Abstract
CBP (CREB-binding protein) is involved in transcriptional activation by a great variety of sequence-specific transcription factors. CBP has been shown to activate transcription through its histone acetyl transferase activity. Acetylation is a common post-translational modification of nucleosomal histone N-terminal tails, which generally correlates with transcriptional activation. Histone N-terminal tails are also modified by methylation but its functional consequences are largely unknown. Here we found that immunoprecipitation of CBP, or of the highly related p300, led to the co-immunoprecipitation of a robust histone methyl transferase (HMT) activity, indicating that CBP physically interacts with an HMT in living cells. The CBP-associated HMT is specific for lysines 4 and 9 of histone H3, which are known to be methylated in living cells. These results suggest that histone methylation could be involved in transcriptional activation. Furthermore, they raise the question of the link between histone methylation and acetylation.
INTRODUCTION
CBP (CREB-binding protein) has been cloned as a protein that specifically associates with PKA-phosphorylated CREB (Chrivia et al., 1993). Subsequent studies have shown that it is involved in transcriptional activation by phosphorylated CREB (Kwok et al., 1994). It is now known that CBP and the highly related E1A-associated p300 protein participate in transcriptional activation by a variety of transcription factors (Giordano and Avantaggiati, 1999). According to the current model, CBP/p300 are recruited to specific promoters through physical interactions with sequence-specific transcription factors. CBP/p300 harbour intrinsic histone acetyl transferase (HAT) activity (Bannister and Kouzarides, 1996; Ogryzko et al., 1996), suggesting that part of their action is mediated through acetylation of nucleosomal histone N-terminal tails.
Acetylation of their protruding N-terminal tails is a prominent modification of histones, which largely correlates with transcriptional activation (Wade et al., 1997). According to the current model, histone acetylation would increase DNA accessibility for transcription factor, thereby allowing transcription. It has also been demonstrated that acetylation can be a signal regulating protein binding to histone N-terminal tails (Edmondson et al., 1996). Consistent with this hypothesis, the bromodomain, a domain shared by many proteins whose function is linked to chromatin, has been shown to interact with acetylated lysines (Dhalluin et al., 1999; Jacobson et al., 2000).
Besides acetylation, nucleosomal histone N-terminal tails are also modified by phosphorylation and methylation (Strahl and Allis, 2000). Histone N-terminal tail methylation has been known for many years. Histone H3 can be methylated on lysine 4, lysine 9 and lysine 27, whereas histone H4 can be methylated on lysine 20 (Strahl and Allis, 2000). Little is known about the functional consequences of histone methylation, due to the lack of specific tools for studying these modifications, such as anti-methylated histone-specific antibodies. Furthermore, methylated histones are biochemically very similar to unmethylated histones. Despite these limitations, a recent study showed that methylation on lysine 4 of histone H3 occurs only in transcriptionally active nuclei in Tetrahymena (Strahl et al., 1999).
The characterization of chromatin-modifying enzymes has been an exploding research field in the last few years. Recent studies have shown that many chromatin-modifying machineries are physically or functionally linked, such as the DNA methyltransferase DNMT1 and histone deacetylases (Fuks et al., 2000), the ATP-dependent chromatin remodelling complex Mi-2 and the histone deacetylase HDAC1 (Tong et al., 1998; Wade et al., 1998; Zhang et al., 1998), histone kinases and acetyl transferases on immediate early gene promoters (Cheung et al., 2000; Lo et al., 2000). These data led us to investigate whether the HAT CBP could be physically associated with other chromatin-modifying activities.
Here, we show that CBP co-immunoprecipitates with a strong histone methyl transferase (HMT) activity. This ability is shared by the highly related p300 protein. The CBP/p300-associated HMT specifically methylates the histone H3 N-terminal tail on lysine residues that are known to be methylated in cells. The involvement of HMTs in transcriptional regulation, as well as the link between histone methylation and acetylation, is discussed.
RESULTS AND DISCUSSION
CBP physically associates with an HMT
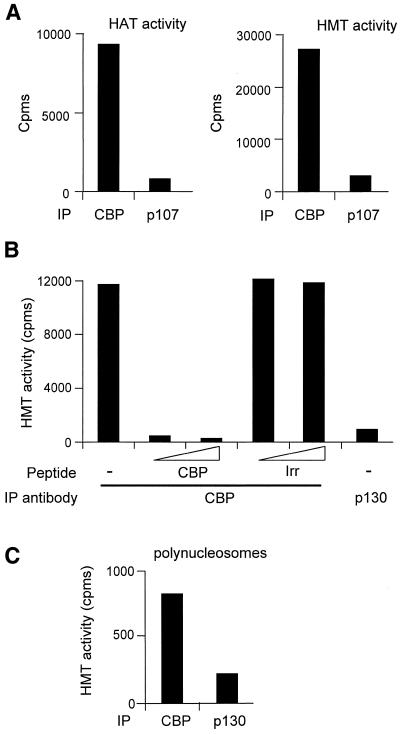
In order to test whether CBP physically interacts with other histone-modifying activities, we immunoprecipitated CBP from HeLa cell nuclear extracts with a polyclonal CBP-specific antibody. As expected, immunoprecipitation of CBP led to the co-immunoprecipitation of HAT activity (Figure 1A, left panel, CBP), which is specific since no HAT activity was co-immunoprecipitated with an irrelevant antibody directed against the p107 protein. We then tested immunoprecipitates for HMT activity by incubating them with purified histones and 3H-labelled S-adenosyl methionine, followed by a standard filter-binding assay. Strikingly, a high level of HMT activity was also found in CBP, but not in p107 immunoprecipitates (Figure 1A, right panel).
Fig. 1. CBP is associated with HMT activity, which methylates free histones and polynucleosomal substrates. (A) HeLa cell nuclear extracts (25 µl) were immunoprecipitated with the indicated antibody (anti-CBP: A-22, Santa Cruz; irrelevant: anti-p107, C-18, Santa Cruz). Immunoprecipitates were then assayed for HAT activity (left panel) or HMT activity (right panel) using purified histones as substrate. (B) As in (A) except that the CBP antibody (A-22) was pre-incubated with an increasing amount (2 and 5 µg) of its specific competitor peptide (peptide A-22 P, Santa Cruz) or of an irrelevant peptide (from P/CAF, Eurogentec) before being used for immunoprecipitation. As a control, immunoprecipitation with the irrelevant anti-p130 antibody (C-20, Santa Cruz) was performed. (C) As in (A) with 17.5 µl of nuclear extracts, except that polynucleosomes were used as substrates instead of histones. The HMT activity measured using purified histones was lower than in (A) due to variation from one batch of HeLa nuclear extracts to the other.
To analyse further the specificity of this interaction, we performed peptide competition experiments (Figure 1B). We found that the peptide against which the antibody was raised (CBP), but not an irrelevant peptide (Irr), could efficiently inhibit the immunoprecipitation of HMT activity by the anti-CBP antibody.
Taken together, these results indicate that in live cells CBP is physically associated with an HMT activity.
Immunoprecipitation of endogenous CBP under stringent conditions (in the presence of 0.1% SDS) immunoprecipitated HAT activity but no HMT activity (data not shown), suggesting that CBP is not an HMT by itself. Indeed, recombinant bacterially produced glutathione S-transferase (GST)–CBP did not harbour any intrinsic HMT activity (data not shown). However, GST–CBP was able to retain specifically HMT activity from HeLa nuclear extracts (data not shown), thus confirming our immunoprecipitation results. We also noticed that the HMT activity recruited by GST–CBP was consistently lower than after immunoprecipitation of endogenous CBP. This probably reflects the fact that the endogenous CBP–HMT complex has to be first disrupted before its reassociation on recombinant CBP, and could suggest that the interaction between CBP and the HMT is not direct.
Another important question to address was to determine whether the CBP-associated HMT activity could methylate nucleosomal histones. To this aim, purified polynucleosomes were prepared (data not shown) and were used as a substrate in an HMT assay. Interestingly, we found that polynucleosomal histones were methylated by the CBP-associated HMT activity (Figure 1C). However, the HMT activity, similarly to HAT activity, was ∼3- to 4-fold lower on nucleosomes than on free histones (data not shown).
Taken together, these results indicate that in live cells, CBP physically associates with an HMT, which can methylate polynucleosomal substrates. Moreover, they suggest the existence of a complex that harbours both HAT and HMT activities. Interestingly, immunoprecipitation of pCAF, which is not associated with CBP/p300 in HeLa cells (Ogryzko et al., 1998), did not result in the co-immunoprecipitation of significant HMT activity (our unpublished result). Thus, the ability to interact with an HMT is specific for CBP/p300 among HATs.
p300 shares with CBP the ability to interact with an HMT
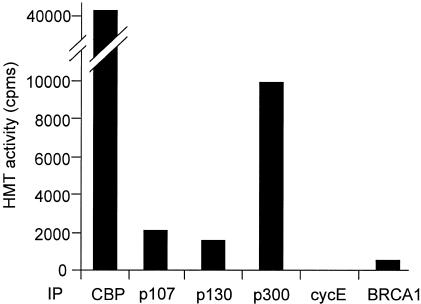
CBP and p300 share most of their biochemical or biological properties known so far. We thus tested whether p300 also interacted with an HMT activity. Immunoprecipitation of p300 with a p300-specific antibody resulted in the co-immunoprecipitation of HMT activity (Figure 2). This co-immunoprecipitation was specific since it was not detected using a panel of other antibodies (p107, p130, cyclin E or BRCA1). We found that the p300-associated HMT activity is consistently lower than the CBP-associated activity (∼4-fold lower, see Figure 2). This could possibly reflect a difference in the relative amount of CBP and p300 immunoprecipitated in our experiments. Alternatively, p300 could associate less efficiently with the HMT than with CBP. Taken together, results from Figures 1 and 2 indicate that the CBP/p300 family of HATs physically associates with an HMT.
Fig. 2. HeLa cell nuclear extracts (50 µl) were immunoprecipitated with the indicated antibody [CBP, p107 and p130 as in Figure 1, p300 (N-15, Santa Cruz), cyclin E (M-20, Santa Cruz), BRCA1 (K-18, Santa Cruz)] and immunoprecipitates were assayed for HMT activity.
The CBP-associated HMT methylates specifically K4 and K9 of histone H3
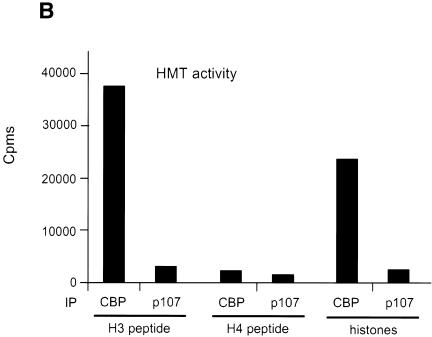
In live cells, methylation of histones has been reported to occur on histone H3 or histone H4 N-terminal tails. In order to test the substrate specificity of the CBP-associated HMT activity, we analysed methylated histones by SDS–PAGE followed by Coomassie blue staining (Figure 3A, left panel) or autoradiography (Figure 3A, right panel). Our results showed that only histone H3 was methylated by the CBP-associated HMT activity (Figure 3A). Remarkably, we could not detect any methylation at all on histone H4, which is also known to be methylated in live cells. When a peptide derived from the histone H3 N-terminal tail was used as a substrate instead of histones, we found that it was very efficiently methylated by the CBP-associated enzyme (Figure 3B, H3 peptide). Again, this methylation was highly specific, as no activity could be seen when a peptide derived from the N-terminal tail of histone H4 was used as a substrate (H4 peptide). Note that both substrates could be acetylated by CBP (data not shown), indicating that the lack of methylation of the histone H4 peptide is not due to a general defect of this peptide.
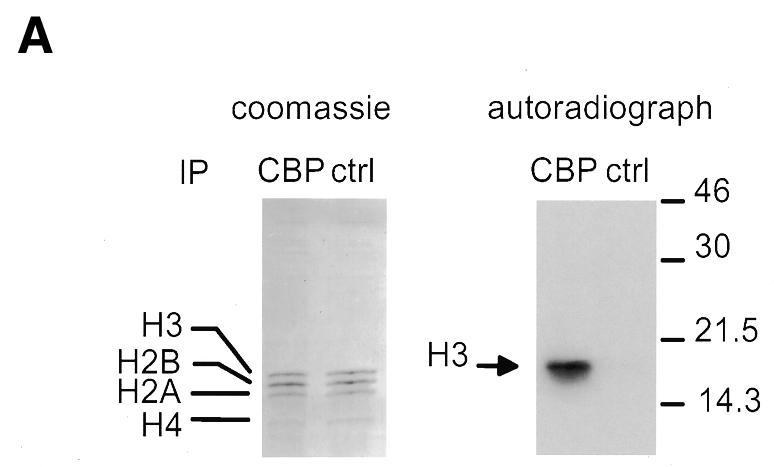
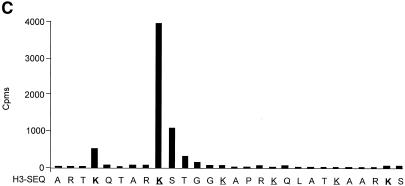
Fig. 3. The CBP-associated HMT methylates the histone H3 N-terminal tail. (A) HeLa nuclear extracts immunoprecipitated with the anti-CBP antibody or the anti-p107 (ctrl) were assayed for the presence of HMT activity using 1 µg of purified histones as substrate. Histones were then separated on an 18% SDS–polyacrylamide gel and detected by Coomassie blue staining (left panel) or by autoradiography (right panel). (B) As in (A), except that a peptide derived from the histone H3 or H4 N-terminal tail was used as a substrate. (C) The CBP-associated HMT methylates lysines 4 and 9, which are methylated in vivo. Histone H3 labelled by CBP-associated HMT activity was subjected to microsequencing analysis and parallel scintillation counting. Lysine residues methylated by the CBP-associated HMT and methylated in vivo are in bold. Acetylated lysines are underlined.
To determine the methylation sites, we undertook Edman degradation followed by sequence analysis in parallel with liquid scintillation counting of radiolabelled methylated histone H3 (Figure 3C). We found that the CBP-associated HMT activity methylates preferentially lysine 9 and to a lesser extent lysine 4 from histone H3. Scintillation counting of the membrane after the degradation of the first 27 amino acids indicated that there was hardly any radioactivity left. Again, this result confirms that the main methylation sites are located within the histone H3 N-terminal tail. Taken together, results from Figures 3 and 4 indicate that the CBP-associated HMT methylates K9 and K4 from histone H3. Interestingly, these sites are found methylated in mammalian cells (Strahl et al., 1999).
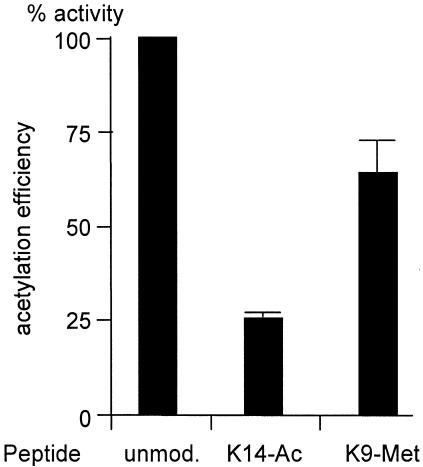
Fig. 4. Acetylation by CBP is not affected by K9 methylation. HAT assays of bacterially produced GST–CBP were performed using either the unmodified histone H3 peptide (unmod.), a peptide methylated on K9 (K9-Met) or a peptide acetylated on K14 (K14-Ac) as substrates. Acetylation efficiency was calculated relative to 100% for the unmodified peptide.
These data also suggest that the CBP-associated enzyme is unlikely to be the known HMT CARM1 (Chen et al., 1999), which methylates arginines (data not shown) or SUV39H1, which has a slightly different substrate specificity (Rea et al., 2000). The possibility still remains, however, that native CARM1 or SUV39H1 could exhibit a different specificity from bacterially produced recombinant proteins, due to post-translational modifications or interactions with other cellular proteins.
K9 methylation does not influence acetylation by CBP
Histone acetylation is generally linked to transcriptional activation. We thus tested whether methylation of H3 tail by the CBP-associated enzyme could interfere with acetylation by CBP (Figure 4). A peptide containing the first 17 amino acids from histone H3 was efficiently acetylated by recombinant bacterially produced GST–CBP (Figure 4, unmod.). As expected, when a peptide acetylated on the major CBP acetylation site (K14-Ac) was used, the acetylation efficiently decreased to 25%. The residual activity probably reflects acetylation of the peptide at K4 or K9, although these lysines are not acetylated by CBP using nucleosomes as substrate (Schiltz et al., 1999). Methylation of K9, the major site methylated by the CBP-associated enzyme (see Figure 3), led to a slight reduction in the efficiency of acetylation (K9-Met), which probably reflects the fact that K9 is not acetylatable in this peptide. This result indicates that methylation on K9 does not significantly affect the efficiency of acetylation by CBP on other lysines. In the converse experiment, we also found that acetylation of histone H3 by CBP did not change the efficiency of methylation by the CBP-associated enzyme (data not shown).
In this paper, we show that the versatile co-activator CBP is physically associated with an HMT (Figure 1). This result raises the question of the possible involvement of histone methylation in transcriptional activation. The functional consequences of histone methylation have been poorly studied so far, due to the absence of anti-methylated histone antibodies and also due to the fact that it is biochemically difficult to separate methylated histones from unmethylated histones. It was recently shown that methylation of lysine 4 of histone H3 occurs only in transcriptionally active macronuclei in Tetrahymena (Strahl et al., 1999). This finding raises the possibility that histone methylation, like acetylation, could be associated with transcriptional activation. The molecular cloning of the enzymes that methylate histones, such as the recently described SUV39H1 (Rea et al., 2000), will undoubtedly be a major breakthrough in the studies on the functional consequences of histone methylation.
The discovery of a putative HMT, named CARM1, which methylates histone H3 in vitro and which can act as a nuclear receptor co-activator (Chen et al., 1999), could provide a potential link between histone methylation and transcriptional activation. CARM1 was found to be homologous to arginine methyl transferases. We found that recombinant bacterially produced CARM1 preferentially methylates arginine 17 of histone H3 in vitro (data not shown). This result confirms that CARM1 is indeed an arginine methyl transferase in vitro. However, methylation of histones on arginines is still a controversial matter and has never been demonstrated to occur in live cells (Gary and Clarke, 1998). These data cast doubt about histones as bona fide substrates for CARM1.
Another interesting question raised by our results deals with the relationship between various histone N-terminal tail modifications.
Strikingly, both histone N-terminal tail acetylation and methylation occur on lysines (Strahl and Allis, 2000). It is thus tempting to speculate that, on a particular lysine, one modification might affect the other. However, the only lysine that can be acetylated or methylated is lysine 9 of histone H3. Pre-acetylation of lysine 9 impairs its methylation by the CBP-associated enzyme (data not shown), as well as by Suv39H1 (Rea et al., 2000), suggesting that both modifications cannot occur at the same time.
Our results (Figure 4) suggest that histone H3 methylation by the CBP-associated enzyme does not significantly change the efficiency of subsequent acetylation by CBP (Rea et al., 2000). However, it was recently shown that methylation at K9 inhibits phosphorylation of histone H3 at S10 and that p300 acetylates more efficiently phosphorylated histone H3 (Cheung et al., 2000; Lo et al., 2000). Thus, histone H3 methylation by the CBP-associated enzyme could indirectly inhibit acetylation by CBP. Interestingly, CBP was shown to physically associate with the putative histone H3 kinases pp90rsk (Nakajima et al., 1996; Sassone-Corsi et al., 1999), raising the possibility that the relative amount of pp90rsk or HMT associated with CBP could be a way to regulate acetylation by CBP.
METHODS
Immunoprecipitations
Twenty-five to 100 µl of HeLa cell nuclear extract (Computer Cell Culture Centre, Belgium) were used depending on the experiment. Extracts were pre-cleared on a rotating wheel for 30 min at 4°C using protein A/G agarose beads (30 µl). Immunoprecipitations were performed overnight at 4°C with 1 µg of antibody (see figure legends for details on the various antibodies) in 1 ml of IPH buffer (50 mM Tris pH 8, 150 mM NaCl, 0.5% NP-40, 5 mM EDTA) supplemented with protease inhibitors (Complete, Roche Diagnostics) in the presence of 10 µl of protein A/G agarose beads. Samples were then washed three times with 1 ml of IPH buffer before being split in half and assayed for HAT and HMT activity in parallel.
HAT and HMT assays
Free histones and polynucleosomes were purified from duck erythrocytes using standard procedures. The histone H3 peptide used in Figure 3 (ARTKQTARKSTGGKAPRKQLATKA) was synthesized by Cybergene (France) and corresponded to the first 24 amino acids of H3. The histone H3 peptides (ARTKQTARKSTGGKAPR either unmodified, acetylated on K14 or tri-methylated on K9) used in Figure 4 were synthesized by Sigma-Genosys or bachem and correspond to the first 17 amino acids from histone H3. The histone H4 peptide has previously been described (Ait-Si-Ali et al., 1998).
HAT assays were performed on filters as described (Brownell and Allis, 1995). [3H]acetyl incorporation into histones (1 µg) was determined by liquid scintillation counting of the histones retained on the filters.
HMT assays
Immunoprecipitates were incubated for 30 min at 30°C in 40 µl of IPH reaction buffer supplemented with 0.8 µM S-adenosyl-l-[3H]methyl-methionine (specific activity 66.0 Ci/mmol; Amersham) with purified histones (1 µg), H3 peptide (30 µM final) or polynucleosomes (corresponding to 1 µg of histones). Labelled histones or peptides were quantified using the filter-binding assay (Brownell and Allis, 1995) or analysed by SDS–PAGE followed by autoradiography (Figure 3A).
Protein microsequencing
Histone H3 (2 µg) labelled through methylation by the CBP-associated HMT activity was resolved on an 18% SDS–PAGE, blotted onto a ‘Problott’ membrane (Applied Biosystems), stained by Coomassie blue and the band corresponding to histone H3 was cut. Histone H3 bound to the membrane was directly sequenced from its N-terminus in an Applied Biosystems model 494A protein sequencer. After conversion, 30% of the samples were transferred to RP-HPLC for amino acid identification and the other 70% were collected for determination of radioactivity by scintillation counting.
Supplementary data
Supplementary data to this paper are available at EMBO reports Online.
Supplementary Material
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to thank Dr A. Harel-Bellan for reagents, and H. Richard-Foy, L. Waltzer and E. Nicolas for helpful discussions. D.T.’s group is an Equipe labellisée Ligue Nationale Contre le Cancer. This work was also supported by grants from the Fondation de la Recherche Médicale and the Association de la Recherche contre le Cancer (ARC). L.V. is a recipient of a fellowship from the ARC.
REFERENCES
- Ait-Si-Ali S., Ramirez, S., Robin, P., Trouche, D. and Harel-Bellan, A. (1998) A rapid and sensitive assay for histone acetyl-transferase activity. Nucleic Acids Res., 26, 3869–3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister A.J. and Kouzarides, T. (1996) The CBP co-activator is a histone acetyltransferase. Nature, 384, 641–643. [DOI] [PubMed] [Google Scholar]
- Brownell J.E. and Allis, C.D. (1995) An activity gel assay detects a single, catalytically active histone acetyltransferase subunit in Tetrahymena macronuclei. Proc. Natl Acad. Sci. USA, 92, 6364–6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Ma, H., Hong, H., Koh, S.S., Huang, S.M., Schurter, B.T., Aswad, D.W. and Stallcup, M.R. (1999) Regulation of transcription by a protein methyltransferase. Science, 284, 2174–2177. [DOI] [PubMed] [Google Scholar]
- Cheung P., Tanner, K.G., Cheung, W.L., Sassoni-Corsi, P., Denu, J.M. and Allis, C.D. (2000) Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol. Cell, 5, 905–915. [DOI] [PubMed] [Google Scholar]
- Chrivia J.C., Kwok, R.P.S., Lamb, N., Hagiwara, M., Montminy, M.R. and Goodman, R.H. (1993) Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature, 265, 855–859. [DOI] [PubMed] [Google Scholar]
- Dhalluin C., Carlson, J.E., Zeng, L., He, C., Aggarwal, A.K. and Zhou, M.M. (1999) Structure and ligand of a histone acetyltransferase bromodomain. Nature, 399, 491–496. [DOI] [PubMed] [Google Scholar]
- Edmondson D.G., Smith, M.M. and Roth, S.Y. (1996) Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev., 10, 1247–1259. [DOI] [PubMed] [Google Scholar]
- Fuks F., Burgers, W.A., Brehm, A., Hughes-Davies, L. and Kouzarides, T. (2000) DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nature Genet., 24, 88–91. [DOI] [PubMed] [Google Scholar]
- Gary J.D. and Clarke, S. (1998) RNA and protein interactions modulated by protein arginine methylation. Prog. Nucleic Acid Res. Mol. Biol., 61, 65–131. [DOI] [PubMed] [Google Scholar]
- Giordano A. and Avantaggiati, M.L. (1999) p300 and CBP: partners for life and death. J. Cell. Physiol., 181, 218–230. [DOI] [PubMed] [Google Scholar]
- Jacobson R.H., Ladurner, A.G., King, D.S. and Tjian, R. (2000) Structure and function of a human TAFII250 double bromodomain module. Science, 288, 1422–1425. [DOI] [PubMed] [Google Scholar]
- Kwok R.P., Lundblad, J.R., Chrivia, J.C., Richards, J.P., Bachinger, H.P., Brennan, R.G., Roberts, S.G., Green, M.R. and Goodman, R.H. (1994) Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature, 370, 223–226. [DOI] [PubMed] [Google Scholar]
- Lo W.-S., Trievel, R.C., Rojas, J.R., Duggan, L., Hsu, J.-Y., Allis, C.D., Marmostein, R. and Berger, S.L. (2000) Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol. Cell, 5, 917–926. [DOI] [PubMed] [Google Scholar]
- Nakajima T., Fukamizu, A., Takahashi, J., Gage, F.H., Fisher, T., Blenis, J. and Montminy, M.R. (1996) The signal-dependent coactivator CBP is a nuclear target for pp90RSK. Cell, 86, 465–474. [DOI] [PubMed] [Google Scholar]
- Ogryzko V.V., Schiltz, R.L., Russanova, V., Howard, B.H. and Nakatani, Y. (1996) The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell, 87, 953–959. [DOI] [PubMed] [Google Scholar]
- Ogryzko V.V., Kotani, T., Zhang, X., Schlitz, R.L., Howard, T., Yang, X.J., Howard, B.H., Qin, J. and Nakatani, Y. (1998) Histone-like TAFs within the PCAF histone acetylase complex. Cell, 94, 35–44. [DOI] [PubMed] [Google Scholar]
- Rea S. et al. (2000) Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature, 406, 593–599. [DOI] [PubMed] [Google Scholar]
- Sassone-Corsi P., Mizzen, C.A., Cheung, P., Crosio, C., Monaco, L., Jacquot, S., Hanauer, A. and Allis, C.D. (1999) Requirement of Rsk-2 for epidermal growth factor-activated phosphorylation of histone H3. Science, 285, 886–891. [DOI] [PubMed] [Google Scholar]
- Schiltz R.L., Mizzen, C.A., Vassilev, A., Cook, R.G., Allis, C.D. and Nakatani, Y. (1999) Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J. Biol. Chem., 274, 1189–1192. [DOI] [PubMed] [Google Scholar]
- Strahl B.D. and Allis, C.D. (2000) The language of covalent histone modifications. Nature, 403, 41–45. [DOI] [PubMed] [Google Scholar]
- Strahl B.D., Ohba, R., Cook, R.G. and Allis, C.D. (1999) Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc. Natl Acad. Sci. USA, 96, 14967–14972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong J.K., Hassig, C.A., Schnitzler, G.R., Kingston, R.E. and Schreiber, S.L. (1998) Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature, 395, 917–921. [DOI] [PubMed] [Google Scholar]
- Wade P.A., Pruss, D. and Wolffe, A.P. (1997) Histone acetylation: chromatin in action. Trends Biochem. Sci., 22, 128–132. [DOI] [PubMed] [Google Scholar]
- Wade P.A., Jones, P.L., Vermaak, D. and Wolffe, A.P. (1998) A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr. Biol., 8, 843–846. [DOI] [PubMed] [Google Scholar]
- Zhang Y., LeRoy, G., Seelig, H.P., Lane, W.S. and Reinberg, D. (1998) The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell, 95, 279–289. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.