Abstract
We have mapped the positions of topoisomerase II binding sites at the centromere of the human Y chromosome using etoposide-mediated DNA cleavage. A single region of cleavage is seen at normal centromeres, spanning ∼50 kb within the centromeric alphoid array, but this pattern is abolished at two inactive centromeres. It therefore provides a marker for the position of the active centromere. Although the underlying centromeric DNA structure is variable, the position of the centromere measured in this way is fixed relative to the Yp edge of the array, and has retained the same position for >100 000 years.
INTRODUCTION
The centromere is essential for faithful chromosome transmission during cell division. Centromeric chromatin is associated with the kinetochore, the key site of attachment between the chromosome and the spindle during mitosis and meiosis (Pluta et al., 1995). Centromere functions include mediating chromosome movement and normal sister-chromatid separation, while the kinetochore provides the site for the metaphase/anaphase checkpoints (Taylor, 1999).
Centromere function in the yeast Saccharomyces cerevisiae is determined by specific DNA sequences. The minimal centromeric DNA is only 125 bp in length, and all 16 centromeres contain the same three elements (CDEs I, II and III), which bind essential centromeric proteins (Hyman and Sorger, 1995). The relationship between centromeric DNA sequence and function is simple: when centromeric DNA is present, an active centromere is formed; when it is absent, there is no active centromere. However, this may be a misleading model for other centromeres where DNA sequence alone does not determine function. No conserved DNA sequence is present and many of the centromeric and kinetochore proteins are also not conserved (Tyler-Smith and Floridia, 2000).
Normal human centromeres contain variably sized arrays of tandemly repeated 170 bp alphoid sequences (Tyler-Smith and Willard, 1993). However, alphoid DNA is not always associated with centromere activity: in stable dicentric chromosomes, one centromere is inactivated despite the presence of alphoid DNA. Moreover, there are chromosomes where centromeric function has been acquired by sequences, called neocentromeres, which do not normally have this function (Choo, 1997). It seems that, in humans, no specific DNA sequence is necessary or sufficient for centromere function, and centromere identity may be determined by a specific ‘higher order structure’ (Karpen and Allshire, 1997). Centromere activation and inactivation seem to be regulated by epigenetic events, heritable changes without a corresponding change in primary DNA sequence. The nature of these epigenetic modifications is now a key question. We know, for example, that there is a particular type of chromatin structure at yeast and mammalian centromeres. In S. cerevisiae, a 220–250 bp region of the centromeric DNA is protected from nuclease digestion, and flanking nucleosomes are precisely positioned with respect to this structure (Schulman and Bloom, 1991). Also, in Schizosaccharomyces pombe, an unusual chromatin structure covers a substantial part of the central domain (Polizzi and Clarke, 1991).
In humans, centromeric chromatin has several characteristic features: it forms a visible primary constriction, is underacetylated (O’Neill and Turner, 1995), contains CENP-A (Warburton et al., 1997) and seems to be late replicating (Warburton, 1999). It has been proposed that satellite sequences surrounding the centromere are utilized to maintain regions of late replication (Csink and Henikoff, 1998), thus ensuring that the centromere is the last region to replicate on a chromosome; a neocentromere may form following chromosome breakage if there is a region of sufficient replication delay. However, while these epigenetic models for centromere function can account for centromere inactivation and neocentromere formation, they raise new questions. Does the centromeric DNA itself have an important role? What determines the location of the centromere? Is it fixed on the chromosome or does it move?
New approaches to studying the epigenetic events involved in centromere function are needed. Topoisomerase II (Topo II) is a chromosomal protein that has the ability to catalyse strand passing of double-stranded DNA. During cell division this enzyme has been found to be essential for chromosome condensation and sister chromatid segregation. It has a dynamic pattern of distribution on the chromosomes, becoming axial as chromosomes condense during prophase and then concentrating at centromeres during metaphase (Rattner et al., 1996; Warburton and Earnshaw, 1997). Topo II has been shown to be the cellular target of several chemotherapeutic agents, including etoposide (Chen et al., 1984), which inhibit Topo II activity by stabilizing a covalent enzyme–DNA intermediate containing double-strand breaks. It is thus possible to investigate the long-range distribution of Topo II binding sites in vivo by treating living cells with these agents and subsequently mapping the positions of the DNA breaks (Gromova et al., 1995).
We set out to identify epigenetic modifications to the chromatin structure of human centromeres associated with centromeric activity or inactivity. We treated living cells with etoposide and analysed the long-range distribution of Topo II cleavages. We chose the Y chromosome centromere for this work because it is haploid, small, has a well understood structure and is available in cell lines in an active or inactive form. Our results reveal a characteristic pattern of Topo II cleavage at the normal Y centromere and a strikingly different pattern at inactive centromeres. These findings allow us to address some novel questions about centromere location and its stability.
RESULTS
Long-range distribution of Topo II cleavage sites in active and inactive human Y chromosome centromeres
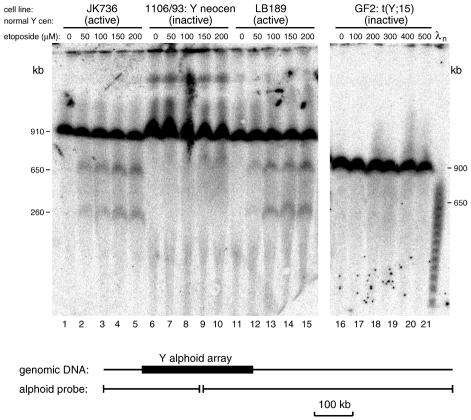
We wished to map the Topo II cleavage sites at the normal Y chromosome centromere and determine whether the pattern was altered in inactive centromeres. We therefore incubated living cells in growth medium at 37°C with etoposide at different concentrations and then digested the DNA with SfiI. Centromeric DNA fragments were detected using a Y alphoid probe. The cell lines were chosen to contain similarly sized Y alphoid arrays (∼280 kb) that generate an SfiI fragment of ∼910 kb containing the flanking sequences (Figure 1). In control cells treated with dimethylsulfoxide (DMSO) alone (Figure 1, tracks 1, 6, 11 and 16), no other bands are seen, even after long exposures of the filters. However, in cells containing normal centromeres treated with etoposide dissolved in DMSO, two prominent new bands are seen at 650 and 260 kb (Figure 1, tracks 2–5 and 12–15). The intensity of these bands increased after treatment with higher etoposide concentrations. In contrast, at the inactive centromeres, there were no specific new bands, although minor changes were observed (Figure 1, tracks 7–10 and 17–21). These results show that there is a specific site of Topo II cleavage at active centromeres, and that this cleavage pattern is absent from inactive centromeres.
Fig. 1. Topo II cleavage at active and inactive human Y chromosome centromeres. Cells (identified at the top of each track) were treated with the etoposide concentration shown, DNA was digested with SfiI, fractionated by pulsed-field gel electrophoresis and probed with a Y alphoid probe. Tracks 1–15 are from one gel run with a 70 s pulse time for 28 h, while tracks 16–21 are from a different gel with a 70 s pulse time and 35 h run time. The fragments detected in active centromeres are shown schematically in the lower part of the figure.
Mapping Topo II cleavage sites at the Y chromosome centromere
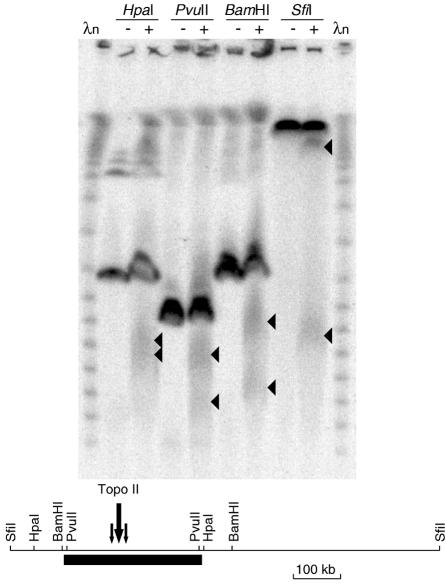
In order to obtain a more detailed map of the Topo II cleavage sites at the Y centromere, DNA samples from the cell line JK736 treated with 0 or 500 µM etoposide were digested with additional enzymes that cleave at known positions near the alphoid array. A single major region of cleavage was detected after etoposide treatment (Figure 2). It spanned ∼50 kb and was centred ∼120 kb from the Yp edge.
Fig. 2. Location of the Topo II cleavage site within the JK736 Y alphoid array. Cells were treated with 0 (–) or 500 µg/ml (+) etoposide, DNA was digested with the enzyme shown, fractionated by pulsed-field gel electrophoresis and probed with a Y alphoid probe. The map is shown in the lower part of the figure.
The size of the Y alphoid array varies considerably between different males, ranging from 130 to >1600 kb (Oakey and Tyler-Smith, 1990 and our unpublished observations). We wished to determine the position of the site of Topo II cleavage in these arrays. Does it vary randomly? Is it fixed in relation to one end of the array or does it always occur at the same relative position, near the centre? Cell lines with array sizes from 240 to 720 kb were treated with DMSO alone or with etoposide, and DNA was digested with PvuII, which cuts close to each edge of the array (Figure 3). As before, no specific cleavage was seen at the inactive centromere. At the active centromeres, pairs of broad bands are seen representing cleavage ∼100–150 kb from one end of the array (shown by SfiI digests to be the Yp end, see supplementary data available at EMBO reports Online). Thus, cleavage remains fixed relative to the Yp edge of the array.
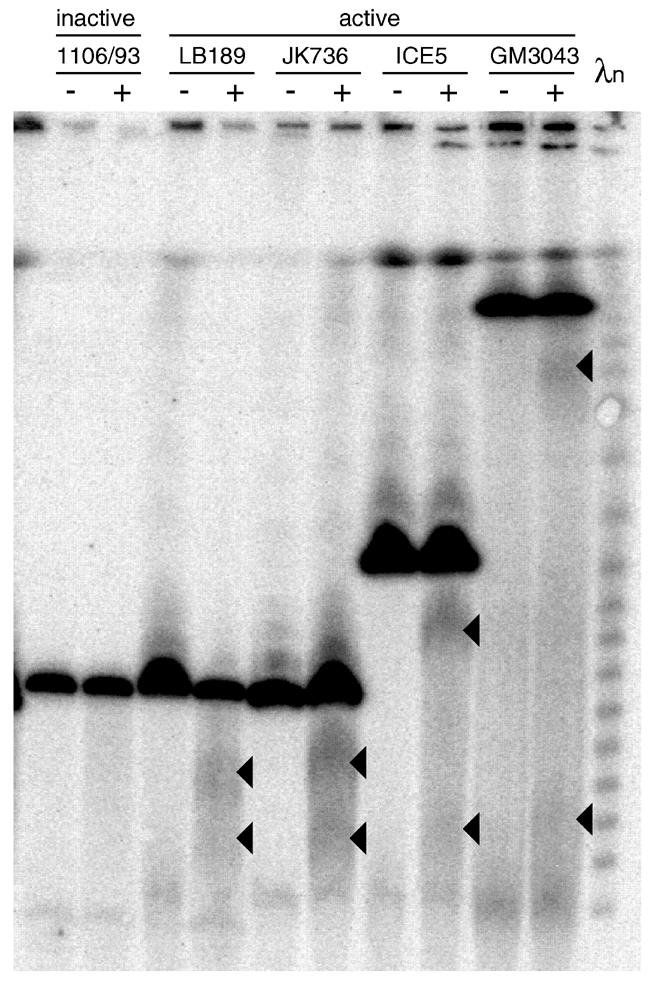
Fig. 3. Topo II cleavage in different-sized alphoid arrays. Cells were treated with 0 (–) or 500 µg/ml (+) etoposide, DNA was digested with PvuII, fractionated by pulsed-field gel electrophoresis and probed with a Y alphoid probe.
Location of centromeric proteins on the Y alphoid array
What is the molecular structure responsible for this cleavage? The most obvious candidate is the centromere itself. We therefore wished to determine whether the centromere was located at the position on the chromatin where the Topo II cleavage occurs. CREST antisera detect a mixture of the centromeric proteins CENP-A and CENP-C on the Y chromosome (Earnshaw and Rothfield, 1985; Pluta et al., 1990). We therefore used a combination of CREST antiserum staining and FISH with a Y alphoid probe on extended chromatin to identify the location of the centromeric proteins within the array (Floridia et al., 2000). In most cells showing the two signals, colocalization to a single round spot was observed. However, in distorted nuclei where the chromatin lies in an extended conformation, the alphoid signal was visible as a line, only part of which was associated with a CREST signal. In an individual who has a small Y alphoid array, the CREST staining showed no consistent sublocalization and could be found at either the end or the middle of the array (Figure 4A; Table I). We interpret this variation as due to the variable extension of the chromatin in these preparations. In contrast, in an individual with a large Y alphoid array, the signal was preferentially located at the end of the array (Figure 4B; Table I). Although variation in extension occurs in the large array as well, it is not sufficient to mask the asymmetrical location of the CREST signal. The two distributions were significantly different (P <0.001, χ2 test). Thus, the location of the centromeric proteins is consistent with the location of the Topo II cleavage site.
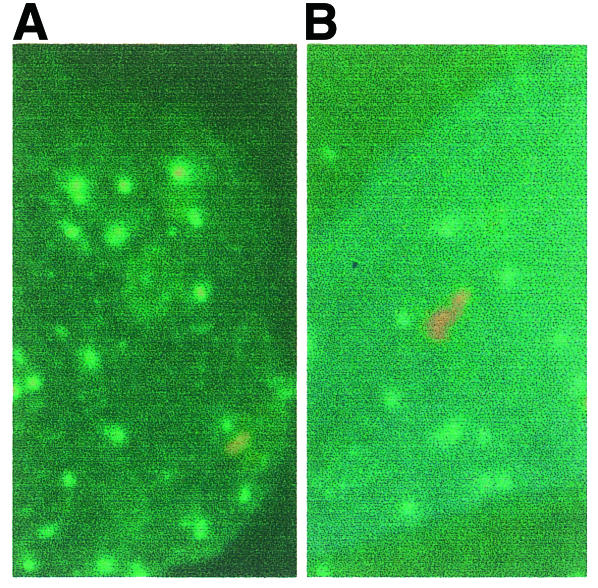
Fig. 4. CREST/FISH analysis of a short array (LB189) (A) or a long array (GM3043) (B). CREST signal: green; FISH signal: red.
Table I. Location of CREST signal on extended Y alphoid arrays.
| CREST signal at enda | CREST signal in middlea | |
|---|---|---|
| Small Y alphoid array (LB189) | 12 | 11 |
| Large Y alphoid array (GM3043) | 20 | 1 |
aNumber of extended arrays with signal at this position.
DISCUSSION
Mammalian centromeres, including neocentromeres, are so strikingly different in their chromatin structure from the rest of the chromosome that they can readily be recognized under the light microscope. It has, however, been difficult to identify centromeric chromatin at the molecular level and so map the position of the centromere onto the DNA. The probable in vivo binding site of CENP-A has been identified as alphoid DNA (Vafa and Sullivan, 1997), but the method used did not allow the sublocalization of the interaction to a particular region within the alphoid array. We therefore set out to investigate the chromatin structure of a human centromere by alternative methods, basing our work on the premise that a difference in chromatin structure between a stretch of DNA carrying an active or an inactive centromere would be due to the presence of the centromere, and would thus allow its location to be determined. We found that DNase I sensitivity and restriction enzyme cleavage in nuclei were not detectably different at active and inactive centromeres (our unpublished observations), but that there was a striking difference in the Topo II cleavage pattern following etoposide treatment.
Two inactive centromeres showed no specific cleavage under the conditions used, but six active centromeres showed cleavage at a single extended position. Since etoposide treatment was carried out on intact living cells, cleavage probably reflects the in vivo location of Topo II. The correlation of cleavage with centromeric activity, and the known concentration of Topo II at centromeres (Rattner et al., 1996), both suggest that the cleavage site is likely to represent the centromere. Complete cleavage was never observed under our conditions (≤1 h incubation), perhaps because Topo II is only concentrated at the centromere for part of the cell cycle. Cytogenetic studies of the locations of the centromere-specific proteins CENP-A and CENP-C have previously revealed that they occupy only a small proportion of the alphoid array (Earnshaw et al., 1989; Warburton et al., 1997; Lo et al., 1999). Our CREST staining combined with YαI FISH showed that the centromeric protein was located near one edge of the array (although it did not reveal which edge), which is consistent with the site of Topo II cleavage.
Previous genetic evidence also supports the idea that the Y centromere is located towards the Yp edge of the array. In a patient carrying a dicentric Y;21 translocation chromosome, three different deletions at inactive Y centromeres all removed sub-portions of the alphoid DNA near the Yp edge (Fisher et al., 1997), a region overlapping the Topo II cleavage site now identified. In a study of the size distribution of 609 Y alphoid arrays (Oakey and Tyler-Smith, 1990; Pandya, 1998), all size classes between 240 and 1600 kb were abundant, but there was a sharp cutoff below this size. The smallest size observed was 130 kb, which would be expected to just include the Topo II cleavage site. Despite the great variation in size, loss of the Topo II cleavage site has not been seen. From these diverse lines of evidence, we conclude that Topo II cleavage occurs at the centromere, and thus provides a marker for one aspect of the functional centromere in the sea of repetitive DNA.
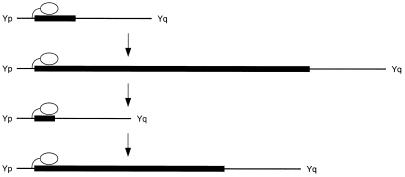
With the ability to map the location of the centromere onto the DNA, we could investigate the stability of its position. Human Y chromosome centromeres do not recombine, and have all descended from a single centromere in an individual who is estimated to have lived ∼150 000 years (>7000 generations) ago (Hammer et al., 1998). The Y chromosomes used in our analysis have been typed with polymorphic markers (Pandya, 1998) and were chosen to include a representative (GM3034) of the most divergent lineage (haplogroup 7), which has therefore maintained its centromere separately from the other chromosomes for >14 000 meioses and perhaps >2 800 000 mitoses. During this time, the underlying alphoid DNA has evolved by expansion and contraction to produce arrays from 130 to >1600 kb in size, but the centromere has moved by <0.1 bp/mitosis. The stability in the location of an epigenetic modification over this period is remarkable and suggests that centromeric location may be determined by the flanking sequence (Figure 5), and thus provides evidence for a novel role for non-alphoid DNA in centromeric function.
Fig. 5. Location of the Y chromosome centromere. The Y centromere (white oval) is located close to the Yp edge of the alphoid array (black box) and remains in the same position relative to Yp as the alphoid array evolves by expansion and contraction over many generations (arrows).
METHODS
Cells and etoposide. The lymphoblastoid cell lines JK736 (Biaka Pygmy), LB189, ICE5, GM3043 (San), ROMUN and DRM were derived from karyotypically normal 46,XY men chosen to show a range of Y-chromosomal alphoid array sizes from 240 to 720 kb. The 1106/93 cell line was derived from a phenotypically normal, fertile, 46,XY male whose normal Y alphoid centromere (280 kb) has been inactivated and functionally replaced by a neocentromere in the long arm heterochromatin (Tyler-Smith et al., 1999). The GF2 cell line was derived from a carrier of a Y;15 translocation, transmitted through four generations of normal fertile men with a karyotype 45,X, t(Y;15)(q11; p11) (Lioniello et al., 1996). The primary constriction was always present at the chromosome 15 centromere, while the Y centromere (280 kb) was always inactive.
Cells were grown in RPMI 1640 medium (Sigma) supplemented with 10% fetal calf serum (Globepharm), streptomycin (20 µg/ml), penicillin (20 U/ml) and 2 mM α-glutamine. Demethylepipodophyllotoxinthenylidene-β-d-glucoside (etoposide; Sigma and Alexis) was dissolved in DMSO at concentrations of 20–500 mM.
Preparation of agarose plugs containing living cells. Cells were resuspended in serum-free RPMI 1640 medium at 37°C, mixed with an equal volume of molten 1.5% low gelling temperature agarose (FMC) at 37°C in RPMI 1640 to produce a final concentration of 4 × 107 cells/ml. The suspension was distributed into a plug mould and left to set at room temperature for 5 min.
Treatment of living cells with etoposide. The agarose plugs containing cells were incubated at 37°C for 0.5 or 1 h in RPMI 1640 medium supplemented with etoposide (0–500 µM) in a final DMSO concentration of 0.5%. After incubation, the plugs were transferred into 0.5 M EDTA pH 9.5/1% N-lauryl-sarcosine/1mg/ml pronase for 2 nights at 50°C with one change. They were then washed in 0.5 M EDTA pH 9.5/1% N-lauryl-sarcosine and stored at 4°C.
Molecular and cytogenetic methods. Variants of standard methods were used. The CREST/FISH protocol was similar to that described previously (Floridia et al., 2000); further details are available from the authors.
Supplementary data. A figure illustrating supplementary data from this study is available at EMBO reports Online.
Supplementary Material
Acknowledgments
ACKNOWLEDGEMENTS
We thank Lina Christopoulou, Lizzie Burns and Arpita Pandya for comments on the manuscript. G.F. was supported by an EMBO short-term fellowship and the CNR (Consiglio Nazionale delle Ricerche). A.Z. thanks the OERC and the Regione Campania. O.Z. was supported by cofin98-MURST (Ministero dell’ Università e della Ricerca Scientifica e Tecnologica) and IRCCS Policlinico San Matteo, Pavia, and C.T.-S. by the CRC.
REFERENCES
- Chen G.L., Yang, L., Rowe, T.C., Halligan, B.D., Tewey, K.M. and Liu, L.F. (1984) Nonintercalative antitumor drugs interfere with the breakage-reunion reaction of mammalian DNA topoisomerase II. J. Biol. Chem., 259, 13560–13566. [PubMed] [Google Scholar]
- Choo K.H. (1997) Centromere DNA dynamics: latent centromeres and neocentromere formation. Am. J. Hum. Genet., 61, 1225–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csink A.K. and Henikoff, S. (1998) Something from nothing: the evolution and utility of satellite repeats. Trends Genet., 14, 200–204. [DOI] [PubMed] [Google Scholar]
- Earnshaw W.C. and Rothfield, N. (1985) Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma, 91, 313–321. [DOI] [PubMed] [Google Scholar]
- Earnshaw W.C., Ratrie, H., III and Stetten, G. (1989) Visualization of centromere proteins CENP-B and CENP-C on a stable dicentric chromosome in cytological spreads. Chromosoma, 98, 1–12. [DOI] [PubMed] [Google Scholar]
- Fisher A.M. et al. (1997) Centromeric inactivation in a dicentric human Y;21 translocation chromosome. Chromosoma, 106, 199–206. [DOI] [PubMed] [Google Scholar]
- Floridia G., Gimelli, G., Zuffardi, O., Earnshaw, W.C., Warburton, P.E. and Tyler-Smith, C. (2000) A neocentromere in the DAZ region of the human Y chromosome. Chromosoma, 109, 318–327. [DOI] [PubMed] [Google Scholar]
- Gromova I.I., Thomsen, B. and Razin, S.V. (1995) Different topoisomerase II antitumor drugs direct similar specific long-range fragmentation of an amplified c-MYC gene locus in living cells and in high-salt-extracted nuclei. Proc. Natl Acad. Sci. USA, 92, 102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer M.F., Karafet, T., Rasanayagam, A., Wood, E.T., Altheide, T.K., Jenkins, T., Griffiths, R.C., Templeton, A.R. and Zegura, S.L. (1998) Out of Africa and back again: nested cladistic analysis of human Y chromosome variation. Mol. Biol. Evol., 15, 427–441. [DOI] [PubMed] [Google Scholar]
- Hyman A.A. and Sorger, P.K. (1995) Structure and function of kinetochores in budding yeast. Annu. Rev. Cell Dev. Biol., 11, 471–495. [DOI] [PubMed] [Google Scholar]
- Karpen G.H. and Allshire, R.C. (1997) The case for epigenetic effects on centromere identity and function. Trends Genet., 13, 489–496. [DOI] [PubMed] [Google Scholar]
- Lioniello A., Renda, S., Floridia, G., Zuffardi, O. and Zatterale, A. (1996) t(Y;15) in 4 generazioni. XI Congresso Nazionale Federazione Italiana Studio Malattie Ereditarie, 219.
- Lo A.W., Liao, G.C., Rocchi, M. and Choo, K.H. (1999) Extreme reduction of chromosome-specific α-satellite array is unusually common in human chromosome 21. Genome Res., 9, 895–908. [DOI] [PubMed] [Google Scholar]
- Oakey R. and Tyler-Smith, C. (1990) Y chromosome DNA haplotyping suggests that most European and Asian men are descended from one of two males. Genomics, 7, 325–330. [DOI] [PubMed] [Google Scholar]
- O’Neill L.P. and Turner, B.M. (1995) Histone H4 acetylation distinguishes coding regions of the human genome from heterochromatin in a differentiation-dependent but transcription-independent manner. EMBO J., 14, 3946–3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandya A. (1998) Human Y-chromosomal DNA variation. D.Phil. thesis. University of Oxford, Oxford, UK.
- Pluta A.F., Cooke, C.A. and Earnshaw, W.C. (1990) Structure of the human centromere at metaphase. Trends Biochem. Sci., 15, 181–185. [DOI] [PubMed] [Google Scholar]
- Pluta A.F., Mackay, A.M., Ainsztein, A.M., Goldberg, I.G. and Earnshaw, W.C. (1995) The centromere: hub of chromosomal activities. Science, 270, 1591–1594. [DOI] [PubMed] [Google Scholar]
- Polizzi C. and Clarke, L. (1991) The chromatin structure of centromeres from fission yeast: differentiation of the central core that correlates with function. J. Cell Biol., 112, 191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattner J.B., Hendzel, M.J., Furbee, C.S., Muller, M.T. and Bazett-Jones, D.P. (1996) Topoisomerase II α is associated with the mammalian centromere in a cell cycle- and species-specific manner and is required for proper centromere/kinetochore structure. J. Cell Biol., 134, 1097–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman I. and Bloom, K.S. (1991) Centromeres: an integrated protein/DNA complex required for chromosome movement. Annu. Rev. Cell Biol., 7, 311–336. [DOI] [PubMed] [Google Scholar]
- Taylor S.S. (1999) Dual control ensures fidelity. Curr. Biol., 9, R562–R564. [DOI] [PubMed] [Google Scholar]
- Tyler-Smith C. and Floridia, G. (2000) Many paths to the top of the mountain: diverse evolutionary solutions to centromere structure. Cell, 102, 5–8. [DOI] [PubMed] [Google Scholar]
- Tyler-Smith C. and Willard, H.F. (1993) Mammalian chromosome structure. Curr. Opin. Genet. Dev., 3, 390–397. [DOI] [PubMed] [Google Scholar]
- Tyler-Smith C., Gimelli, G., Giglio, S., Floridia, G., Pandya, A., Terzoli, G., Warburton, P.E., Earnshaw, W.C. and Zuffardi, O. (1999) Transmission of a fully functional human neocentromere through three generations. Am. J. Hum. Genet., 64, 1440–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafa O. and Sullivan, K.F. (1997) Chromatin containing CENP-A and α-satellite DNA is a major component of the inner kinetochore plate. Curr. Biol., 7, 897–900. [DOI] [PubMed] [Google Scholar]
- Warburton P.E. (1999) Making CENs of mammalian artificial chromosomes. Mol. Genet. Metab., 68, 152–160. [DOI] [PubMed] [Google Scholar]
- Warburton P.E. and Earnshaw, W.C. (1997) Untangling the role of DNA topoisomerase II in mitotic chromosome structure and function. BioEssays, 19, 97–99. [DOI] [PubMed] [Google Scholar]
- Warburton P.E. et al. (1997) Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr. Biol., 7, 901–904. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.