Abstract
The land plant Arabidopsis thaliana contains three closely related nuclear genes encoding phage-type RNA polymerases (RpoT;1, RpoT;2 and RpoT;3). The gene products of RpoT;1 and RpoT;3 have previously been shown to be imported into mitochondria and chloroplasts, respectively. Here we show that the transit peptide of RpoT;2 possesses dual targeting properties. Transient expression assays in tobacco protoplasts as well as stable transformation of Arabidopsis plants demonstrate efficient targeting of fusion peptides consisting of the N-terminus of RpoT;2 joined to green fluorescent protein to both organelles. Thus, RpoT;2 might be the first RNA polymerase shown to transcribe genes in two different genomes. RNA polymerase activity of recombinant RpoT;2 is uneffected by the inhibitor tagetin, qualifying the gene product of RpoT;2 as a phage-type polymerase.
INTRODUCTION
Plants possess two extranuclear transcriptionally active compartments, mitochondria and plastids, relying on nucleus-encoded proteins for transcription. Plastids contain at least two RNA polymerases (RNAPs) termed PEP (plastid-encoded polymerase) and NEP (nuclear-encoded polymerase) (Hess and Börner, 1999). PEP contains the plastid-encoded subunits of a eubacterial-type core RNAP, but nuclear encoded sigma-like factors seem to confer promoter recognition to the enzyme. Based on its biochemical properties, NEP was suggested to be an RNAP related to polymerases of phages like T7 and T3 (Lerbs-Mache, 1993). In yeast mitochondria, the core RNAP is a bacteriophage-type enzyme encoded by a nuclear gene (Masters et al., 1987). Recently, a number of nuclear genes belonging to the family of genes encoding phage-type RNAPs were cloned from a variety of organisms. In land plants, genes encoding RNA polymerases of this type (RpoT genes) were identified in Chenopodium album (Weihe et al., 1997), Arabidopsis thaliana (Hedtke et al., 1997), maize (Young et al., 1998; Chang et al., 1999) and wheat (Ikeda and Gray, 1999).
In Arabidopsis which has been studied more intensively three closely related genes were found and designated RpoT;1, RpoT;2 and RpoT;3 (Hedtke et al., 1997; Sanchez and Schuster, DDBJ/EMBL/GenBank accession No. AJ001037). The subcellular localization of RpoT;1 and RpoT;3 has previously been investigated (Hedtke et al., 1997, 1999). RpoT;1 was identified as a mitochondrially targeted enzyme, whereas RpoT;3 showed exclusively plastid (chloroplast) targeting and thus represents a nuclear encoded plastid RNA polymerase (NEP). Mitochondrial and chloroplast targeting has also been demonstrated for the maize RpoTm and RpoTp, respectively (Chang et al., 1999).
We have now investigated the targeting properties of the putative transit peptide of Arabidopsis RpoT;2. Our data indicate that RpoT;2 is targeted to both mitochondria and plastids, i.e. this RNAP might be involved in the transcription of two different genomes.
RESULTS
RpoT;2 targeting studies in tobacco protoplasts
The cDNA sequence of Arabidopsis RpoT;2 of 3784 nucleotides (nt) (DDBJ/EMBL/GenBank accession No. AJ278248) was obtained by aligning overlapping RT–PCR and RACE (rapid amplification of cDNA ends) products. The 5′-RACE reaction gave rise to a single product; the untranslated leader sequence of 231 nt contains one in-frame stop codon. The 3′-UTR was found to be variable, 435 or 493 nt, indicating the presence of at least two polyadenylation sites.
Mitochondrial and plastid transit peptides exhibit primary sequence features such as richness in hydroxylated amino acids and a low abundance of acidic residues as well as the ability to form α-helices and amphiphilic β-sheets, respectively (Glaser et al., 1998; Soll and Tien, 1998). Analysis of RpoT;2 using common targeting prediction programs (http://psort.nibb.ac.jp/) did not yield conclusive results, but detected a consensus sequence pattern for the cleavage of mitochochondrial targeting sequences at residue 104.
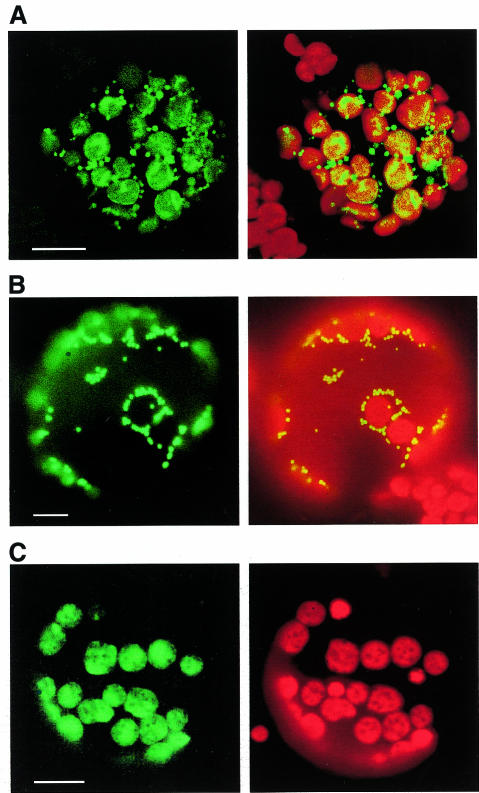
To investigate the cellular localization of RpoT;2 129 N-terminal amino acids were translationally fused to the green fluorescent protein (GFP) coding region and used to transform tobacco protoplasts. Transient expression was examined using confocal laser scanning microscopy (CLSM). The RpoT;2–GFP construct gave rise to bright-green GFP fluorescence both in typical mitochondrial structures of punctate morphology and in chloroplasts (Figure 1A, green channel). Combination of green and red (chlorophyll autofluorescence) channels confirmed co-localization of red and green fluorescence in chloroplasts which appeared in the merged image greenish-yellow to orange (Figure 1A, merged image). A control construct with known mitochondrial targeting showed the expected punctate GFP localization (Figure 1B). Using a plastid-targeted control construct, the green fluorescence was coincident with the chloroplast structures (Figure 1C).
Fig. 1. Transient expression of GFP fusions in wild-type tobacco protoplasts. (A) Expression of construct RpoT;2–GFP examined by CLSM. GFP fluorescence was detected in the green channel (left) and merged (right) with chlorophyll autofluorescence visible in the red channel. (B) Mitochondrial targeting control CoxIV–GFP (Akashi et al., 1998). (C) Plastid targeting control RecA–GFP (Akashi et al., 1998). The control images were taken by epifluorescence microscopy using GFP- (left) and FITC- (right) filter sets, respectively. Scale bars indicate 10 µm.
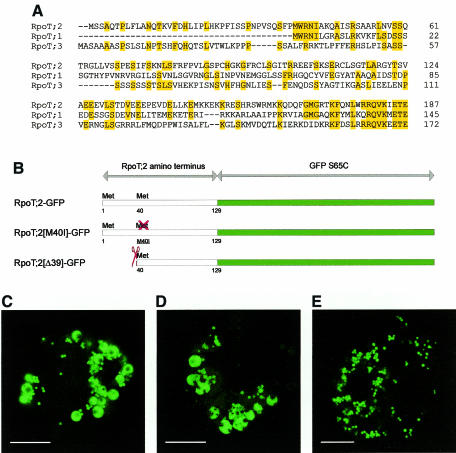
Although the complete sequence of Arabidopsis RpoT;2 is more similar to the mitochondrial polymerase RpoT;1 (Börner et al., 1999), its putative transit peptide exhibits features of the N-termini of both RpoT;1 and RpoT;3. While the 39 N-terminal amino acids of RpoT;2 show limited similarity to the N-terminus of RpoT;3, the extreme N-terminus of RpoT;1 aligns to a similar region in RpoT;2 (conserving the methionine start codon of RpoT;1 and 4 subsequent amino acids) (Figure 2A). Hence, a potential second translation initiation site is present in the N-terminal sequence of RpoT;2. Two further constructs were generated to investigate both sites independently (Figure 2B). To show that dual targeting is not due to concurrent initiation from both methionines, the in-frame AUG at codon position 40 was mutated to encode isoleucine instead of methionine (RpoT;2[M40I]–GFP). The other construct contained a shortened transit peptide starting at the second methionine (RpoT;2[Δ39]–GFP). To enhance the sensitivity of GFP visualization by avoiding chlorophyll autofluorescence, protoplasts were prepared from white leaves of tobacco plants lacking the plastid rpoB gene (De Santis-Maciossek et al., 1999). Figure 2 shows a comparison of the GFP localization employing the different constructs. Both RpoT;2–GFP (Figure 2C) and RpoT;2[M40I]–GFP (Figure 2D) exhibited dual targeting with GFP fluorescence occuring in both mitochondria and plastids. The latter are characterized by the occurence of typical membrane-bounded vesicles inside the organelles (De Santis-Maciossek et al., 1999). However, when we used the shortened transit peptide starting at the second in-frame AUG (RpoT;2[Δ39]–GFP), plastids showed no fluorescence, leaving mitochondria as the sole target of GFP (Figure 2E). We conclude that initiation at the first methionine is essentially required for dual targeting of RpoT;2 into plastids and mitochondria.
Fig. 2. Functional analysis of the two methionine residues in the N-terminus of RpoT;2. (A) Sequence comparison of of the three Arabidopisis RpoT gene products. Only the variable N-termini up to the motif RQVKXETE are shown. The alignment was performed using Multalin (Corpet, 1988); identical amino acid positions are highlighted. (B) Schematic presentation of the three different GFP fusions used in transient expression experiments (for details see Results). (C–E) Expression of RpoT;2–GFP (C), RpoT;2[M40I]–GFP (D), and RpoT;2[Δ39]–GFP (E) in protoplasts of the rpoB– mutant of N. tabacum. Pictures were taken by CLSM in the green channel. Scale bars indicate 10 µm.
Stable expression of RpoT;2–GFP in Arabidopsis
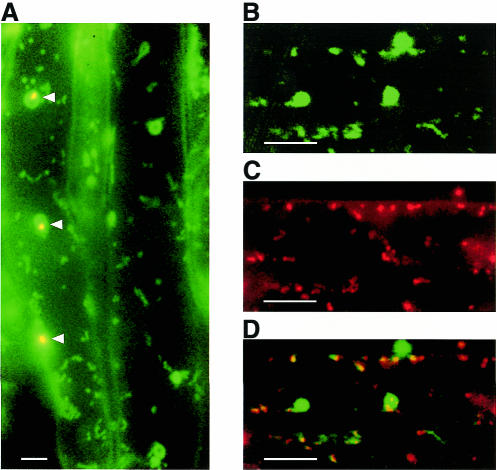
To confirm the data obtained by transient expression with tobacco protoplasts in a homologous system, a construct containing the full-length N-terminus of RpoT;2 was used for stable transformation of Arabidopsis. In root tissue of transgenic plants GFP fluorescence was detected in typical punctate mitochondria as well as in much larger structures being qualified as plastids by chlorophyll autofluorescence in some pigment containing chloroplasts (indicated by arrowheads, Figure 3A). Plastid targeting was corroborated unequivocally by observations in green tissue of the transformants (data not shown). Counterstaining with Mitotracker-Red, a mitochondrial specific dye, proved co-localization of GFP fluorescence within mitochondria (Figure 3B–D). Plastid GFP fluorescence was not superimposed by Mitotracker-Red. Thus, subcellular localization of the GFP fusion constructs in stably transformed Arabidopsis plants was the same as in transiently expressing tobacco protoplasts, confirming that dual targeting is indeed a general property of the RpoT;2 transit peptide in Arabidopsis.
Fig. 3. Expression of the RpoT;2-mGFP5 in stably transformed Arabidopsis plants. (A) Fluorescence in root hair cells using the FITC-filter set. Under tissue culture conditions, some of the root plastids (arrowheads) accumulate photosynthetic pigments as evident by their yellowish color. (B–D) Counterstain of root tissue with MitoTracker-Red. GFP (B), and red MitoTracker (C) fluorescence were detected separately and merged (D) using Photoshop 5.0 software. Images were taken by epifluorescence microscopy using filter sets 488013 (B) and 488015 (C). Bars equal 10 µm.
Activity of recombinant RpoT;2 protein
Since an enzymatic activity has not been demonstrated yet for an Arabidopsis RpoT gene product, we fused the coding region of the gene with exception of 104 N-terminal amino acid residues coding for the putative transit peptide to a thioredoxin-His-patch sequence and expressed the construct (Trx-RpoT;2) in Escherichia coli. A 112 kDa recombinant protein was found to be expressed in soluble form. The polypeptide was partially purified using a nickel agarose matrix and a two-step dialysis procedure (Figure 4A). The recombinant protein revealed unspecific RNA polymerase activity in an in vitro assay with activated calf thymus DNA as a template. To prove that recombinant Trx-RpoT;2 preparations were free of E. coli RNAP, we tested the effects of different transcription inhibitors (Figure 4B). Most remarkably, tagetin, a potent inhibitor of eubacterial-type polymerases which has been also used to discriminate between the plastid-encoded eubacterial-type RNAP and the nuclear-encoded RNAP in chloroplast transcription (Kapoor et al., 1997; Liere and Maliga, 1999), did not inhibit the activity of the recombinant protein, implying that RpoT;2 indeed encodes a functional phage-type RNA polymerase. Actinomycin D, however, did not completely suppress RNA synthesis in the Trx-RpoT;2 preparations.
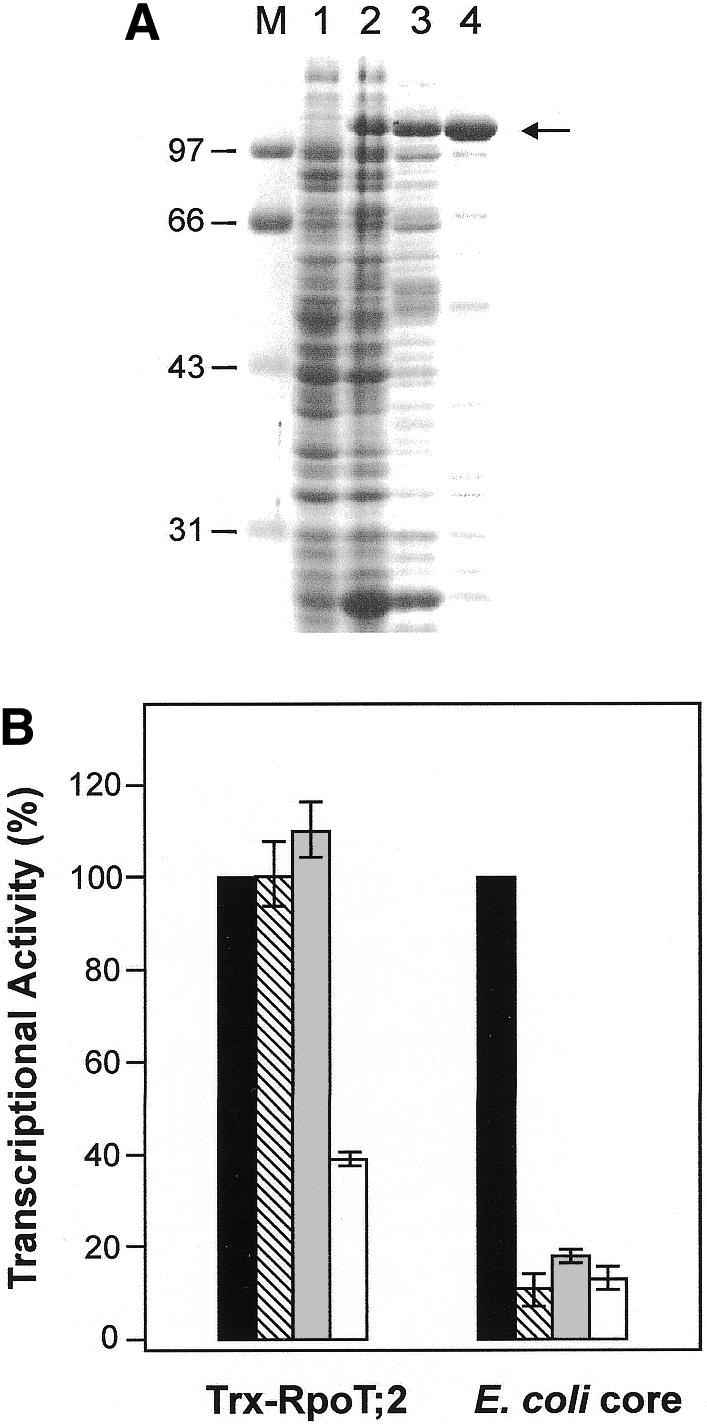
Fig. 4. Purification and RNAP activity of recombinant RpoT;2. (A) SDS–PAGE of partially purified Trx-RpoT;2. Lanes: M, marker proteins, molecular masses in kDa; 1, uninduced E. coli lysate, soluble fraction; 2, induced cells, soluble fraction; 3, Ni-NTA-agarose purified fraction; 4, purified recombinant RNA polymerase after two-step dialysis purification. Arrow: 112 kDa recombinant Trx-RpoT;2. (B) RNA polymerase activity of Trx-RpoT;2 as incorporation of [32P]UTP in vitro. Bars indicate relative RNA polymerase activity: black, no inhibitor; hatched, tagetin; gray, rifampicin; white, actinomycin D. Control reactions were performed using E. coli core RNA polymerase.
DISCUSSION
Our data provide clear-cut evidence for dual targeting properties of the N-terminus of RpoT;2, one of the three phage-type RNA polymerases encoded in the nuclear genome of A. thaliana. In contrast, for the other two RNAPs, distinct mitochondrial (RpoT;1) and chloroplast (RpoT;3) import pathways have been demonstrated (Hedtke et al., 1999). Dual targeting to mitochondria and chloroplasts has been shown only for a few other enzymes so far (reviewed in Small et al., 1998; Peeters et al., 2000).
Activity assays performed with partially purified Trx-RpoT;2 protein demonstrate for the first time an enzymatic activity for a recombinant phage-type RNA polymerase from higher plants. As expected, RNA synthesis by Trx-RpoT;2 is not influenced by tagetin. We have no explanation yet for the observed incomplete inhibition by actinomycin D.
According to their sequence and conservation of intron positions, RpoT;1, RpoT;2 and RpoT;3 are paralogous genes which most likely arose by duplication from an ancestral gene for a mitochondrial RNAP (Börner et al., 1999; Hess and Börner, 1999). Our data suggest that RpoT;2 aquired during evolution a bipartite transit peptide with the first 39 amino acids being required for chloroplast targeting and the remaining C-terminal part being sufficient for mitochondrial targeting (Figure 2).
The nuclear genome of tobacco harbours genes homologous to the Arabidopsis genes RpoT;1, RpoT;2 and RpoT3 coding also for similar putative transit sequences (B. Hedtke, J. Legen and R.G. Herrmann, in preparation). In the case of cereals, however, only two RpoT genes have been detected so far (Chang et al., 1999; Ikeda and Gray, 1999), and it remains to be shown if a third phage-type RNAP exists. Sequence information on RpoT genes from more plants including also algae, mosses and ferns will provide a more detailed insight into the evolution of this gene family in general and specifically into the evolution of the targeting peptides.
Taken together, our data demonstrate that the nuclear Arabidopsis RpoT;2 gene encodes a phage-type RNA polymerase which is targeted to both mitochondria and chloroplasts. Therefore, the results strongly suggest that one and the same RNA polymerase functions in the transcription of genes residing in two different genomes. Eubacteria, to which also the potential ancestors of mitochondria and plastids belong, use for transcription only one RNA polymerase. Although having much smaller genomes than their eubacterial ancestors, mitochondria of dicotyledonous plants seem to use at least two (RpoT;1 and RpoT;2) and plastids at least three (the eubacterial-type plastid-ecoded RNAP and the nuclear-encoded RpoT;2 and RpoT;3) different RNA polymerases. Recent data on the transcription of rDNA in spinach chloroplasts suggest an even more complex situation, namely the occurrence of another nuclear encoded plastid RNAP activity which seems to be different from both eubacterial-type and phage-type RNAPs (Bligny et al., 2000). Our data indicate a highly specific path for the evolution of transcriptional machineries in the mitochondria and plastids of higher plants. To explore why these organelles possess such a highly sophisticated transcriptional apparatus will be a challenging task of future research.
METHODS
Cloning of RpoT;2 cDNA. RpoT;2 (DDBJ/EMBL/GenBank accession No. AJ001037; H. Sanchez and W. Schuster, unpublished) was amplified in the course of the present study as a composite full-length cDNA sequence (DDBJ/EMBL/GenBank accession No. AJ278248) of 3784 nt. 5′- and 3′-RACE reactions were performed with the gene-specific primers GCAGCTCTTTCCCATTCTTCAGTCTCG and AGACATCCCTCCAGACCCTATCGC, respectively, using the SMART RACE Kit (Clontech). A cDNA fragment spanning the entire CDS was amplified by RT–PCR with primers ATGTCCAGTGCTCAAACCCC and GTACAGCTCAAGCCTTGGGG from total RNA of A. thaliana (ecotype Columbia) leaves. All PCR products were cloned into vector pGEM-T (Promega) and sequenced on an ABI373 (Applied Biosystems).
Generation of targeting constructs. The RpoT;2 sequence in construct RpoT;2–GFP was amplified using primers F1: tctagaATGTCCAGTGCTCAAACCC and R1: gtcgacAGACCTCCTCTTCGGCTACAC. In construct RpoT;2[M40I]–GFP, amino acid Met40 was substituted by Ile using primers Fmut: TCCTTCCCCATcTGGAGAAACATTG and Rmut: CAATGTTTCTCCAgATGGGGAAGGA in combination with primers R1 and F1, respectively, to introduce the base change. The shortened leader peptide of construct RpoT;2[Δ39]–GFP was generated using F2: tctagaATGTGGAGAAACATTGCTAAACA and R1. PCR products were ligated into vector pGEM-T and cutted using XbaI and SalI. Fragments were inserted into Vector pOL–GFP S65C (Peeters et al., 2000) digested with SpeI and SalI.
To generate plasmid pGPTV-RpoT;2-mGFP5, the 129 N-terminal amino acids of RpoT;2 were amplified using primers ggatccaaggagatataacaATGTCCAGTGCTCAAACCCCAC and gaattcGACCTCTTCGGCTACACTCG. Fusion with mGFP5 and insertion into vector pGPTV-bar were performed as described previously (Hedtke et al., 1999).
Transient expression in tobacco protoplasts and microscopy. Protoplasts were prepared from green wild-type leaves of Nicotiana tabacum (var. SNN) or white leaves of a mutant of N. tabacum lacking PEP activity due to targeted disruption of the plastid rpoB gene (De Santis-Maciossek et al., 1999). Preparation of protoplasts and transformation followed the protocol of Morgan and Ow (1995).
CLSM was done using an LSM 510 (Zeiss) with excitation at 488 nm. Fluorescence was detected in green (BP 505–530) and red channels (LP 560). Images were merged using LSM 510 software. Epifluorescence microscopy was done on an Axioscope (Zeiss) using GFP- (Zeiss filter set 488013; excitation 470/20, emission 505–530) and FITC- (Zeiss filter set 488009; excitation 450–490, emission LP 520) filter sets. MitoTracker-Red (Molecular Probes) staining was performed as described previously (Hedtke et al., 1999) and visualized using Zeiss filter set 488015 (excitation 546/12, emission LP 590).
Stable Transformation of Arabidopsis. Arabidopsis thaliana (ecotype Columbia) plants were transformed according to Bechtold et al. (1993) using Agrobacterium tumefaciens strain EHA105 harbouring pGPTV-RpoT;2-mGFP5. Plants were selected on plates containing 30 mg/l phosphinotricine (Sigma). Integration of RpoT;2-mGFP5 was confirmed by PCR and Southern hybridization (data not shown).
Expression of recombinant RpoT;2 in E. coli. A cDNA sequence of RpoT;2 was amplified by RT-PCR using primers GAGTTTTCCAAGAGCGAGAG and TCAGTTGAAGAAATAAGGTGAATC and inserted in-frame with an N-terminal thioredoxin His-patch sequence in vector pBAD/Thio-TOPO (Invitrogen). Expression was induced by adding arabinose to a final concentration of 0.02%. Cells were harvested by centrifugation and lysed in 0.1 M Tris–HCl, pH 7.8; 300 mM NaCl, 0.1 mM DTT in a mill with glass pearls. The soluble fraction, obtained by centrifugation at 18000 g for 15 min, was allowed to bind to a nickel NTA-agarose matrix (Qiagen). After washing the resin with 0.1 M Tris–HCl, pH 6.0, 300 mM NaCl, the bound protein was eluted with 100 mM imidazole, 100 mM Tris–HCl, 300 mM NaCl, pH 6.0. The recombinant RNAP was further enriched by a two-step dialysis procedure (Zawadzki and Gross, 1991).
RNAP activity assays. RNAP activity was measured as incorporation of [32P]UTP using activated calf thymus DNA as a template. 20 µl reactions contained 1 µl of partially purified recombinant RNAP (1.5 µg protein), 1× reaction buffer (40 mM Tris–HCl pH 7.9, 6 mM MgCl2, 10 mM NaCl, 2 mM spermidine), 0.5 mM each ATP, CTP and GTP, 25 µM UTP, 5 µCi [32P]UTP, 10 units RNase inhibitor (MBI Fermentas), and 1 µg activated calf thymus DNA. Inhibitor test reactions additionally contained 50 units of tagetin (Epicentre), 1 µg/ml of rifampicin or 50 µg/ml actinomycin D, respectively. Control reactions with 0.5 units of E. coli core RNAP (Epicentre) were performed under the same conditions. Aliquots were spotted onto DE81 paper (Whatman) and washed three times with 7.5% sodium phosphate. Radioactivity of the dried filters was counted in a liquid scintillation counter LS6000SC (Beckman).
Acknowledgments
ACKNOWLEDGEMENTS
We gratefully acknowledge the support of C. Emanuel, K. Kuehn, and C. Stock. We thank Dr I. Small for providing vector pOL–GFP, Dr J. Haseloff for vector mGFP4, and Dr. R.G. Herrmann for rpoB-deficient tobacco plants. CLSM was performed at the Fraunhofer Institut für Mikroelektronik und Zuverlässigkeit through the generous support of Dr S. Fiedler (BMBF-FKZ 01M2989D). This work was supported by the Deutsche Forschungsgemeinschaft (SFB 429).
REFERENCES
- Akashi K., Grandjean, O. and Small, I. (1998) Potential dual targeting of an Arabidopsis archaebacterial-like histidyl-tRNA synthetase to mitochondria and chloroplasts. FEBS Lett., 431, 39–44. [DOI] [PubMed] [Google Scholar]
- Bechtold N., Ellis, J. and Pelletier, G. (1993). In planta Agrobacterium gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris, 316, 1194–1199. [Google Scholar]
- Bligny M., Courtois, F., Thaminy, S., Chang, C.C., Lagrange, T., Baruah-Wolff, J., Stern, D.B. and Lerbs-Mache, S. (2000) Regulation of plastid rDNA transcription by interaction of CDF2 with two different RNA polymerases. EMBO J., 191851–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börner T., Hedtke, B., Hess, W.R., Legen, J., Herrmann, R.G. and Weihe, A. (1999) Phage-type RNA polymerases in higher plants. In Argyroudi-Akoyunoglou, J.H. and Senger, H. (eds) The Chloroplast: From Molecular Biology to Biotechnology, Kluwer Academic Publishers, Dordrecht, The Netherlands, pp. 73–78.
- Chang C., Sheen, J., Bligny, M., Niwa, Y., Lerbs-Mache, S. and Stern, D.B. (1999) Functional analysis of two maize cDNAs encoding T7-like RNA polymerases. Plant Cell, 11, 911–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F. (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res., 16, 10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santis-Maciossek G., Kofer, W., Bock, A., Schoch, S., Maier, R.M., Wanner, G., Rüdiger, W., Koop, H.U. and Herrmann, R.G. (1999) Targeted disruption of the plastid RNA polymerase genes rpoA, B and C1: molecular biology, biochemistry and ultrastructure. Plant J., 18, 477–489. [DOI] [PubMed] [Google Scholar]
- Glaser E., Sjoling, S., Tanudji, M. and Whelan, J. (1998) Mitochondrial protein import in plants. Plant Mol. Biol., 38, 311–338. [DOI] [PubMed] [Google Scholar]
- Hedtke B., Börner, T. and Weihe, A. (1997) Mitochondrial and chloroplast phage-type RNA polymerases in Arabidopsis. Science, 277, 809–811. [DOI] [PubMed] [Google Scholar]
- Hedtke B., Meixner, M., Gillandt, S., Richter, E., Börner, T. and Weihe, A. (1999) Green fluorescent protein as a marker to investigate targeting of organellar RNA polymerases of higher plants in vivo. Plant J., 17, 557–561. [DOI] [PubMed] [Google Scholar]
- Hess W.R. and Börner, T. (1999) Organellar RNA polymerases of higher plants. Int. Rev. Cytol., 190, 1–59. [DOI] [PubMed] [Google Scholar]
- Ikeda T.M. and Gray, M.W. (1999) Identification and characterization of T3/T7 bacteriophage-like RNA polymerase sequences in wheat. Plant Mol. Biol., 40, 567–578. [DOI] [PubMed] [Google Scholar]
- Kapoor S., Suzuki, J.Y. and Sugiura, M. (1997) Identification and functional significance of a new class of non-consensus-type plasmid promoters. Plant J., 11, 327–337. [DOI] [PubMed] [Google Scholar]
- Lerbs-Mache S. (1993) The 110-kDa polypeptide of spinach plastid DNA-dependent RNA polymerase: Single-subunit enzyme or catalytic core of multimeric enzyme complexes? Proc. Natl Acad. Sci. USA, 90, 5509–5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liere K. and Maliga, P. (1999) In vitro characterization of the tobacco rpoB promoter reveals a core sequence motif conserved between phage-type plastid and plant mitochondrial promoters. EMBO J., 18, 249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters B.S., Stohl, L.L. and Clayton, D.A. (1987) Yeast mitochondrial RNA polymerase is homologous to those encoded by bacteriophages T3 and T7. Cell, 51, 89–99. [DOI] [PubMed] [Google Scholar]
- Morgan M.K. and Ow, D.W. (1995) Polyethylene glycol-mediated transfection of tobacco leaf mesophyll protoplasts: An experiment in the study of cre-lox recombination. In Maliga, P., Klessig, D.F., Cashmore, A.R., Gruissem, W. and Varner, J.E. (eds), Methods in Plant Molecular Biology: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 1–17.
- Peeters N.M., Chapron, A., Giritch, A., Grandjean, O., Lancelin, D., Lhomme, T., Vivrel, A. and Small, I. (2000) Duplication and quadruplication of Arabidopsis thaliana cysteinyl- and asparaginyl-tRNA synthetase genes of organellar origin. J. Mol. Evol., 50, 413–423. [DOI] [PubMed] [Google Scholar]
- Small I., Wintz, H., Akashi, K. and Mireau, H. (1998) Two birds with one stone: genes that encode products targeted to two or more compartments. Plant Mol. Biol., 38265–277. [PubMed] [Google Scholar]
- Soll J., and Tien, R. (1998) Protein translocation into and across the chloroplastic envelope membranes. Plant Mol. Biol., 38, 191–207. [PubMed] [Google Scholar]
- Weihe A., Hedtke, B. and Börner, T. (1997) Cloning and characterization of a cDNA encoding a bacteriophage-type RNA polymerase from the higher plant Chenopodium album. Nucleic Acids Res., 25, 2319–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young D.A., Allen, R.L., Harvey, A.J. and Lonsdale, D.M. (1998) Characterization of a gene encoding a single-subunit bacteriophage-type RNA polymerase from maize which is alternatively spliced. Mol. Gen. Genet., 260, 30–37. [DOI] [PubMed] [Google Scholar]
- Zawadzki V. and Gross, H.J. (1991) Rapid and simple purification of T7 RNA polymerase. Nucleic Acids Res., 19, 1948. [DOI] [PMC free article] [PubMed] [Google Scholar]