Abstract
The poly(A) tail of influenza virus mRNAs is synthesized by the viral RNA polymerase by reiterative copying of a U5–7 sequence near the 5′ end of the viral RNA (vRNA) template. We have engineered a vRNA molecule by replacing its viral U6 poly(A) site with a negative-sense eukaryotic polyadenylation signal. The vRNA was transcribed by the viral RNA polymerase and the transcription product was processed by the cellular 3′ end processing machinery in vivo. According to the current model, 3′ end processing of eukaryotic pre-mRNAs is coupled to cellular RNA polymerase II (pol II) transcription; thus only RNAs synthesized by pol II are believed to be polyadenylated efficiently. Our results show that the cellular polyadenylation machinery is nevertheless able to recognize and process RNA transcripts that are not synthesized by pol II, indicating that synthesis by pol II is not an absolute requirement for 3′ end processing in vivo.
INTRODUCTION
The maturation of eukaryotic mRNAs that are synthesized by RNA polymerase II (pol II) is a complex, multi-step process (Barabino and Keller, 1999; Hirose and Manley, 2000; Proudfoot, 2000). RNA processing events include (i) the addition of a 7-methylguanosine and 2′-O-methylation to form a cap structure at the 5′ end; (ii) the removal of intron sequences by splicing; and (iii) 3′ end formation. The 3′ end is generated by endonucleolytic cleavage of the mRNA precursors 10–30 nt downstream of a conserved AAUAAA sequence (Proudfoot and Brownlee, 1976), followed by the addition of a poly(A) tail to the upstream cleavage product. There is emerging evidence that all these processing events are linked and effectively coordinated in vivo (Bentley, 1999; Minvielle-Sebastia and Keller, 1999). The C-terminal domain (CTD) of the largest subunit of pol II has emerged as a key determinant in coupling pol II transcription and RNA processing (McCracken et al., 1997). The CTD specifically binds the capping enzyme, splicing factors and 3′ end processing factors, helping to target mRNA processing factors to pol II transcripts. In addition, the CTD facilitates 3′ end RNA cleavage reactions even in the absence of transcription in vitro, suggesting that the CTD itself might be a cofactor for 3′ end processing (Hirose and Manley, 1998). The involvement of the CTD in RNA processing effectively couples pol II transcription and RNA processing events.
Like cellular mRNAs, influenza virus mRNA transcripts are capped at their 5′ end and polyadenylated at their 3′ end, but the mechanism by which these features are obtained differs from the mechanism by which cellular pol II transcripts are capped and polyadenylated (Krug et al., 1989). The cap structure at the 5′ end of influenza transcripts is derived from cellular pre-mRNAs. The influenza RNA polymerase cleaves pol II transcripts ∼9–17 nt downstream of the cap and it uses the generated capped RNA fragments as primers to initiate viral mRNA synthesis. The poly(A) tail of influenza mRNAs is synthesized by reiterative copying of a 5–7 nt long U sequence ∼16 nt from the 5′ end of the viral RNA (vRNA) template (Robertson et al., 1981). The U sequence acts directly as a template for poly(A) synthesis, since when replaced by an A sequence it serves as a template for poly(U) synthesis both in vitro and in vivo (Poon et al., 1999, 2000). During the replication cycle of influenza virus, vRNA is transcribed into another positive-sense transcript, cRNA. cRNA synthesis occurs in the absence of a primer and cRNA is a full-length copy of vRNA acting as a template for the synthesis of more vRNA. cRNA represents <5% of the total positive-sense transcripts. Both mRNA and cRNA synthesis are performed by the viral RNA-dependent RNA polymerase complex, which consists of three subunits, PB1, PB2 and PA, in the nucleus of infected cells (Herz et al., 1981).
We have engineered the influenza vRNA template coding for the neuraminidase protein by replacing its viral poly(A) site, the U6 sequence, with a negative-sense eukaryotic poly(A) signal that was derived from the rabbit β-globin gene. Although 3′ end processing of RNA transcripts is believed to be coupled to pol II transcription, we show that RNA transcripts synthesized from this modified vRNA template by the influenza virus RNA polymerase can be cleaved and polyadenylated by the cellular 3′ end processing machinery in vivo.
RESULTS
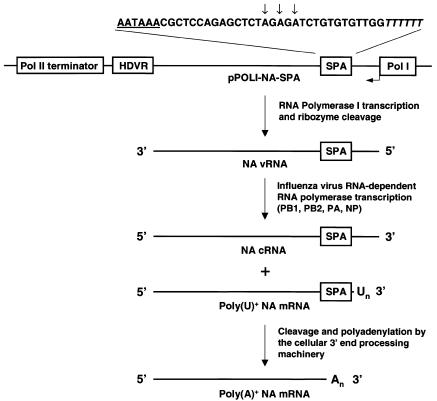
To assess whether RNAs synthesized by the influenza RNA polymerase could undergo cleavage and polyadenylation by the cellular 3′ end processing machinery, we used a plasmid-based assay developed for studying influenza virus RNA transcription and replication in vivo (Pleschka et al., 1996). We generated pPOLI-NA-SPA, which encodes the vRNA for the viral neuraminidase (NA) (Figure 1). The NA vRNA transcribed by RNA polymerase I (pol I) from pPOLI-NA-SPA contains a negative-sense and therefore non-functional eukaryotic polyadenylation signal (SPA) instead of the viral U6 poly(A) site 16 nt from its 5′ end. This vRNA can be transcribed by the vRNA polymerase into two types of RNA transcripts: a major mRNA product that is capped and poly(U)-tailed and a minor cRNA product that is a full-length exact copy of vRNA. The poly(U) tail of mRNA is added by reiterative copying of the A6 sequence at the 5′ end of the SPA in the vRNA by the influenza RNA polymerase (Figure 1) (Poon et al., 1999, 2000).
Fig. 1. Schematic representation of the pPOLI-NA-SPA plasmid and RNA transcripts. The sequence of the SPA (modified from Levitt et al., 1989) inserted into the NA cDNA is shown. The conserved hexamer of the SPA is underlined. Potential cleavage sites in the SPA are indicated by arrows. The T6 sequence of the SPA, transcribed into an A6 sequence in the vRNA, which serves as a template for poly(U) addition to influenza RNA transcripts, is shown in italics. HDVR, hepatitis delta virus ribozyme; SPA, synthetic poly(A) site; Pol I, RNA polymerase I promoter.
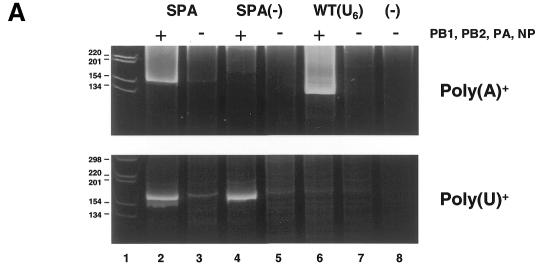
To test whether the RNAs with the eukaryotic poly(A) site, which were transcribed by the influenza virus RNA polymerase from the NA vRNA, were substrates for cleavage and polyadenylation in vivo, pPOLI-NA-SPA was co-transfected with pcDNA-PB1, -PB2, -PA or -NP into 293T cells. Cellular RNA was isolated at 48 h post-transfection and analysed for the presence of polyadenylated NA-specific RNA transcripts by an RT–PCR assay. The results show that poly(A)+ NA transcripts were present in the transfected cells (Figure 2A, top panel, lane 2). The strong DNA smear characteristic of PCR products containing poly(A) sequences of various length was absent if an empty pcDNA3 plasmid was co-transfected with pPOLI-NA-SPA (lane 3). This demonstrates that poly(A)+ NA RNA molecules were synthesized in the transfected cells and that their synthesis was dependent on the presence of influenza RNA polymerase and NP. If the conserved AATAAA hexamer in the SPA (see Figure 1) was mutated into TCTACG, no signal for poly(A)+ NA transcripts was detected, either in the presence or absence of influenza RNA polymerase and NP (lanes 4 and 5, respectively). This shows that the synthesis of poly(A)+ NA transcripts is dependent on the eukaryotic poly(A) signal. As a positive control, poly(A)+ NA transcripts were detected in cells transfected with pPOLI-NA-RT, which produced a vRNA with a viral poly(A) site [WT(U6)] (lane 6). No signal was detected if RNA from control non-transfected cells was used in the RT–PCR assay (lane 8).
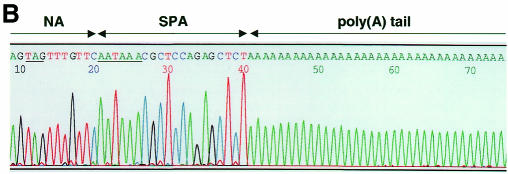
Fig. 2. RNA transcripts synthesized by the influenza RNA polymerase are substrates for cleavage and polyadenylation by the cellular 3′ end processing machinery. (A) RT–PCR assay for NA-specific poly(A)+ and poly(U)+ RNA transcripts. Total RNA from cells transfected with pPOLI-NA-SPA (lanes 2 and 3), pPOLI-NA-SPA(–) (lanes 4 and 5), pPOLI-NA-RT[WT(U6)] (lanes 6 and 7) or non-transfected cells (–) (lane 8), in the presence (lanes 2, 4 and 6) or absence (lanes 3, 5, 7 and 8) of influenza PB1, PB2, PA and NP, was tested by RT–PCR for NA-specific poly(A)+ and poly(U)+ transcripts (top and bottom panel, respectively). Expected size of RT–PCR products containing poly(A)+ sequences: SPA ≥158 bp, WT(U6) ≥137 bp; or poly(U)+ sequences: SPA ≥174 bp, SPA(–) ≥174 bp. The faint band in lane 3 (top panel) might represent pol II transcripts from cryptic pol II promoters in the pPOLI-NA-SPA plasmid. The band present in lane 3 (bottom panel) is presumably due to non-specific priming. Lane 1, DNA size markers. (B) Influenza RNA polymerase transcripts are cleaved and polyadenylated at the eukaryotic poly(A) site. A sequence of a clone derived from an NA-specific poly(A)+ mRNA, cleaved and polyadenylated 14 nt downstream of the conserved hexamer AAUAAA (see Figure 1), is shown. The NA sequence, SPA and poly(A) tail are indicated. The stop codon for NA translation and the conserved hexamer of the poly(A) signal are underlined.
Analysis of the same RNA samples as above for the presence of poly(U)+ NA transcripts by RT–PCR resulted in positive signal for both pPOLI-NA-SPA and pPOLI-NA-SPA(–) (Figure 2A, lower panel, lanes 2 and 4, respectively). The positive signal for pPOLI-NA-SPA(–) (lane 4) demonstrates that poly(U)+ NA transcripts were present in the sample. Thus the lack of positive signal for poly(A)+ transcripts (top panel, lane 4) in the same RNA sample is not due to the absence of influenza RNA transcripts from this sample. Rather, poly(U)+ influenza transcripts could not be polyadenylated due to the mutation of the conserved hexamer in the SPA. Overall, these results suggest that influenza RNA polymerase transcripts could be cleaved and polyadenylated by the cellular 3′ end processing machinery.
To determine whether cleavage and polyadenylation of influenza transcripts occurred at the expected eukaryotic poly(A) site, poly(A)+ NA-specific RT–PCR products were isolated from agarose gels, cloned and sequenced. Nine clones were obtained which showed that cleavage and polyadenylation occurred at three different sites, either 14, 16 or 18 nt downstream of the AAUAAA hexamer (see Figure 1), as expected (Levitt et al., 1989). An example of a clone is shown in Figure 2B.
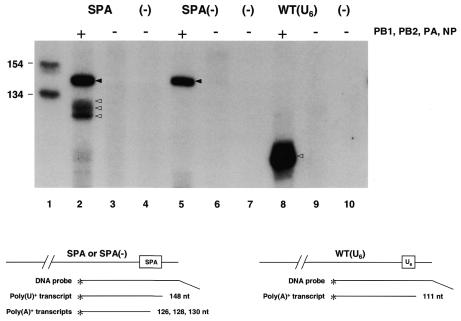
To characterize the RNA transcripts further, the RNA samples as above were analysed by S1 nuclease mapping with probes specific for the 3′ end of the positive-sense NA RNA transcripts. In RNA isolated from cells co-transfected with pPOLI-NA-SPA and pcDNA-PB1, -PB2, -PA or -NP, signals for both poly(U)+ and poly(A)+ transcripts were detected (Figure 3, lane 2). The signal for the poly(A)+ transcripts appears as a triplet due to the multiple sites at which cleavage and polyadenylation of NA RNA transcripts can occur (see above). Phosphoimager analysis showed that at 48 h post-transfection ∼40% of the transcripts were processed into poly(A)+ molecules (lane 2). When the conserved hexamer AATAAA in the SPA was mutated into TCTACG, only a signal for poly(U)+ transcripts was observed (lane 5). The positive control, pPOLI-NA-RT, showed a band of the expected size (111 nt) (lane 8). No specific signals were observed in the absence of influenza RNA polymerase and NP with any of the three constructs tested (lanes 3, 6 and 9). None of the three DNA probes produced a signal with RNA isolated from non-transfected cells (lanes 4, 7 and 10). We conclude that poly(U)+ NA influenza transcripts were cleaved at the eukaryotic poly(A) site.
Fig. 3. Analysis of the 3′ ends of influenza RNA polymerase transcripts by S1 nuclease protection. Total RNA from cells transfected with pPOLI-NA-SPA (lanes 2 and 3), pPOLI-NA-SPA(–) (lanes 5 and 6), pPOLI-NA-RT[WT(U6)] (lanes 8 and 9) or non-transfected cells (lanes 4, 7 and 10), in the presence (lanes 2, 5 and 8) or absence (lanes 3, 4, 6, 7, 9 and 10) of influenza PB1, PB2, PA and NP, was tested by S1 nuclease protection. Black arrowheads indicate DNA products protected by poly(U)+ RNA, white arrowheads indicate DNA products protected by poly(A)+ RNA. Lane 1, DNA size markers. The levels of cRNA are too low to detect under the experimental conditions used.
To assess whether the influenza RNA polymerase transcripts cleaved and polyadenylated by the eukaryotic 3′ end processing machinery could be translated, extracts from cells transfected with the plasmids as above were tested for the presence of NA. We found ∼33% NA-activity in cells transfected with pPOLI-NA-SPA compared with pPOLI-NA-RT (Figure 4). No significant NA-activity was observed either when the conserved hexamer AATAAA of the SPA was mutated or in the absence of viral polymerase and NP. Overall, these results demonstrate that poly(A)+ NA transcripts synthesized by the influenza RNA polymerase and cleaved and polyadenylated by the cellular 3′ end processing machinery were translated into a functional neuraminidase glycoprotein.
Fig. 4. NA protein expression in transfected cells. The NA-activities are expressed as a percentage relative to pPOLI-NA-RT. For symbols refer to Figure 2A. The activities shown are an average of five experiments. Standard deviations are indicated.
DISCUSSION
In this study we demonstrated that RNAs transcribed by the influenza virus RNA polymerase can be processed by means of the RNA maturation pathway that is used by RNA pol II transcripts. We found that influenza RNA polymerase transcripts that contain sequences necessary and sufficient to direct 3′ cleavage and polyadenylation are substrates for these processing events. The presence of influenza RNA polymerase and NP in cells was essential for efficient synthesis of RNA transcripts cleaved and polyadenylated at the cellular poly(A) site. This excludes the possibility that the observed cleaved and polyadenylated RNAs were synthesized by pol II from potential cryptic pol II promoters in pPOLI-NA-SPA. Although we were unable to detect any specific RNA transcripts from pPOLI-NA-SPA in the absence of PB1, PB2, PA and NP in the S1 protection assay (see Figure 3, lane 3), the RT–PCR test showed a very faint signal, which could be derived from pol II transcripts transcribed directly from the transfected pPOLI-NA-SPA (Figure 2A, top panel, lane 3). However, pol II transcription could not account for the strong signals observed in the presence of influenza RNA polymerase and NP (Figure 2A, top panel, lane 2 and Figure 3, lane 2). The synthesis of polyadenylated influenza RNA polymerase transcripts was also dependent on the presence of the conserved hexamer AAUAAA in the RNA transcript, confirming that polyadenylation occurred through the cellular polyadenylation machinery.
Although early in vitro studies indicated that accurate cleavage and polyadenylation of purified precursor RNAs can be unlinked from pol II transcription in nuclear extracts (Hart et al., 1985; Moore and Sharp, 1985), later studies showed that in vivo polyadenylation is linked to pol II transcription (McCracken et al., 1997). Not surprisingly, when ‘mRNAs’ were engineered to be transcribed by RNA pol I, RNA pol III or bacteriophage T7 RNA polymerase, the transcripts were not substrates for RNA processing (Smale and Tjian, 1985; Sisodia et al., 1987; McCracken et al., 1998). In the work reported here, however, we have demonstrated that RNA transcripts, which were synthesized by the influenza RNA polymerase, could be cleaved and polyadenylated by the factors normally involved in the 3′ end formation of pol II transcripts. We have found that ∼40% of the influenza RNA polymerase transcripts were cleaved. Although a figure of 40% efficiency is probably lower than might have been expected with a pol II transcript, it nevertheless shows that a significant proportion of the influenza transcripts were 3′ end processed.
In the light of the evidence that 3′ end processing is linked to pol II transcription we might ask how cleavage and polyadenylation of influenza RNA polymerase transcripts containing a cellular poly(A) signal are achieved? Influenza vRNA transcription requires the continuous functioning of host pol II since it is inhibited in the presence of α-amanitin, a specific inhibitor of pol II (Krug et al., 1989). Influenza RNA polymerase utilizes capped RNA primers cleaved from newly synthesized pol II transcripts to initiate viral transcription. In addition, two of the influenza transcripts are spliced (Krug et al., 1989) and it is likely that influenza mRNAs use the cellular splicing factors. Here we have shown that by inserting a eukaryotic poly(A) signal into influenza transcripts, they become substrates for cleavage and polyadenylation by the cellular 3′ end processing machinery. Taken together these observations suggest that influenza transcription might be more intimately coupled to pol II transcription than was previously suspected. Although none of the viral proteins that participate in transcription (PB1, PB2, PA and NP) are known to interact with cellular proteins involved in pol II transcription and RNA processing, we might speculate that viral ribonucleoproteins are specifically targeted to the sites of pol II transcription, to ‘mRNA factories’ (Iborra et al., 1996). We cannot exclude the possibility that pol II participates indirectly in influenza RNA polymerase transcription, by interacting directly or indirectly with the influenza RNA polymerase complex or NP. In such a situation, pol II could recruit, via its CTD, the factors involved in 3′ end processing, and thus be required for the processing of influenza RNA polymerase transcripts. Nevertheless the results presented in this paper demonstrate that a catalytically active pol II on the nascent pre-mRNA transcript is not required for 3′ end processing.
In summary, we have shown that an RNA transcript synthesized by the influenza virus RNA-dependent RNA polymerase was cleaved and polyadenylated by the eukaryotic 3′ end processing machinery. We conclude that synthesis of RNA transcripts by pol II is not an absolute requirement for efficient 3′ end processing in vivo.
METHODS
Plasmids. pPOLI-NA-SPA was generated from pPOLI-NA-RT (Pleschka et al., 1996; Fodor et al., 1999) by an inverse PCR technique using mutagenic primers. The viral poly(A) site in the NA cDNA was deleted and replaced by a eukaryotic poly(A) site (SPA) as specified in Figure 1. In order to minimize the possibility of pol II readthrough into the NA gene across the ribozyme sequence from potential pol II cryptic promoters in the vector sequence, a pol II terminator [consisting of a strong poly(A) site, SPA and four copies of a pol II pause site, MAZ] from pMLPIII (Yonaha and Proudfoot, 1999) was inserted into pPOLI-NA-SPA. In the pPOLI-NA-SPA(–) plasmid the conserved hexamer AATAAA in the SPA (underlined in Figure 1) was mutated into TCTACG. The protein expression plasmids for the three subunits of the influenza A/WSN/33 virus RNA polymerase and NP, pcDNA-PB1, pcDNA-PB2, pcDNA-PA and pcDNA–NP are based on pcDNA3 (Invitrogen).
Transfections and RNA isolation. DNA transfections were performed in human kidney 293T cells in 35 mm dishes using LipofectAMINE 2000 (Gibco-BRL) and 1 µg of each of the plasmids according to the manufacturer’s instructions. At 48 h post-transfection, the cells were lysed in 1 ml of TRIzol Reagent (Gibco-BRL) and total RNA was isolated as instructed by the manufacturer.
RT–PCR analysis and cloning of transcription products. Analysis of poly(A)+ and poly(U)+ NA-specific RNA transcripts was performed using a 5′ GC-clamped T20 and A20 primer, respectively, as described (Poon et al., 2000). RT–PCR products were analysed on 12% native acrylamide or 1% agarose gels, cloned into pGEM-T (Promega) and sequenced.
S1 nuclease protection analysis. To assess RNAs transcribed by the influenza RNA polymerase from pol I transcripts derived from pPOLI-NA-SPA, pPOLI-NA-SPA(–) and pPOLI-NA-RT, three S1 nuclease probes were prepared by digesting the corresponding plasmids with SpeI and HincII restriction enzymes. DNA fragments were gel-isolated and labelled with [α-32P]dATP and Klenow DNA polymerase. The DNA probe was mixed with ∼20 µg of total RNA isolated from transfected cells (see above) and hybridized in a total volume of 30 µl of 80% formamide, 40 mM PIPES pH 7.4, 0.4 M NaCl and 1 mM EDTA for 14 h at 40°C. Samples were then diluted with 300 µl of ice-cold S1 nuclease buffer (50 mM sodium acetate pH 4.5, 280 mM NaCl, 4.5 mM ZnSO4) containing 0.75 U of S1 nuclease (Amersham) per microlitre and digested at 30°C for 1.5 h. Protected DNA fragments were precipitated and analysed on 6% polyacrylamide–7 M urea gels.
NA-activity assay. Transfected 293T cells in 35 mm dishes (see above) were harvested 48 h post-transfection and lysed in 100 µl of 10 mM Tris–HCl pH 7.4, 10 mM CaCl2, 2% Triton X-100 by three cycles of freeze-thawing. Ten microlitres of cell extracts were incubated in a total reaction volume of 100 µl of 100 mM potassium phosphate pH 6.0, 100 mM KCl, 1 mM CaCl2 and 0.5 mM 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid (Sigma) at 37°C for 1 h. Reactions were terminated by adding 2 ml of 0.5 M glycine–NaOH pH 10.4, and the released 4-methylumbelliferone was assayed by using a spectrofluorophotometer (Shimadzu, RF-540) (Poon et al., 2000).
Acknowledgments
ACKNOWLEDGEMENTS
We thank Nick Proudfoot for providing the pMPLIII plasmid and for advice, Alice Taylor for DNA sequencing, and Shona Murphy, Masatomo Yonaha and David Pritlove for advice. This work was supported by the MRC (programme grant G9523972 to G.G.B.) and the FEMS (fellowship to A.M.).
REFERENCES
- Barabino S.M. and Keller, W. (1999) Last but not least: regulated poly(A) tail formation. Cell, 99, 9–11. [DOI] [PubMed] [Google Scholar]
- Bentley D. (1999) Coupling RNA polymerase II transcription with pre-mRNA processing. Curr. Opin. Cell Biol., 11, 347–351. [DOI] [PubMed] [Google Scholar]
- Fodor E., Devenish, L., Engelhardt, O.G., Palese, P., Brownlee, G.G. and García-Sastre, A. (1999) Rescue of influenza A virus from recombinant DNA. J. Virol., 73, 9679–9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart R.P., McDevitt, M.A. and Nevins, J.R. (1985) Poly(A) site cleavage in a HeLa nuclear extract is dependent on downstream sequences. Cell, 43, 677–683. [DOI] [PubMed] [Google Scholar]
- Herz C., Stavnezer, E., Krug, R. and Gurney, T., Jr (1981) Influenza virus, an RNA virus, synthesizes its messenger RNA in the nucleus of infected cells. Cell, 26, 391–400. [DOI] [PubMed] [Google Scholar]
- Hirose Y. and Manley, J.L. (1998) RNA polymerase II is an essential mRNA polyadenylation factor. Nature, 395, 93–96. [DOI] [PubMed] [Google Scholar]
- Hirose Y. and Manley, J.L. (2000) RNA polymerase II and the integration of nuclear events. Genes Dev., 14, 1415–1429. [PubMed] [Google Scholar]
- Iborra F.J., Pombo, A., Jackson, D.A. and Cook, P.R. (1996) Active RNA polymerases are localized within discrete transcription ‘factories’ in human nuclei. J. Cell Sci., 109, 1427–1436. [DOI] [PubMed] [Google Scholar]
- Krug R.M., Alonso-Caplen, F.V., Julkunen, I. and Katz, M.G. (1989) Expression and replication of the influenza virus genome. In Krug, R.M. (ed.), The Influenza Viruses. Plenum Press, New York, NY, pp. 89–152.
- Levitt N., Briggs, D., Gil, A. and Proudfoot, N.J. (1989) Definition of an efficient synthetic poly(A) site. Genes Dev., 3, 1019–1025. [DOI] [PubMed] [Google Scholar]
- McCracken S., Fong, N., Yankulov, K., Ballantyne, S., Pan, G., Greenblatt, J., Patterson, S.D., Wickens, M. and Bentley, D.L. (1997) The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature, 385, 357–361. [DOI] [PubMed] [Google Scholar]
- McCracken S., Rosonina, E., Fong, N., Sikes, M., Beyer, A., O’Hare, K., Shuman, S. and Bentley, D. (1998) Role of RNA polymerase II carboxy-terminal domain in coordinating transcription with RNA processing. Cold Spring Harb. Symp. Quant. Biol., 63, 301–309. [DOI] [PubMed] [Google Scholar]
- Minvielle-Sebastia L. and Keller, W. (1999) mRNA polyadenylation and its coupling to other RNA processing reactions and to transcription. Curr. Opin. Cell Biol., 11, 352–357. [DOI] [PubMed] [Google Scholar]
- Moore C.L. and Sharp, P.A. (1985) Accurate cleavage and polyadenylation of exogenous RNA substrate. Cell, 41, 845–855. [DOI] [PubMed] [Google Scholar]
- Pleschka S., Jaskunas, S.R., Engelhardt, O.G., Zürcher, T., Palese, P. and García-Sastre, A. (1996) A plasmid-based reverse genetics system for influenza A virus. J. Virol., 70, 4188–4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon L.L., Pritlove, D.C., Fodor, E. and Brownlee, G.G. (1999) Direct evidence that the poly(A) tail of influenza A virus mRNA is synthesized by reiterative copying of a U track in the virion RNA template. J. Virol., 73, 3473–3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon L.L., Fodor, E. and Brownlee, G.G. (2000) Polyuridylated mRNA synthesized by a recombinant influenza virus is defective in nuclear export. J. Virol., 74, 418–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. (2000) Connecting transcription to messenger RNA processing. Trends Biochem. Sci., 25, 290–293. [DOI] [PubMed] [Google Scholar]
- Proudfoot N.J. and Brownlee, G.G. (1976) 3′ non-coding region sequences in eukaryotic messenger RNA. Nature, 263, 211–214. [DOI] [PubMed] [Google Scholar]
- Robertson J.S., Schubert, M. and Lazzarini, R.A. (1981) Polyadenylation sites for influenza mRNA. J. Virol., 38, 157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisodia S.S., Sollner-Webb, B. and Cleveland, D.W. (1987) Specificity of RNA maturation pathways: RNAs transcribed by RNA polymerase III are not substrates for splicing or polyadenylation. Mol. Cell. Biol., 7, 3602–3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smale S.T. and Tjian, R. (1985) Transcription of herpes simplex virus tk sequences under the control of wild-type and mutant human RNA polymerase I promoters. Mol. Cell. Biol., 5, 352–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonaha M. and Proudfoot, N.J. (1999) Specific transcriptional pausing activates polyadenylation in a coupled in vitro system. Mol. Cell, 3, 593–600. [DOI] [PubMed] [Google Scholar]