7–1 target sequence (RNAi-00148): GCTCACCGTGAAGATCATGAA.
Human shSYT7–2 (RNAi-00149) target sequence: CTGGAACGAGACCTTCCTCTT.
Human shSYT7–3 (RNAi-00150) target sequence: GCTCTTGTCTCTCTGCTACAA.
2.4. MTT assay
After trypsinization, the cells were resuspended and counted. Next, 2000 cells in 100 μL of complete media were introduced to individual wells of 96-well plates (Corning, NT, USA, Cat. No: 3596). Subsequently, 20 μL/well of MTT solution (3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl tetrazolium bromide) (Sangon Biotech, Shanghai, China; # A600799–0250) (5 mg/mL) was introduced to the cells. Upon a 4-h incubation, the culture medium with MTT was slowly aspirated and replaced with 100 μL dimethyl sulfoxide (DMSO), with shaking on a calibrated shaker for 2–5 min at RT. Absorbance was recorded at 490/570 nm with a microplate reader to analyze cell survivability.
2.5. Apoptosis assay
We employed stable shCtrl- or shSYT7-incorporated HeLa and SiHa cells, which were plated and incubated in a 6-well plate. Upon reaching 70 % confluency, cells underwent trypsin-based digestion and resuspension in a 200 μL 1 × binding buffer. Then, 10 μL of each Annexin V-APC and PI were introduced to cells, followed by a 10–15 min staining without light. Following staining, 400–800 μL 1 × binding buffer was introduced, and flow cytometry was performed. Flow analysis was conducted via the flow cytometry software guava InCyte.
2.6. Colony formation assay
Cells (800 cells/well) were cultured for five days after transfection. Subsequently, they underwent resuspension in 2 mL of complete media followed by seeding into 6-well plates. After eight days of culture, when most of the colonies were in a cluster of over 50 cells under a microscope, we stained cells for 10–20 min using 500 μL of GIEMSA staining solution (DingGuo Biotech, Shanghai, China, #KGA229) at RT. Next, plates were ddH2O rinsed multiple times and air-dried to photograph and count the number of colonies.
2.7. Wound-healing assay
HeLa and SiHa cell lines were employed for the wound-healing assay. Approximately 50,000 cells/well were plated in each well of a 96-well plate to achieve 90 % confluence within one day. The following day, a scratch device was used to create a gentle wound. Subsequently, we performed three gentle washes with serum-free medium. The old medium was then replaced with a complete medium having medium or low serum concentration (0.5 % FBS). The wound closure distance was monitored at the indicated time points and photographed using an IX73 Olympus inverted microscope with 100X magnification.
2.8. Transwell migration assay
Cell invasion was assessed using a 6.5 mm Transwell® with an 8.0 μm Pore Polycarbonate Membrane Insert (Corning, Cat. 3422). Cells (50,000 cells/well) were resuspended in 100 μL serum-free DMEM media before being plated onto the upper insert, while the lower chamber received 600 μL DMEM with 30 % FBS as a chemoattractant. Cells that successfully penetrated the upper 8.0 μm pore polycarbonate membrane insert underwent a 5-min 0.5 % crystal violet staining at RT after 24 h of incubation. The membrane was then photographed and examined under an IX73 Olympus inverted microscope.
2.9. Tumorigenic assay
The Animal Care & Welfare Committee approved the animal experiment at Guangxi Medical University with approval number: 201906283. BALB/c female nude mice (4 weeks old) were obtained from Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China) and kept under special pathogen-free (SPF) conditions. They were fed on demand and housed in 12-h light/12-h dark environments. Stable transfected HeLa cells with shCtrl or shSYT7 were subcutaneously injected into female BALB/c BALB/c nude mice (4 × 106 cells/mouse, 10 mice/group). The tumor's dimensions were measured every three days. On day 28, after CO2 inhalation to anesthetize the mice, xenografts were obtained through cervical dislocation. The xenografts were then measured and imaged.
Tumor tissues from shSYT7 and shCtrl were collected, and tissue slices were subjected to primary antibody Ki67 (1:200, Abcam, USA, Cat. # ab16667). The secondary antibody IgG (1:400, Abcam, USA, Cat. # ab6721) was applied, followed by a 1-h incubation at RT after overnight (ON) incubation with the primary antibody at 4 °C. After DAB staining, hematoxylin staining was performed on the tissue slices. A photomicroscope was utilized to capture and examine the images.
2.10. Transcriptomic analysis
GeneChip PrimeView Human PathArray™ was conducted with SiHa cells with stable shCtrl or shSYT7 expression. Triplicate samples were used to minimize sample variations. After examining the signal histogram and the relative log expression graph for quality control, we performed correlation analysis, principal component analysis, and differential expression analysis, such as volcano plot, hierarchical clustering, etc. We uploaded our database to GEO and obtained the GSE200526 database. The website link is https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc =GSE200526.
2.11. qRT–PCR
The extraction of total RNA from cell lysates and reverse transcription PCR were carried out according to the protocols of TRIzol (Sigma, USA) and HiScript III-RT SuperMix for qPCR (+g DNA wiper) Kit (Vazyme, Nanjing, China), respectively. Quantitative RT-PCR was carried out with AceQ qPCR SYBR Green Master Mix (Vazyme, Nanjing, China). GAPDH served as the internal control.
2.12. Western blotting
HeLa and SiHa cells were trypsinized and washed in ice-cold PBS. After centrifugation and removal of the supernatant, the cell pellets were lysed in ice-cold 1 × lysis buffer containing 1 × protease and phosphatase inhibitors. Protein quantification was performed using the BCA protein assay kit (HyClone-Pierce, MA, USA, # 23225). Subsequently, protein separation was carried out on a 10 % SDS-PAGE gel, and the transfer was conducted onto a polyvinylidene difluoride membrane (PVDF) using a current of 300 mA for 150 min at 4 °C. Following this, the membrane was blocked for 1 h with 5 % BSA dissolved in 1 × TBST. It is important to wash the membrane with TBST three times (10 min each) before and after adding antibodies. Primary antibodies were diluted in 5 % nonfat powdered milk (BBI-A-600669–0250) before being incubated overnight with the membrane at 4 °C. Corresponding secondary antibodies were applied, followed by a 1-h incubation at room temperature. The blot was developed using the ECL-PLUS Kit (Amersham, Chalfont, UK, # RPN2232).
2.13. Human Apoptosis Antibody Array
Total lysate proteins (400 μg/sample) were incubated overnight with a Human Apoptosis Antibody Array Membrane (Abcam, USA, Cat. # ab134001) pre-spotted with antibodies for 43 human apoptosis markers. The procedure, including incubations, washes, and chemiluminescence detection, strictly followed the kit directions. The final signal intensity value of each dot was quantified using ImageJ (NIH, USA).
2.14. Statistical analysis
Data are provided as mean ± Standard Error Mean (SEM), (n ≥ 3). Pairwise comparisons employed Student's t-test, one-way ANOVA with subsequent Tukey's multiple comparison. Table 3 data analysis was with Spearman's test. P-value <0.05 was set as the significance threshold (*P < 0.05; **P < 0.01, ***P < 0.001, ****P < 0.0001). The qRT-PCR information was calculated using the 2−ΔΔCT formula. The t-test assessed statistical differences. The graphs were statistically analyzed and generated via GraphPad Prism 8 (GraphPad Software Inc., San Diego, CA, USA).
Table 3.
Link between SYT7 content and tumor profiles of cervical cancer patients.
| SYT7 | ||
|---|---|---|
| Grade | Spearman correlation | 0.177 |
| Significance (two-tailed) | P < 0.001 | |
| N | 485 |
3. Results
3.1. Enhanced SYT7 protein levels in CESC tissues and its clinical significance
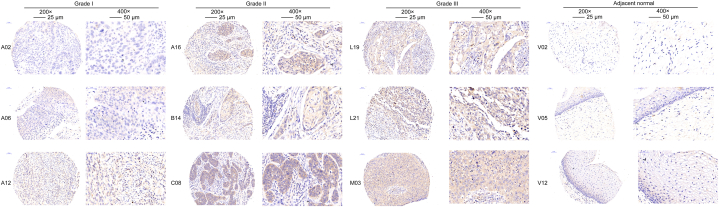
IHC examination of the tissue microarray revealed that SYT7 expression was considerably elevated in CESC versus nearby normal tissues (P 0.0001) (Table 1). The Grade III tumor tissues had much greater levels of SYT7 expression, as seen in Fig. 1 (Fig. 1). Additionally, we discovered, by the Mann-Whitney U test (Table 2), a strong link between SYT7 content and CESC pathological grading. According to Spearman's correlation analysis, the clinical grade of CESC was positively correlated with SYT7 expression (Table 3). According to bioinformatic analysis, SYT7 was not connected to the prognosis for the 306 CC patients (Supplementary Fig. 1). Collectively, SYT7 may be associated with the progression of CESC.
Table 1.
Gene expression profiles in cervical cancer and para-carcinoma tissues, as evidenced via IHC assessment.
| SYT7 | Tumor tissue | Para-carcinoma tissue | p-value | ||
|---|---|---|---|---|---|
| content | Cases | Percentage | Cases | Percentage | |
| Reduced | 265 | 48.5 % | 24 | 100 % | P < 0.001 |
| Enhanced | 281 | 51.5 % | 0 | 0 % | |
Fig. 1.
Overexpression of SYT7 in CESC tissues compared to the adjoining normal tissues. SYT7 protein expression in CESC tissues and adjoining normal tissues were determined via IHC with three representative cases in each category for presentation (200 × , left, 400 × , right).
Table 2.
Link between SYT7 content and tumor profiles of cervical cancer (CC) patients.
| Features | No. of patients | SYT7 level |
p-value |
|
|---|---|---|---|---|
| low | high | |||
| All patients | 546 | 265 | 281 | |
| Age(years) | 0.264 | |||
| <48 | 271 | 125 | 146 | |
| ≥48 | 275 | 140 | 135 | |
| Grade | P < 0.001 | |||
| 1 | 58 | 36 | 22 | |
| 2 | 313 | 145 | 168 | |
| 3 | 114 | 36 | 78 | |
| Primary Tumor(T) | 0.062 | |||
| T1 | 497 | 248 | 249 | |
| T2 | 42 | 16 | 26 | |
| T3 | 5 | 0 | 2 | |
| T4 | 2 | 0 | 1 | |
| Regional Lymph Nodes (N) | 0.241 | |||
| N0 | 515 | 253 | 262 | |
| N1 | 29 | 11 | 18 | |
| Distant Metastasis (M) | 0.152 | |||
| I | 468 | 234 | 234 | |
| II | 37 | 14 | 23 | |
| III | 31 | 11 | 20 | |
| IV | 6 | 5 | 1 | |
3.2. Verification of the cancer-promoting role of SYT7 in CESC in vitro
qRT-PCR assessed the knockdown efficiencies of shRNA in the HeLa cell line (Supplementary Fig. 2A), where shSYT7 (RNAi-00148) demonstrated an 80 % knockdown efficiency and was chosen to establish SYT7-knockdown cell lines based on HeLa and SiHa cell lines. To confirm successful transfection, the GFP signal was examined under a fluorescence microscope following 72 h of infection (Supplementary Fig. 2B). The establishment of a stable SYT7-knockdown cell line was further validated by conducting qRT-PCR, revealing more than 60 % and 80 % knockdown efficiencies in HeLa and SiHa cells (both P < 0.001), respectively (Supplementary Fig. 2C). Western blot results demonstrated consistent knockdown in both cell lines (Supplementary Fig. 2D). All these data confirmed the successful establishment of the SYT7-knockdown cell model in HeLa and SiHa cell lines.
3.3. Silencing of SYT7 suppresses cell growth and colony formation
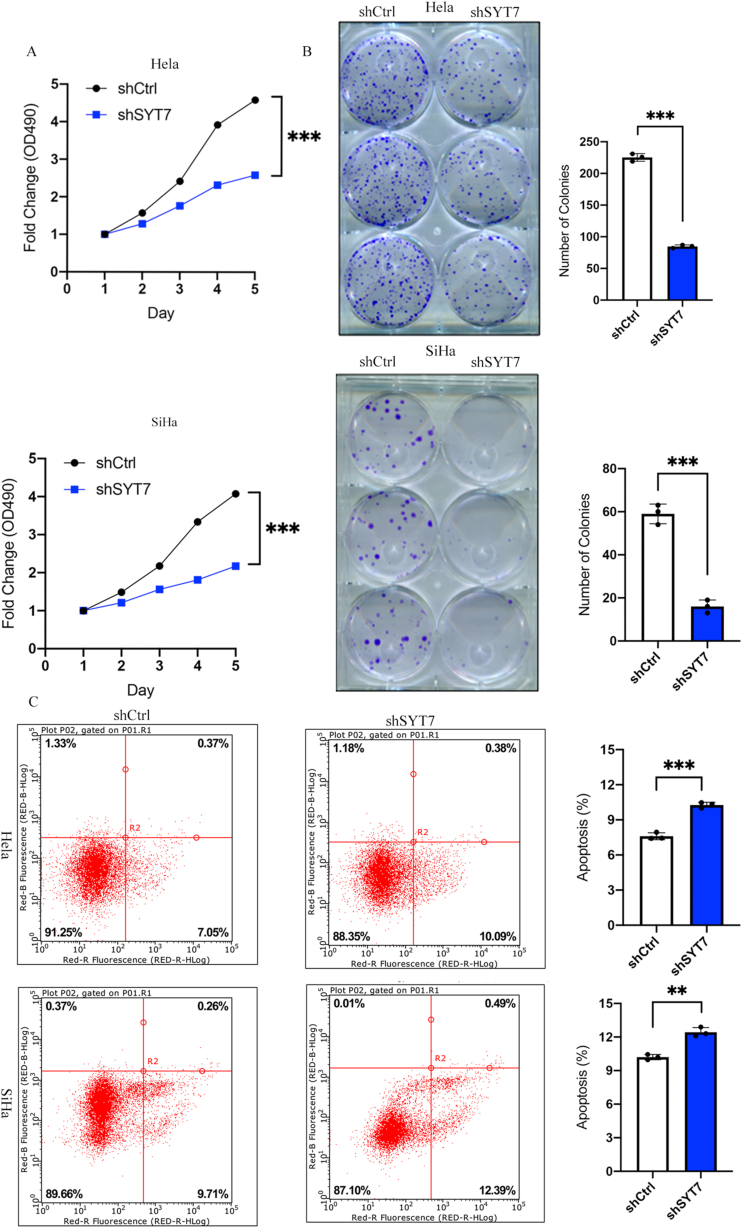
shSYT7-or shCtrl-transfected HeLa and SiHa cells were incubated for five days, and cell growth was monitored using the MTT assay. As depicted in Fig. 2A, cell proliferation was significantly inhibited in SYT7-deficient cells (P < 0.001). Similarly, the SiHa cell line with SYT7 silencing also exhibited notable suppression of cell proliferation (P < 0.001) (Fig. 2A). In line with the MTT assay, the colony formation assay also displayed a reduction in cell proliferation, as evidenced by the decrease in colony size and number of HeLa and SiHa cells (both P < 0.001) (Fig. 2B). The presented data strongly indicate that SYT7 deficiency profoundly suppresses CESC cell proliferation.
Fig. 2.
SYT7 silencing suppressed cell growth and promoted cell apoptosis. A. The SYT7 knockdown inhibited cell proliferation during MTT assay (HeLa: P < 0.001; SiHa: P < 0.001); B. The SYT7 knockdown suppressed colony formation ability (HeLa: P < 0.001; SiHa: P < 0.001); C. The SYT7 silencing induced apoptosis in HeLa (P < 0.001) and SiHa (P < 0.001) cell lines. (*P < 0.05; **P < 0.01, ***P < 0.001, ****P < 0.0001).
3.4. Silencing of SYT7 induces apoptosis in CESC cells
To evaluate the modulatory impact of SYT7 on CESC cell apoptosis, we assessed cell apoptosis using flow cytometry. Knockdown of SYT7 in both HeLa and SiHa cells led to increased apoptosis compared to shCtrl cells (Fig. 2C). Quantification revealed a significant rise in the percentage of apoptotic cells in SYT7-knockdown CESC cells relative to shCtrl cells (HeLa P < 0.0001, SiHa P < 0.001). Altogether, SYT7 knockdown significantly induced apoptosis in CESC cells.
3.5. SYT7 silencing prohibits CESC cell migration and invasion
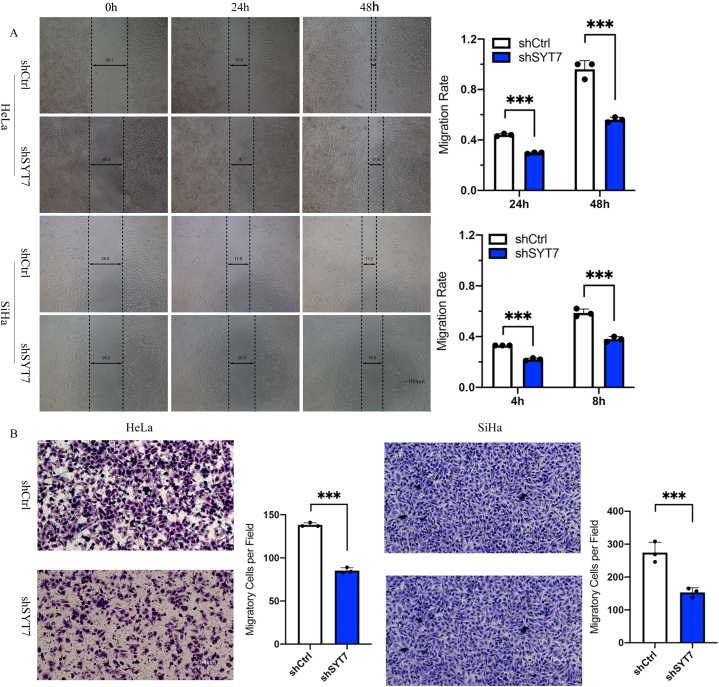
Metastasis poses a significant challenge to the curative efficacy of various strategies employed for treating CESC. Therefore, we examined the role of SYT7 in cell migration and invasion using wound healing and Transwell migration assays, respectively. Suppressed cell migration due to SYT7 knockdown was demonstrated by reduced wound closure efficiency in both HeLa and SiHa cells (Fig. 3A). The wound width was quantified at 24 h and 48 h for HeLa and 4 h and 8 h for SiHa, showing a significantly decreased wound closure (P < 0.001). Similarly, upon silencing the SYT7 gene, the Transwell assay exhibited decreased cell invasion abilities in both HeLa and SiHa cell lines (P < 0.001) (Fig. 3B). The data presented strongly suggest that SYT7 silencing may inhibit CESC cell migration and invasion, which are representative phenotypes of tumor metastasis.
Fig. 3.
SYT7 knockdown inhibited cell migration and cell invasion. A. SYT7 silencing inhibited cell migration as determined by wound healing assay in HeLa (P < 0.001) and SiHa (P < 0.001) cells; B. The SYT7 knockdown in HeLa and SiHa cells inhibited cell invasion (both P < 0.001). (*P < 0.05; **P < 0.01, ***P < 0.001, ****P < 0.0001).
3.6. SYT7 gene silencing in CESC cells suppresses tumorigenesis in vivo
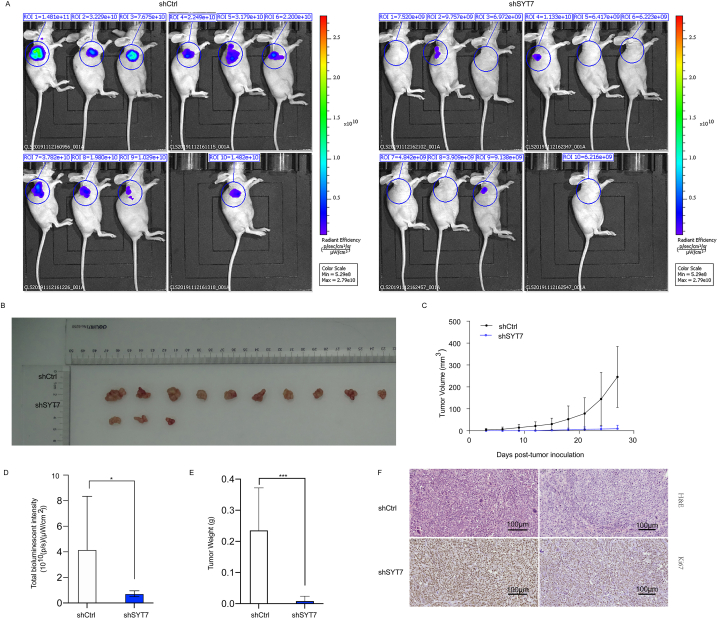
Building upon the aforementioned in vitro findings, we further investigated the impact of SYT7 on tumor growth in vivo. The multiple in vitro assays confirmed that SYT7 silencing suppresses cell proliferation, migration, and invasion while concurrently enhancing cell apoptosis. Therefore, we examined whether SYT7 knockdown influenced tumorigenesis in vivo. To accomplish this, we established a subcutaneous xenograft model by injecting stably transfected HeLa cells with shCtrl or shSYT7. Transfection efficiencies of both cell groups were validated before injection (Supplementary Fig. 3A). Consistent with in vitro results, the time-dependent monitoring of tumor volume in the shSYT7 group displayed a significant reduction in tumor growth rate compared to the shCtrl group. Particularly, seven out of ten mice injected with shSYT7 cells did not develop observable solid tumors (Fig. 4A, Supplementary Fig. 3B). Similar outcomes were observed through measurements of tumor weight and total in vivo fluorescence intensity, revealing substantial inhibition of tumor development in the shSYT7 cells compared to the shCtrl cells (Fig. 4B–E). Ki-67 staining further validated reduced cell proliferation in xenografts generated by the shSYT7 group (Fig. 4F). These findings indicated that silencing SYT7 led to suppressed tumor progression in vivo.
Fig. 4.
SYT7 silencing inhibited tumorigenesis in a subcutaneous xenograft mouse model. A. Representative bioluminescence imaging of a subcutaneous xenograft mouse model and solid tumor at harvest; B Representative tumor growth of subcutaneous xenograft mouse model. C. Growth curve of tumor volume measured periodically (V = ½ (Length × Width2)); D. Total bioluminescent intensity measured at harvest (P < 0. 0.05). E. Tumor weight (g) measured at harvest (P < 0.001); F. H&E and Ki67 staining of tumor sections derived from xenograft mouse models; (*P < 0.05; **P < 0.01, ***P < 0.001, ****P < 0.0001).
3.7. Exploration of possible SYT7 downstream genes in CESC
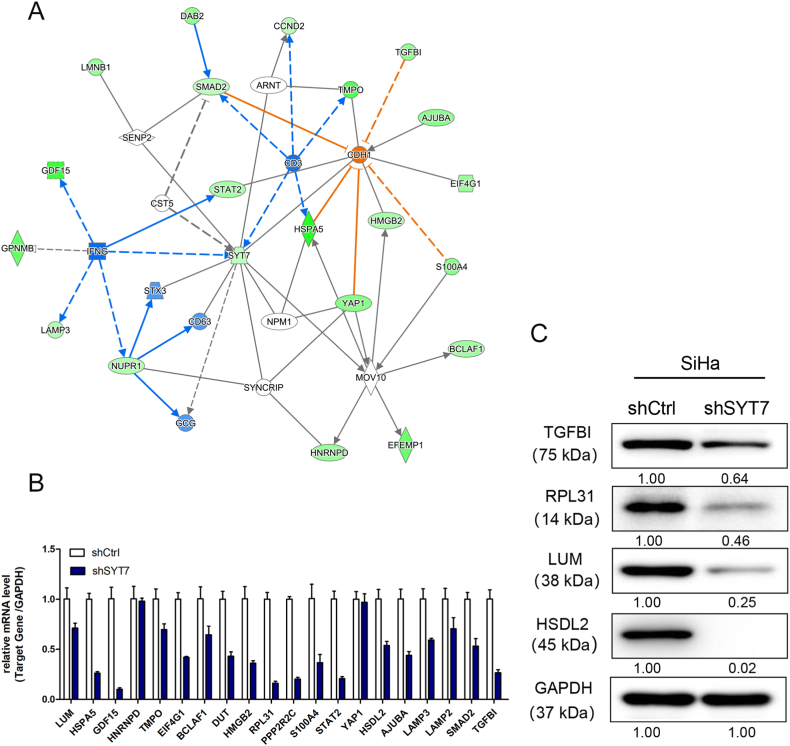
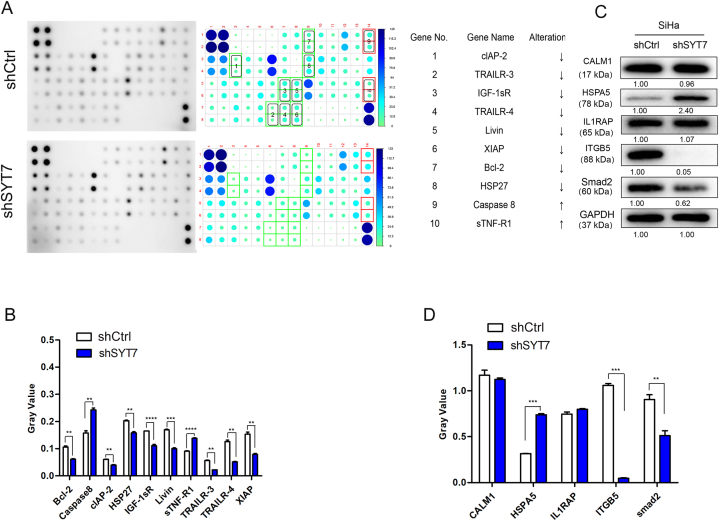
To elucidate the associated mechanism behind the SYT7-deficient CESC tumor suppressor phenotypes, we employed the GeneChip PrimeView Gene Expression Array. This array profiled expression patterns of over 36,000 transcripts and variants using SiHa-shCtrl and shSYT7-cDNAs (Supplementary Fig. 4). Through bioinformatics and literature-based interaction network analysis, we identified that SYT7 could directly interact with IFNG, STX3, and CD63 to regulate downstream targets, including AJUBA, BCLAF1, CCND2, DAB2, EFEMP1, EIF4G1, GDF15, GPNMB, HMGB2, HNRNPD, HSPA5, LAMP3, LMNB1, NUPR1, S100A4, SMAD2, STAT2, TGFBI, TMPO, and YAP1 (Fig. 5A). Twenty downstream targets were chosen for further validation through qPCR. As shown in Fig. 5B, SYT7 knockdown strongly suppressed most of the selected targets (Fig. 5B). Similarly, Western blot results demonstrated a similar inhibition of TGFBI, RPL31, LUM, and HSDL2 in the SYT7-silenced group (Fig. 5C). Additionally, since SYT7 was implicated in the regulation of cell apoptosis, we conducted the Human Apoptosis Antibody Array to elucidate how cell apoptosis was affected after SYT7 knockdown. Post SYT7 knockdown, Caspase8 and sTNF-R1 exhibited up-regulation (Fig. 6A), whereas Bcl-2, cLAP-2, HSP27, IGF-1sR, Livin, TRAILR-3, TRAILR-4, and XIAP displayed down-regulation (Fig. 6A). Quantification of the grey value data indicated significant changes (Fig. 6B). Western blot data revealed that ITGB5 and Smad2 were downregulated, while HSPA5 was up-regulated. However, CALM1 and IL1RAP showed no differences in expression (Fig. 6C and D). In sum, these data tentatively identified potential downstream targets of SYT7 essential for modulating cell growth, migration, invasion, and apoptosis.
Fig. 5.
Screening of downstream targets of SYT7 by the GeneChip PrimeView PathArray™. A. The interaction network between the target gene SYT7 and differentially expressed genes. B. Quantitative PCR validation of PathArray identified the differentially expressed genes, which were downstream of SYT7; C. Western blot validation of downstream targets.
Fig. 6.
Identification of downstream targets of SYT7 by Human Apoptosis Antibody Array. A. Comparison between the shCtrl and shSYT7 proteins (SiHa) using Human Apoptosis Antibody array with densitometry quantification is shown in the right panel (Top 10 candidates listed); B. Grey value of top 10 candidates identified in the Human Apoptosis Antibody Array; C. Western blot validation of potential downstream targets. D. The quantification of the bands of CALM1, HSPA5, IL1RAP, ITGB5 and smad2 (*P < 0.05; **P < 0.01, ***P < 0.001, ****P < 0.0001).
4. Discussion
SYT7 has been suggested to participate in the etiology of several forms of cancer. Nevertheless, the SYT7 role in CESC remains elusive. In addition to the multiple reported functions of SYT7 in mediating cell apoptosis/differentiation, vesicular transportation, and lysosome-regulated repair, increasing evidence has also shown its significant relevance in cancer occurrence and progression [10,11]. Bing et al. reported that SYT7 down-regulation suppressed glioblastoma development by inducing cell apoptosis [12]. Fei et al. reported that the SYT7 content was elevated in lung cancer, further illustrating the association between SYT7 and p53, which enhances the link between p53 and MDM2. Thus, its role in promoting lung cancer cell proliferation and inhibiting cell senescence was revealed [13]. Similarly, another study confirmed the up-regulation of SYT7 protein contents in non-small cell lung cancer (NSCLC) relative to the adjoining paired normal tissue. A higher SYT7 content was linked to poorer outcomes among NSCLC patients. Further investigations indicated that SYT7 functioned as a crucial promoter of epithelial-mesenchymal transition (EMT) and the NSCLC development and progression [6]. SYT7 stabilizes HMGB3 and promotes thyroid cancer progression [14]. Although accumulating studies have proven the carcinogenic role of SYT7 in the cancer types mentioned above, its role in CC remains elusive.
Our study demonstrated a substantial increase in SYT7 protein content within CESC tissue specimens in comparison to adjacent noncancerous counterparts. Furthermore, elevated SYT7 levels in CESC tissues exhibited a significant correlation with advanced tumor grade. TCGA data indicated that SYT7 was not associated with prognosis. However, it has been linked to CC progression, suggesting that it might not act as an independent prognostic factor. Furthermore, our investigation revealed that SYT7 knockdown markedly diminishes CESC cell proliferation, migration, colony formation, and tumorigenicity, while concurrently promoting apoptosis. These findings parallel previous observations in other types of cancer cells [4,6,15].
To better understand the cancer-promoting mechanism of SYT7, we attempted to identify potential downstream targets of SYT7 in CESC. GeneChip PrimeView PathArray qPCR and Human Apoptosis Antibody array were performed employing the SiHa (shCtrl, shSYT7) cell line. Here, we identified potential downstream targets that showed statistical significance. Caspase 8 and sTNF-R1 were up-regulated, while Bcl-2, cLAP-2, HSP27, IGF-1sR, Livin, TRAILR-3, TRAILR-4, XIAP, TGFBI, RPL31, LUM, HSDL2, etc., were down-regulated. Most of these genes are related to signaling pathways that play an important function in mediating cell proliferation, cell apoptosis, and cell cycle. For example, TGFβ/Smads signaling pathway exhibited close participation in regulating cancer development and progression. Specifically, Smads participate in TGFβ signaling as a class of intracellular molecules that act during cancer metastases. Most studies have revealed dual roles for Smad2, Smad3, and Smad4, while only a few insights have been obtained for Smad5 [16,17]. Li et al. demonstrated that miR-195-induced downregulation of Smad5 could lead to suppressed EMT in nasopharyngeal carcinoma (NPC), implying that Smad5 could be a potential therapeutic target for treating NPC [17]. We observed the down-regulation of TGFBI and Smad2 in the SYT7-silenced group, which can finally be correlated with a reduction in cell migration and invasion.
Another potent regulator of cancer is the ribosomal protein L31 (RPL31), which has been demonstrated as critical for prostate and colorectal cancer cell proliferation and cell cycle via regulation of p53 and p21 levels [18,19]. In our study, RPL31 was significantly down-regulated in both GeneChip PrimeView PathArray™ and Western blot validation, further confirming the potential of the RPL31 gene as a downstream target of SYT7.
Likewise, the hydroxysteroid dehydrogenase-like 2 (HSDL2) gene has also been implicated in exerting oncogenic roles in ovarian cancer by promoting cell growth and metastasis [20]. Specifically, the down-regulation of HSDL2 has been shown to inhibit cell proliferation and motility while facilitating apoptosis and cell cycle arrest [21]. Similar findings have been observed across various cancer types. For instance, Bing et al. identified that HSDL2 depletion hindered cell growth but facilitated cell cycle arrest in breast cancer [22]. Furthermore, Shi et al. demonstrated that HSDL2 played an oncogenic role in promoting lung cancer cell proliferation and metastasis via AKT2 up-regulation [23]. In our study, HSDL2 was reduced to an undetectable level in the SYT7-silenced group, thereby linking it to the suppression of cell proliferation, invasion, and tumorigenesis. Integrins (ITGs) are transmembrane heterodimers with one α chain and one β chain. These chains interact with the ITG receptors and further mediate extracellular signal transductions to regulate cell survival, apoptosis, cell growth, and motility [[24], [25], [26]]. Akira et al. reported that ITGB5 downregulation was intricately linked to distant metastases and poor prognosis in squamous cell carcinoma of the tongue [27]. However, we found that the ITGB5 level decreased in the SYT7 silenced group, contradicting the abovementioned findings. Therefore, further investigations are needed to reveal the underlying mechanisms.
5. Conclusion
In this study, we are the first to report on the tumor-promoting functions of SYT7 in CESC. Silencing SYT7 expression robustly inhibited cell proliferation, colony formation, migration, and invasion, while concurrently promoting apoptosis. Moreover, SYT7 knockdown cells demonstrated compromised tumorigenic potential and suppressed tumor growth. Mechanistically, the silencing of SYT7 led to the upregulation of Caspase 8, TNF-R1, and HSPA5 genes, while the downregulation of TGFBI, RPL31, LUM, HSDL2, ITGB5, and Smad5 genes was observed. Nevertheless, this study had several limitations. Firstly, the quantity of normal tissues was small in comparison to tumor tissues, resulting in limited statistical power. The findings from bioinformatics analysis should be corroborated with different cohorts. The potential downstream targets of SYT7 offer initial insights into the cancer-promoting mechanism of SYT7. In the future, more samples from diverse cohorts will be included to further authenticate the results obtained from bioinformatics analysis. Furthermore, the regulatory mechanism of SYT7 warrants further investigation.
Ethics approval
The collection and usage of the clinical samples were performed upon receiving ethical approval from The First Affiliated Hospital of Guangxi Medical University [Approve number: 2021(KY-E−347)] and conducted with informed consent from participants. The animal experimental protocols also received ethical approval from The Animal Care & Welfare Committee of Guangxi Medical University. [Approve number: 201906283].
Funding statement
Research funding was granted by the National natural science foundation of China (grant no.81860457), China Postdoctoral Science Foundation General Project (grant no.2019M663411), Guangxi Science and Technology Project – Key research and development plan (grant no. Guike AB23026003), medical and health appropriate technology development and promotion application project of Guangxi Province (grant no.S2022073), and the Guangxi Medical University Training Program for Distinguished Young Scholars.
Data availability statement
All analyzed data are provided within the manuscript and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE200526.
CRediT authorship contribution statement
Jinbing Huang: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing – original draft. Wensheng Xu: Conceptualization, Data curation, Formal analysis, Writing – review & editing. Qiaoqiao Huang: Investigation, Methodology, Writing – original draft. Erling Chen: Data curation, Formal analysis, Writing – review & editing. Junying Chen: Conceptualization, Data curation, Formal analysis, Writing – original draft, Writing – review & editing, Project administration.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:Junying Chen reports financial support was provided by National Natural Science Foundation of China. Junying Chen reports financial support was provided by China Postdoctoral Science Foundation General Project. Junying Chen reports financial support was provided by Guangxi Medical University Training Program for Distinguished Young Scholars. Junying Chen reports financial support was provided by medical and health appropriate technology development and promotion application project of Guangxi Province. Junying Chen reports financial support was provided by Guangxi Science and Technology Project.
Acknowledgements
We thank Medgy Technology Co., Ltd. for editing and proofreading the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e24806.
List of abbreviations
- SYT7
synaptotagmin 7
- CESC
Cervical squamous cell carcinoma
- SCC
squamous cell carcinoma
- EMT
epithelial-mesenchymal
- HR
transition hazard ratio
- CI
confidence interval
- DMSO
dimethyl sulfoxide
Appendix A. Supplementary data
The following are the Supplementary data to this article.
References
- 1.Singh D., Vignat J., Lorenzoni V., Eslahi M., Ginsburg O., Lauby-Secretan B., Arbyn M., Basu P., Bray F., Vaccarella S. Global estimates of incidence and mortality of cervical cancer in 2020: a baseline analysis of the WHO Global Cervical Cancer Elimination Initiative. Lancet Global Health. 2023;11(2):e197–e206. doi: 10.1016/S2214-109X(22)00501-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rahangdale L., Mungo C., O'Connor S., Chibwesha C.J., Brewer N.T. Human papillomavirus vaccination and cervical cancer risk. BMJ. 2022;379 doi: 10.1136/bmj-2022-070115. [DOI] [PubMed] [Google Scholar]
- 3.Ferrara P., Dallagiacoma G., Alberti F., Gentile L., Bertuccio P., Odone A., Prevention Diagnosis and treatment of cervical cancer: a systematic review of the impact of COVID-19 on patient care. Prev. Med. 2022;164 doi: 10.1016/j.ypmed.2022.107264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jin H., Pang Q., Fang M., Wang Y., Man Z., Tan Y., Liu H. Syt-7 overexpression predicts poor prognosis and promotes cell proliferation in hepatocellular carcinoma. Future Oncol. 2020;16(34):2809–2819. doi: 10.2217/fon-2020-0262. [DOI] [PubMed] [Google Scholar]
- 5.Kanda M., Tanaka H., Shimizu D., Miwa T., Umeda S., Tanaka C., Kobayashi D., Hattori N., Suenaga M., Hayashi M. SYT7 acts as a driver of hepatic metastasis formation of gastric cancer cells. Oncogene. 2018;37(39):5355–5366. doi: 10.1038/s41388-018-0335-8. [DOI] [PubMed] [Google Scholar]
- 6.Liu X., Li C., Yang Y., Liu X., Li R., Zhang M., Yin Y., Qu Y. Synaptotagmin 7 in twist-related protein 1-mediated epithelial–Mesenchymal transition of non-small cell lung cancer. EBioMedicine. 2019;46:42–53. doi: 10.1016/j.ebiom.2019.07.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang W., Long J., Tang P., Chen K., Guo G., Yu Z., Lin J., Liu L., Zhan R., Xu Z. SYT7 regulates the progression of chronic lymphocytic leukemia through interacting and regulating KNTC1. Biomark. Res. 2023;11(1):58. doi: 10.1186/s40364-023-00506-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Z., Xu X., Chen D., Zhang L., Pan Y., Liu D., Shen M., Chen M. Circ_0022340 promotes colorectal cancer progression via HNRNPC/EBF1/SYT7 or miR-382-5p/ELK1 axis. Cell Mol Biol (Noisy-le-grand) 2022;68(7):107–116. doi: 10.14715/cmb/2022.68.7.18. [DOI] [PubMed] [Google Scholar]
- 9.Goldman M.J., Craft B., Hastie M., Repečka K., McDade F., Kamath A., Banerjee A., Luo Y., Rogers D., Brooks A.N. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020;38(6):675–678. doi: 10.1038/s41587-020-0546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dlugosz A., Muschiol S., Zakikhany K., Assadi G., D'Amato M., Lindberg G. Human enteroendocrine cell responses to infection with Chlamydia trachomatis: a microarray study. Gut Pathog. 2014;6(1):24. doi: 10.1186/1757-4749-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Divangahi M., Chen M., Gan H., Desjardins D., Hickman T.T., Lee D.M., Fortune S., Behar S.M., Remold H.G. Mycobacterium tuberculosis evades macrophage defenses by inhibiting plasma membrane repair. Nat. Immunol. 2009;10(8):899–906. doi: 10.1038/ni.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao B., Li J., Fan Y., Ye M., Lv S., Xu B., Chai Y., Zhou Z., Wu M., Zhu X. Downregulation of SYT7 inhibits glioblastoma growth by promoting cellular apoptosis. Mol. Med. Rep. 2017;16(6):9017–9022. doi: 10.3892/mmr.2017.7723. [DOI] [PubMed] [Google Scholar]
- 13.Fei Z., Gao W., Xie R., Feng G., Chen X., Jiang Y. Synaptotagmin-7, a binding protein of P53, inhibits the senescence and promotes the tumorigenicity of lung cancer cells. Biosci. Rep. 2019;39(2) doi: 10.1042/BSR20181298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong S., Pan J., Shen Y.B., Zhu L.X., Chen L., Zhu F., Li H., Shen H.X., Xia Q., Wu Y.J., et al. SYT7 plays a role in promoting thyroid cancer by mediating HMGB3 ubiquitination. Endocr. Relat. Cancer. 2022;29(4):175–189. doi: 10.1530/ERC-21-0146. [DOI] [PubMed] [Google Scholar]
- 15.Wu Z., Sun Z., Huang R., Zang D., Wang C., Yan X., Yan W. Silencing of synaptotagmin 7 regulates osteosarcoma cell proliferation, apoptosis, and migration. Histol. Histopathol. 2019;35(3):303–312. doi: 10.14670/HH-18-174. [DOI] [PubMed] [Google Scholar]
- 16.Han G., Wang X.-J. Roles of TGFβ signaling Smads in squamous cell carcinoma. Cell Biosci. 2011;1(1):41. doi: 10.1186/2045-3701-1-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li S., Zhao B., Zhao H., Shang C., Zhang M., Xiong X., Pu J., Kuang B., Deng G. Silencing of long non-coding RNA SMAD5-AS1 reverses epithelial mesenchymal transition in nasopharyngeal carcinoma via microRNA-195-dependent inhibition of SMAD5. Front. Oncol. 2019;9:1246. doi: 10.3389/fonc.2019.01246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maruyama Y., Miyazaki T., Ikeda K., Okumura T., Sato W., Horie-Inoue K., Okamoto K., Takeda S., Inoue S. Short hairpin RNA library-based functional screening identified ribosomal protein L31 that modulates prostate cancer cell growth via p53 pathway. PLoS One. 2014;9(10) doi: 10.1371/journal.pone.0108743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai M.D., Xu J. Ribosomal proteins and colorectal cancer. Curr. Genom. 2007;8(1):43–49. doi: 10.2174/138920207780076938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X., Wang Y. Identification of hub genes and key pathways associated with the progression of gynecological cancer. Oncol. Lett. 2019;18(6):6516–6524. doi: 10.3892/ol.2019.11004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun Q., Zhang Y., Su J., Li T., Jiang Y. Role of hydroxysteroid dehydrogenase-like 2 (HSDL2) in human ovarian cancer. Med. Sci. Mon. Int. Med. J. Exp. Clin. Res. 2018;24:3997–4008. doi: 10.12659/MSM.909418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong B., Yang Y., Han A., Zhang S., Lin Z., Wang Y., Piao J. Ectopic expression of HSDL2 is related to cell proliferation and prognosis in breast cancer. Cancer Manag. Res. 2019;11:6531–6542. doi: 10.2147/CMAR.S205316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi Y., Mao Z., Huang Y., Sun Y., Cao Q., Yin X., Huang J., Zhang Q. Knockdown of HSDL2 inhibits lung adenocarcinoma progression via down-regulating AKT2 expression. Biosci. Rep. 2020;40(4) doi: 10.1042/BSR20200348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Danen E.H., Sonnenberg A. Integrins in regulation of tissue development and function. J. Pathol. 2003;200(4):471–480. doi: 10.1002/path.1416. [DOI] [PubMed] [Google Scholar]
- 25.Calderwood D.A., Shattil S.J., Ginsberg M.H. Integrins and actin filaments: reciprocal regulation of cell adhesion and signaling. J. Biol. Chem. 2000;275(30):22607–22610. doi: 10.1074/jbc.R900037199. [DOI] [PubMed] [Google Scholar]
- 26.Stupack D.G., Cheresh D.A. Get a ligand, get a life: integrins, signaling and cell survival. J. Cell Sci. 2002;115(19):3729–3738. doi: 10.1242/jcs.00071. [DOI] [PubMed] [Google Scholar]
- 27.Kurokawa A., Nagata M., Kitamura N., Noman A.A., Ohnishi M., Ohyama T., Kobayashi T., Shingaki S., Takagi R. Diagnostic value of integrin α3, β4, and β5 gene expression levels for the clinical outcome of tongue squamous cell carcinoma. Cancer. 2008;112(6):1272–1281. doi: 10.1002/cncr.23295. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All analyzed data are provided within the manuscript and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE200526.