Abstract
The haloarchaeon Haloferax volcanii degrades D-glucose via the semiphosphorylative Entner-Doudoroff pathway and D-fructose via a modified Embden-Meyerhof pathway. Here, we report the identification of GfcR, a novel type of transcriptional regulator that functions as an activator of both D-glucose and D-fructose catabolism. We find that in the presence of D-glucose, GfcR activates gluconate dehydratase, glyceraldehyde-3-phosphate dehydrogenase and pyruvate kinase and also acts as activator of the phosphotransferase system and of fructose-1,6-bisphosphate aldolase, which are involved in uptake and degradation of D-fructose. In addition, glyceraldehyde-3-phosphate dehydrogenase and pyruvate kinase are activated by GfcR in the presence of D-fructose and also during growth on D-galactose and glycerol. Electrophoretic mobility shift assays indicate that GfcR binds directly to promoters of regulated genes. Specific intermediates of the degradation pathways of the three hexoses and of glycerol were identified as inducer molecules of GfcR. GfcR is composed of a phosphoribosyltransferase (PRT) domain with an N-terminal helix-turn-helix motif and thus shows homology to PurR of Gram-positive bacteria that is involved in the transcriptional regulation of nucleotide biosynthesis. We propose that GfcR of H. volcanii evolved from a PRT-like enzyme to attain a function as a transcriptional regulator of central sugar catabolic pathways in archaea.
Keywords: archaea, D-glucose and D-fructose catabolism, electrophoretic mobility shift assay (EMSA), Haloferax volcanii, phosphoribosyltransferase (PRT) protein family, transcriptional regulation
1 |. INTRODUCTION
Comparative analyses of sugar degradation pathways indicate that archaea degrade D-glucose via modifications of the classical Embden-Meyerhof (EM) and Entner-Doudoroff (ED) pathways operative in bacteria and eukarya. Hyperthermophilic archaea predominantly utilize modified EM pathways, whereas thermoacidophilic and halophilic archaea utilize modified ED pathways (Bräsen et al., 2014; Siebers & Schönheit, 2005). Transcriptional regulation of archaeal sugar degradation pathways has been studied in some detail for the degradation of the D-glucose oligomers maltose, trehalose and maltodextrin in the hyperthermophilic Thermococcales Pyrococcus furiosus and Thermococcus kodakarensis and in thermoacidophilic Sulfolobus species (Kanai et al., 2007; Lee et al., 2008; Wagner et al., 2014). The transcriptional regulators involved belong to the TrmB protein family and function as both repressors and activators, e.g., Tgr from Thermococcus kodakarensis represses genes involved in sugar transport and glycolysis, and activates genes involved in gluconeogenesis (Kanai et al., 2007). The TrmB-like regulator MalR from Sulfolobus acidocaldarius is a transcriptional activator for maltose uptake and degradation (Wagner et al., 2014) and a TrmB-like regulator has also been reported for the haloarchaeon Halobacterium salinarum, functioning as a regulator of diverse metabolic pathways in response to D-glucose (Schmid et al., 2009; Todor et al., 2014). In this organism, a role of TrmB in gluconeogenesis rather than in D-glucose degradation has been indicated (Todor et al., 2014).
In recent years, sugar degradation pathways have begun to be studied in detail in haloarchaea; in particular, in the model organism Haloferax volcanii. This haloarchaeon utilizes hexoses and pentoses, as well as the deoxysugar L-rhamnose, as carbon and energy sources and the degradation pathways have been elucidated. Pentoses are degraded by oxidative pathways, and L-rhamnose via the diketo-hydrolase pathway (Johnsen et al., 2009; Johnsen et al., 2013; Johnsen et al., 2020; Reinhardt et al., 2019; Sutter et al., 2017). The hexoses D-glucose, D-fructose and D-galactose are degraded via the semiphosphorylative Entner-Doudoroff (spED) pathway, a modified Embden-Meyerhof pathway and the DeLey-Doudoroff pathway as follows: (i) D-Glucose is converted to gluconate and then to 2-keto-3-deoxygluconate (KDG) by glucose dehydrogenase and gluconate dehydratase, respectively. KDG is then phosphorylated by KDG kinase, forming 2-keto-3-deoxy-6-phosphogluconate (KDPG) that is subsequently cleaved by KDPG aldolase to yield pyruvate and glyceraldehyde-3-phosphate (GAP) (Figure 1) (Pickl et al., 2014; Sutter et al., 2016); (ii) D-Fructose is taken up by a phosphoenolpyruvate-dependent phosphotransferase system (PTS), generating fructose-1-phosphate, which is then converted to two molecules of GAP via the intermediate fructose-1,6-bisphosphate (FBP) in reactions catalyzed by fructose-1-phosphate kinase, FBP aldolase and triosephosphate isomerase (Figure 1) (Pickl et al., 2012); and (iii) D-Galactose is converted to pyruvate and GAP via the DeLey-Doudoroff pathway enzymes galactose dehydrogenase, galactonate dehydratase, 2-keto-3-deoxygalactonate kinase and 2-keto-3-deoxy-6-phosphog alactonate aldolase (Pickl et al., 2014; Tästensen et al., 2020). GAP is the common product of all three pathways and is also formed by oxidation of the non-sugar substrate glycerol in H. volcanii involving glycerol kinase, glycerol-3-phosphate dehydrogenase and triosephosphate isomerase (Rawls et al., 2011; Sherwood et al., 2009). GAP is then oxidized to 3-phosphoglycerate (3PG), catalyzed by GAP dehydrogenase and 3PG kinase (Tästensen & Schönheit, 2018), and 3PG is further converted to pyruvate by 3PG mutase, enolase and pyruvate kinase (Johnsen et al., 2019).
FIGURE 1.

Proposed pathways of D-glucose and D-fructose degradation in Haloferax volcanii via semiphosphorylative Entner-Doudoroff (spED) and modified Embden-Meyerhof (EM) pathways. 1,3-BPG, 1,3-bisphosphoglycerate; DHAP, dihydroxyacetone phosphate; ENO, enolase; F1P, fructose-1-phosphate; FBA, FBP aldolase; FBP, fructose-1,6-bisphosphate; GAD, gluconate dehydratase; GAP, glyceraldehyde-3-phosphate; GAPDH, GAP dehydrogenase; GDH, glucose dehydrogenase; KDG, 2-keto-3-deoxygluconate; KDGK, KDG kinase; KDPG, 2-keto-3-deoxy-6-phosphogluconate; KDPGA, KDPG aldolase; PEP, phosphoenolpyruvate; PFK, 1-phosphofructokinase; PGK, phosphoglycerate kinase; PGM, phosphoglycerate mutase; PK, pyruvate kinase; PTS, PEP-dependent-phosphotransferase system; TIM, triosephosphate isomerase.
To date, transcriptional regulators in H. volcanii have been identified for the degradation pathways of pentoses, D-galactose and L-rhamnose, designated as XacR, GacR and RhaR, respectively (Johnsen et al., 2015; Reinhardt et al., 2019; Tästensen et al., 2020). These regulators are members of the bacterial-type IclR regulator family and function as activators of genes in their respective pathways. Furthermore, the transcriptional regulator GlpR has been reported to repress genes of D-glucose and D-fructose catabolism in the presence of glycerol (Martin et al., 2018; Rawls et al., 2010).
In this communication, we report the identification and characterization of a novel type of transcriptional regulator of H. volcanii, designated GfcR (for glucose and fructose catabolism regulator), that activates genes involved in D-glucose and D-fructose degradation. GfcR also functions as a transcriptional activator for the degradation of D-galactose and glycerol by activating GAP dehydrogenase and pyruvate kinase. Finally, we have identified intermediates of the catabolic pathways as inducer molecules of GfcR.
2 |. RESULTS
2.1 |. GfcR, a member of PRT protein family, is functionally involved in D-glucose degradation
Analysis of Haloferax volcanii genes involved in glucose degradation reveals that the genes gdh (HVO_1083, encoding glucose dehydrogenase) and gfcR (HVO_1082) form a transcriptional unit (Figure 2a), where four nucleotides are overlapping. Consistent with this, Northern blots using a probe against gdh revealed signals of about 1900 and 1100 nucleotides in cells grown on D-glucose but not in those grown on the gluconeogenic casamino acids (Figure 2b). The 1900 nucleotide transcript corresponds to a cotranscript of gfcR (633 nucleotides) and gdh (1074 nucleotides), and that of 1100 nucleotides to a single transcript of gdh. The co-expression of gfcR with gdh suggests a potential function for gfcR in D-glucose catabolism.
FIGURE 2.

Transcriptional analysis of the gdh-gfcR operon and growth of the Haloferax volcanii ΔgfcR mutant on D-glucose. (a) Genomic view of the gdh-gfcR operon. (b) Northern blot analysis with RNA from cells grown on casamino acids (Cas) or D-glucose (G) using a probe against gdh (HVO_1083). 16S rRNA was used as loading control. (c) Growth on 25 mM D-glucose of the ΔgfcR mutant (square), wild-type (circle) and the deletion mutant that was complemented with gfcR (triangle), gfcR-Δα1 (diamond) or gfcR-Δα1α2 (asterisk).
Examination of its amino acid sequence suggests that GfcR, which was formerly named PyrE1, is a member of the phosphoribosyltransferase (PRT) superfamily. This superfamily comprises enzymes involved in biosynthesis of pyrimidine and purine, e.g., orotate phosphoribosyltransferase (PyrE), and also the transcriptional regulators of their respective pathways, e.g., PurR from Bacillus subtilis (Hove-Jensen et al., 2017; Sinha & Smith, 2001). The hallmark of the PRT domain is a conserved motif involved in the binding of phosphoribosyl pyrophosphate (PRPP). Transcriptional regulators of the PurR-type contain an additional N-terminal DNA-binding domain with a helix-turn-helix (HTH) motif. PurR is reported to be a transcriptional regulator of purine biosynthesis in some Gram-positive bacteria, including Bacillus subtilis and Lactococcus lactis (Hove-Jensen et al., 2017; Kilstrup & Martinussen, 1998; Sinha et al., 2003). In addition to GfcR, H. volcanii has been reported to contain a second member of the PRT superfamily designated as PyrE2. PyrE2 was found to be catalytically active as orotate phosphoribosyltransferase, whereas GfcR lacks this activity and its function therefore remains unknown (Bitan-Banin et al., 2003).
A sequence comparison of GfcR with haloarchaeal homologs and with the bacterial regulators of the PRT superfamily PurR from B. subtilis and L. lactis is shown in Figure 3. The alignment includes secondary-structure assignments of GfcR based on an AlphaFold model (see below) and those for PurR from B. subtilis based on its crystal structure (Sinha et al., 2003). Interestingly, GfcR contains several alpha helices in the N-terminal region that are analogous to the winged-helix DNA-binding domain present in PurR. One of these (α2 in GfcR and α3 in PurR) exhibits a high degree of sequence similarity. The large insertion in PurR sequences present after the helical region, however, suggests a different position for the respective N-terminal domains of PurR and GfcR relative to the PRT domain. Most notably, the alignment also shows that residues in the conserved motif of PurR that binds PRPP are generally conserved in GfcR (Figure 3).
FIGURE 3.

Amino acid sequence alignment of GfcR from Haloferax volcanii, haloarchaeal homologs and PurR from Bacillus subtilis and Lactococcus lactis. The alignment was calculated with Clustal Omega X (Sievers et al., 2011) and then ESPript 3.0 (Buchan & Jones, 2019; Robert & Gouet, 2014) was used to plot the secondary structure of GfcR based on the AlphaFold model and PurR based on its crystal structure (PDB code 1057). Invariant residues are shaded red, and highly conserved residues are colored red with blue borders. The PRPP motif of the PRT domain is highlighted by a frame (Prosite documentation, PDOC00096). The sequences used (and their UniProt accession codes) are as follows: Haloferax volcanii GfcR (Q8NKQ2), Haloterrigena turkmenica PyrE-like protein (D2RW31), Haloarcula marismortui PyrE-like protein (Q5V3K1), Halorubrum lacusprofundi PyrE-like protein (B9LUS0), Lactococcus lactis PurR (P0A3Z9), and Bacillus subtilis PurR (P37551).
To determine whether GfcR functions in D-glucose catabolism, a gfcR deletion mutant of H. volcanii was generated (Figure S1). The ΔgfcR mutant lost its ability to grow on D-glucose (Figure 2c), whereas growth on casamino acids was unaffected (not shown). Growth on D-glucose could be recovered by complementation of ΔgfcR with gfcR in trans (Figure 2c). To test whether the N-terminal HTH domain is essential for GfcR function, the ΔgfcR mutant was also complemented with two versions of gfcR in which either the predicted α1 helix or both of the α1 and α2 helices at the N terminus were absent. Growth on D-glucose of the ΔgfcR mutant could not be restored with either of these constructs (Figure 2c), suggesting that GfcR functions as a DNA-binding transcriptional regulator in D-glucose catabolism.
2.2 |. GfcR acts as transcriptional activator of D-glucose catabolism via the spED pathway
The potential role of GfcR as a transcriptional regulator of genes involved in D-glucose degradation was then examined by using RT-PCR to measure transcript levels of the gad gene in the wild-type and ΔgfcR mutant of H. volcanii. The gad gene encodes gluconate dehydratase, which is the second step of the upper half of the spED pathway (Figure 1). When cells were grown on medium supplemented with casamino acids, only low levels of the gad transcript were evident in both wild-type and ΔgfcR mutant cells (Figure 4). Two hours after addition of D-glucose, however, a strong signal for the gad transcript was detected in wild-type cells, but significantly less in the ΔgfcR mutant (Figure 4). Wild-type levels of the gad transcript were restored in the deletion mutant upon complementation with gfcR in trans. These data indicate that GfcR functions as a transcriptional activator of gluconate dehydratase in the presence of D-glucose.
FIGURE 4.

GfcR dependent transcription of gad encoding gluconate dehydratase of Haloferax volcanii analyzed by RT-PCR. Cells of wild-type, ΔgfcR mutant and the mutant complemented with gfcR in trans (ΔgfcR+gfcR) were pre-grown on 0.2% casamino acids and expression of gad was induced by 5 mM D-glucose. Formation of gad transcripts was monitored before (0 h) and 2 h after the addition of D-glucose (2 h). The ribL gene encoding the ribosomal protein L10 was used as constitutive expression control.
The transcriptional regulation of gad, and also of enzymes of the lower half of the pathway, GAP dehydrogenase (GAPDH), pyruvate kinase (PK) and phosphoglycerate mutase (PGM), encoded by gapI, pykA and pgmI, respectively, was then determined by using a β-galactosidase-based reporter-gene system to measure promoter activities (Johnsen et al., 2015; Patenge et al., 2000). Transformants of wild-type and ΔgfcR mutant carrying the plasmid-based promoter-reporter gene constructs were first grown on casamino acids to determine basal promoter activities. Promoter activities were then measured after the addition of 5 mM D-glucose. As shown in Table 1, promoter activities of gad (9.8-fold), gapI (2.7 fold) and pykA (2.2 fold) were all upregulated by D-glucose in the wild-type strain but not in the ΔgfcR mutant. This indicates a function for GfcR as a transcriptional activator of gapI and pykA, in addition to gad. By contrast, the promoter activity of pgmI was not induced by D-glucose in wild-type cells. The constitutive expression of this gene is consistent with a role for PGM in both glycolysis and gluconeogenesis. Together, the data indicate that GfcR functions as a transcriptional activator of genes of the upper (gad) and the lower halves (gapI and pykA) of the pathway and thus GfcR represents the first identified transcriptional regulator of the spED pathway of H. volcanii.
TABLE 1.
Promoter activities of selected genes involved in D-glucose degradation via the spED pathway in Haloferax volcanii.
| β-galactosidase activity (mU/mg) | Induction (fold) | |||
|---|---|---|---|---|
| Promotor of | Strain | CAS CAS+D-glucose | ||
| gad (HVO_1488) | wt | 48 ±18 | 471 ± 84 | 9.8 |
| ΔgfcR | 47 ±22 | 58 ± 30 | 1.2 | |
| gapl (HVO_0481) | wt | 218 ± 21 | 585±191 | 2.7 |
| ΔgfcR | 220 ± 44 | 221 ± 60 | 1.0 | |
| pykA (HVO_0806) | wt | 65 ± 24 | 141 ± 26 | 2.2 |
| ΔgfcR | 58 ± 15 | 72 ± 8 | 1.2 | |
| pgml (HVO_2516) | wt | 28 ± 10 | 26 ± 4 | 0.9 |
| ΔgfcR | 17 ± 1 | 13 ± 3 | 0.8 | |
Note: Promotor activities were measured in a β-galactosidase assay in wild-type and ΔgfcR cells grown on casamino acids (CAS, 0.2%) and after induction by 5 mM D-glucose (CAS+D-glucose). All experiments were carried out in triplicates.
2.3 |. GfcR is a transcriptional activator of D-fructose uptake and degradation via the modified EM pathway
The GfcR-dependent activation of genes encoding GAPDH and PK by D-glucose raises the prospect that GfcR also regulates the catabolism of other substrates. This is because both enzymes are within the lower half of the degradation pathways of D-fructose, D-galactose and of the non-sugar substrate glycerol. In accordance with this prediction, the ΔgfcR mutant did not grow on any of these three substrates (Figure 5), whereas growth on the pentose sugar D-xylose (as a control) was not affected.
FIGURE 5.

Growth of Haloferax volcanii H555 and the ΔgfcR mutant on D-fructose, D-galactose and glycerol. Wild-type (circles) and ΔgfcR mutant (squares) were grown in synthetic medium with 25 mM D-fructose (a), 25 mM D-galactose (b) or 20 mM glycerol (c).
We also examined whether GfcR regulates expression of gapI and pykA in the presence of D-fructose. The promoter activities of gapI and pykA in wild-type cells were 3.4-and 1.4-fold higher, respectively, in cells induced by D-fructose compared to casamino acid-grown cells. No induction was observed in the ΔgfcR mutant, indicating that GfcR functions as a transcriptional activator of genes encoding GAPDH and PK in the presence of both D-fructose and D-glucose (Tables 1 and 2).
TABLE 2.
Promoter activities of selected genes involved in D-fructose degradation via modified EM pathway in Haloferax volcanii.
| β-galactosidase activity (mU/mg) | ||||
|---|---|---|---|---|
| Promotor of | Strain | CAS | CAS+D-fructose | Induction (fold) |
| ptfC (HVO_1499) | wt | 43 ± 19 | 214 ± 98 | 5.0 |
| ΔgfcR | 39 ± 10 | 44 ± 7 | 1.1 | |
| fba (HVO_1494) | wt | 22.0 ± 1a | 52 ± 7a | 2.4 |
| ΔgfcR | 7.0 ± 0a | 8 ± 2a | 1.1 | |
| gapl (HVO_0481) | wt | 218 ± 21 | 736 ± 159 | 3.4 |
| ΔgfcR | 220 ± 44 | 185 ± 55 | 0.8 | |
| pykA (HV0_0806) | wt | 65 ± 24 | 91b | 1.4 |
| ΔgfcR | 58 ± 15 | 41b | 0.7 | |
Note: Promotor activities were measured in a β-galactosidase assay in wild-type and ΔgfcR cells grown on casamino acids (CAS, 0.2%) and after induction by 5 mM D-fructose (CAS+D-fructose).
Basal promotor activity of fba was measured in cells grown on 5 mM D-glucose rather than on casamino acids. Experiments were carried out in triplicates.
Single measurements.
We then analyzed whether GfcR regulates steps in the upper part of the modified EM pathway for D-fructose uptake and degradation by measuring the promoter activities of ptfC and fba, encoding the PTS component EIIC and fructose-1,6-bisphosphate aldolase (FBA), respectively, in both wild-type and ΔgfcR mutant cells. The promoter activity of ptfC was five-fold higher in wild-type cells grown in the presence of D-fructose compared to those grown on casamino acids (Table 2). This D-fructose-specific induction was absent in the ΔgfcR mutant. Since fba is essential for gluconeogenesis (Pickl et al., 2012), we measured the basal promoter activity of fba using D-glucose instead of casamino acids. We found that promoter activity was 2.4-fold higher in D-fructose-grown cells compared to those grown in D-glucose and induction was not observed in ΔgfcR cells. Together, these experiments indicate that GfcR transcriptionally activates the degradation of D-fructose by activating genes of the upper half of the EM pathway, in addition to gapI and pykA in the lower half (Table 2).
2.4 |. PtfC is independently regulated by GfcR and GlpR
We also investigated whether GfcR affects ptfC transcription in the presence of glucose. A two-fold induction of ptfC was observed in wild-type cells but not in ΔgfcR cells, indicating that GfcR activates ptfC in the presence of glucose as well as fructose (Tables 2 and 3). It has been reported that in H. volcanii, the ptfC gene is repressed by the transcriptional regulator GlpR during growth on glucose (Martin et al., 2018). To elucidate the interplay between GfcR and GlpR in ptfC regulation, we generated ΔglpR single-and ΔgfcRΔglpR double-deletion mutants and measured ptfC promoter activity on casamino acids and after addition of glucose, i.e., under conditions in which ptfC is repressed by GlpR and activated by GfcR (Table 3). When cells were grown on casamino acids, basal promoter activity was about 10-fold higher in both mutants compared to wild-type and ΔgfcR cells (Table 3), likely reflecting derepression of ptfC caused by the glpR deletion. This higher level of ptfC activity was also found in the presence of glucose. For the ΔglpR mutant, the addition of glucose resulted in a 2.6-fold increase of ptfC activity and was similar to wild-type. This induction was abolished in the ΔgfcRΔglpR double mutant. These results show that both regulators, GfcR and GlpR, independently affect the promoter activity of ptfC.
TABLE 3.
Promoter activities of the ptfC gene measured in ΔgfcR, ΔglpR mutants and in the respective double deletion mutant in comparison to the wild type.
| β-galactosidase activity (mU/mg) | ||||
|---|---|---|---|---|
| Promotor of | Strain | CAS | CAS+D-glucose | Induction (fold) |
| ptfC (HVO_1499) | wt | 49.2 ± 6.3 | 98.6 ± 13.2 | 2.0 |
| ΔgfcR | 55.0 ± 6.3 | 43.6 ± 5.7 | 0.8 | |
| ΔglpR | 409.1 ± 27.8 | 1047.6 ± 51.6 | 2.6 | |
| ΔgfcRΔglpR | 424.1 ± 24.4 | 397.6± 29.5 | 0.9 | |
Note: Promotor activities were measured in a β-galactosidase assay in cells of wild-type and of mutants grown on casamino acids (CAS, 0.2%) and after induction by 5 mM D-glucose (CAS+D-glucose). Experiments were carried out in triplicates.
2.5 |. GfcR regulates D-galactose and glycerol degradation by activating GAP dehydrogenase and pyruvate kinase
It has been shown that H. volcanii degrades D-galactose via the DeLey-Doudoroff pathway (Tästensen et al., 2020) (Figure 6). To test whether GfcR activates genes of the DeLey-Doudoroff pathway, we analyzed the promoter activity of HVO_A0330, which encodes galactose dehydrogenase. Activity increased by about 3.5-fold in both wild-type and ΔgfcR mutant cells in the presence of D-galactose but not casamino acids, demonstrating that GfcR is not involved in the activation of galactose dehydrogenase (Table 4). For gapI and pykA, promoter activities increased 3.8-and 1.6-fold, respectively, in wild-type cells after addition of D-galactose, but not in the ΔgfcR mutant (Table 4). These data show that GfcR functions as a transcriptional activator of gapI and pykA in the presence of D-galactose in the same way as it does with D-glucose and D-fructose (Tables 1 and 2).
FIGURE 6.

Pathways of D-glucose, D-fructose, D-galactose and glycerol catabolism of Haloferax vocanii transcriptionally regulated by GfcR. Enzymes encoded by genes activated by GfcR are highlighted in green, while enzymes encoded by non-activated genes are marked in gray. Inducer molecules of GfcR are boxed. See Figure 1; G3PDH, glycerol-3-phosphate dehydrogenase; GalD, galactonate dehydratase; GalDH, galactose dehydrogenase; GK, glycerol kinase; KDGal, 2-keto-3-deoxygalactonate; KDGalK, KDGal kinase; KDPGal, 2-keto-3-deoxy-6-phosphogalactonate; KDPGalA, KDPGal aldolase.
TABLE 4.
Promoter activities of selected genes involved in D-galactose and glycerol degradation in Haloferax volcanii.
| β-galactosidase activity (mU/mg) | Induction (fold) | |||||
|---|---|---|---|---|---|---|
| Promotor of | Strain | CAS | CAS+D-galactose | CAS+glycerol | D-galactose | Glycerol |
| HV0_A0330 | wt | 76 ± 14 | 260 ± 54 | n.d. | 3.4 | |
| ΔgfcR | 78 ± 18 | 287 ± 38 | n.d. | 3.7 | ||
| glpK (HVO_1541) | wt | 106a | n.d. | 202a | 1.9 | |
| ΔgfcR | 121a | n.d. | 180a | 1.5 | ||
| gapl (HV0_0481) | wt | 218 ± 21 | 822 ± 128 | 344 ± 48 | 3.8 | 1.6 |
| ΔgfcR | 220 ± 44 | 263 ± 37 | 147 ± 29 | 1.2 | 0.7 | |
| pykA (HV0_0806) | wt | 65 ± 24 | 102a | n.d. | 1.6 | |
| ΔgfcR | 58 ± 15 | 35a | n.d. | 0.6 | ||
Note: Promotor activities were measured in a β-galactosidase assay in wild-type and ΔgfcR cells grown on casamino acids (CAS, 0.2%) and after induction by 5 mM D-galactose (CAS+D-galactose) or 5 mM glycerol (CAS+glycerol). Experiments were carried out in triplicates. Abbreviation: n.d., not determined.
Single measurements.
The DeLey-Doudoroff enzymes, e.g., galactose dehydrogenase, have been shown to be transcriptionally activated by the IclR-like regulator GacR (Tästensen et al., 2020). To determine whether GacR provides additional regulatory control to the expression of GAPDH, an enzyme of the lower part of the pathway, we made a ΔgacR mutant and examined the promoter activation of gapI in the presence of D-galactose. The promoter activity increased to the same extent, about 2-fold, in both wild-type and ΔgacR mutant strains, thus excluding GacR as an activator of GAPDH (Table S1).
To investigate whether GfcR is also a regulator of glycerol catabolism, we next analyzed the effect of its deletion on the promoter activity of glpK (encoding glycerol kinase). Promoter activity was stimulated to the same extent in both wild-type and the ΔgfcR mutant strains after addition of glycerol (Table 4), indicating that GfcR does not regulate glpK transcription. However, analyses of promoter activity of gapI revealed 1.6-fold higher activity in the wild-type as compared to the mutant, thus indicating a transcriptional activation of gapI by GfcR during growth on glycerol. This adds glycerol to the list of growth substrates that activate GAPDH expression in a GfcR-dependent fashion.
2.6 |. GfcR binds intermediates of sugar and of glycerol degradation pathways
To identify possible inducer molecules of GfcR, we performed thermal shift assays for the purified protein (Figure S2) with intermediates of the degradation pathways for D-glucose, D-fructose, D-galactose and glycerol. In the absence of putative ligands, the melting temperature (Tm) of GfcR was 70.0°C. Among the intermediates tested, only the addition of 2-keto-3-deoxy-6-phosphogluconate (KDPG), fructose-1,6-bisphosphate (FBP), 2-keto-3-deoxy-6-phosphogalact onate (KDPGal) and glycerol-3-phosphate (G3P) resulted in an increase of the melting temperature of 1.5 to 3.7 in comparison to the Tm values of GfcR without effector (70.0°C) and with other intermediates tested (Table 5). This indicates a complex formation of GfcR with these four metabolites.
TABLE 5.
Identification of inducer molecules of GfcR by differential scanning fluorimetry (DSF).
| Effector | ΔTm (°C) |
|---|---|
| No effector | 0 ± 0.4 |
| D-Glucose | −0.8 ± 1.4 |
| D-Gluconate | −0.01 ± 0.8 |
| 2-Keto-3-deoxygluconate | −1.0 ± 1.7 |
| 2-Keto-3-deoxy-6-phosphogluconate | +3.1 ± 0.7 |
| D-Fructose | −0.2 ± 0.3 |
| Fructose-1-phosphate | +0.06a |
| Fructose-1,6-bisphosphate | +1.5 ± 0.6 |
| D-Galactose | +0.4 ± 0.26 |
| 2-Keto-3-deoxy-6-phosphogalactonate | +3.7a |
| Glycerol | +0.01 ± 0.02 |
| Glycerol-3-phosphate | +2.8 ± 0.25 |
| Glyceraldehyde-3-phosphate | −0.4a |
| DHAP | +0.3 ± 0.06 |
| 3-Phosphoglycerate | −0.15 ± 0.07 |
| PEP | −0.7a |
| Pyruvate | −0.3 ± 0.2 |
Note: GfcR (1–5 μM) and 5 mM of ligand were used. Melting temperature (Tm value) of GfcR without ligand corresponds to 70.0 ± 0.4°C. ΔTm values indicate a shift of melting temperature of GfcR in the presence of ligand; values are given with standard deviations (SD).
Single measurements.
2.7 |. GfcR binds to promoters of gdh-gfcR, gad and ptfC
Our observation that GfcR increases expression of several genes of sugar degradation pathways, as well as the presence of a putative helix-turn-helix (HTH) DNA-binding motif at the N-terminal end of the protein, led us to examine whether GfcR can bind directly to promoters of the regulated genes. To do this, we performed electrophoretic mobility shift assays (EMSAs) using the promoters of genes implicated in glucose (gdh-gfcR and gad) and fructose (ptfC) degradation. For these analyses, GfcR was purified as fusion protein containing a C-terminal StrepII tag. Size exclusion chromatography indicated the protein is a homotetramer in solution (Figure S3).
The promoters tested were gdh-gfcR, gad and ptfC; the rad3b promoter served as negative control. The experiments were conducted in the presence and absence of 10 μM KDPG and increasing concentrations of GfcR-StrepII (Figure 7a). At concentrations of GfcR-StrepII higher than 31 nM gel shift bands appeared in the sugar regulated promoters rather than in the control rad3b.
FIGURE 7.
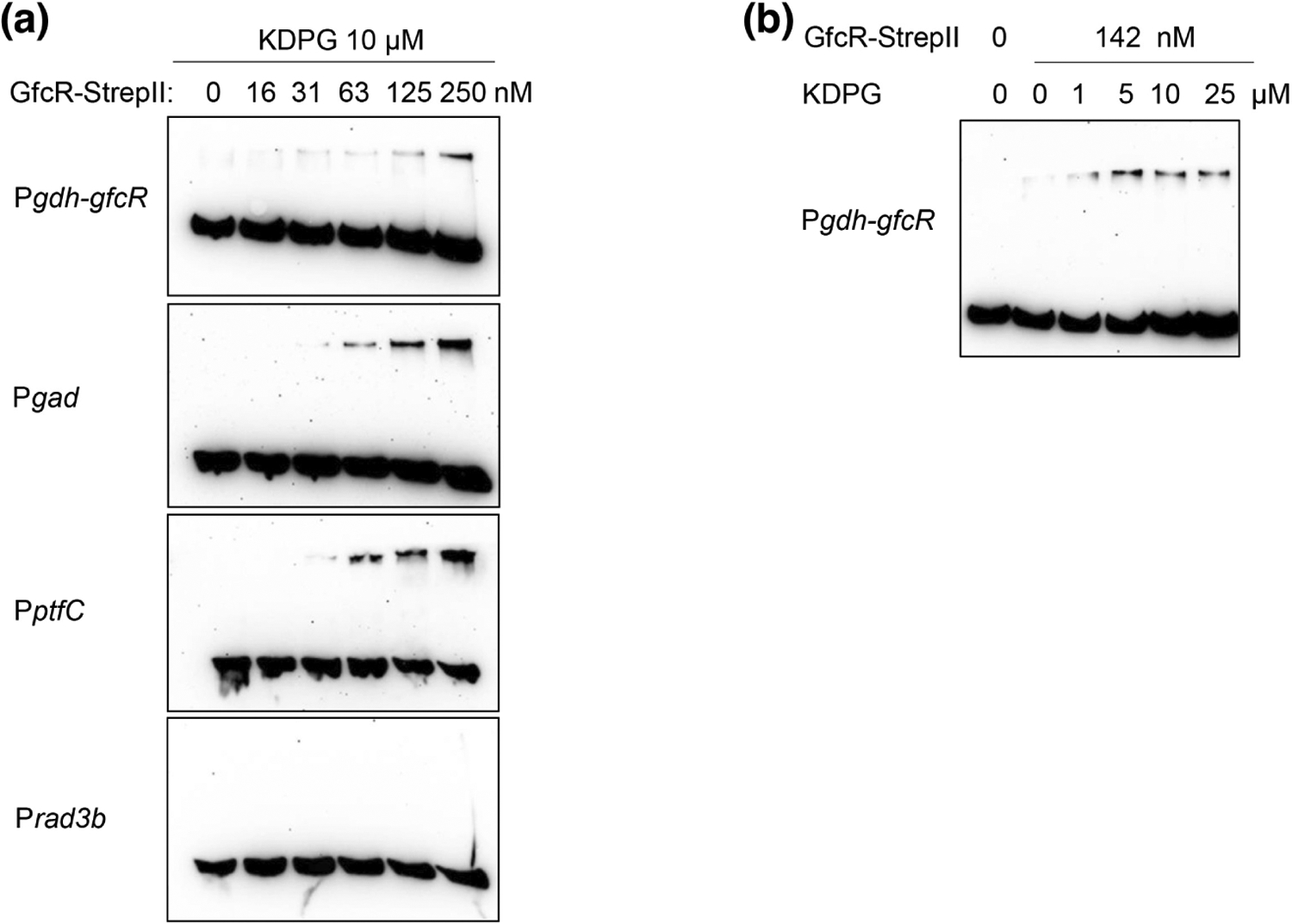
GfcR from Haloferax volcanii binds directly and specifically to promoters of gad, ptfC and gdh-gfcR demonstrated by EMSA. (a) GfcR-StrepII binds to gdh-gfcR, gad and ptfC promoters in the presence of 10 μM KDPG; rad3b was used as a negative control. (b) Effect of KDPG (1–25 μM) on gdh-gfcR promoter GfcR complex formation using 142 nM GfcR-StrepII.
We also examined the dependence of promoter binding in the presence of increasing concentrations of KDPG. Commensurate with the role of this intermediate as an inducer that binds GfcR, we observed that the intensity of the gel-shifted band increased with KDPG concentration (Figure 7b). Overall, these data indicate direct binding of GfcR to the promoters of at least three genes involved in glucose or fructose catabolism and that binding increases in the presence of the inducer KDPG.
2.8 |. Homology modeling of GfcR
GfcR is a member of the PRT-like superfamily and shows homology to orotate phosphoribosyltransferases (OPRTs). These enzymes catalyze the formation of orotidine-5′-monophosphate (OMP) from orotate and phosphoribosyl pyrophosphate (PRPP) as part of the pyrimidine biosynthesis pathway. To understand how ligands might bind the protein, we downloaded a model of GfcR from the AlphaFold Protein Structure Database server (code AF-Q8NKQ2-F1) (Jumper et al., 2021; Varadi et al., 2021) and superimposed onto the crystal structure of orotidine-5′-monophosphate synthase (OMPS) from Salmonella typhimurium in complex with PRPP and orotate (PDB 1LH0; Grubmeyer et al., 2012). This structure was selected because it has the highest sequence identity with GfcR (23% for residues 48–208 of OMPS). The superimposition shows that many residues of OMPS that are involved in binding PRPP are also present in GfcR (Figure S4). These include a stretch of residues (159–162, GfcR numbering) that form amide-mediated contacts with the 5′-phosphate (Thr-Ser-Gly-Thr in GfcR vs Thr-Ala-Gly-Thr in OMPS) and two aspartates (155–156, GfcR numbering) that form hydrogen bonds with the ribose hydroxyls. By contrast, residues that contact the pyrophosphate of PRPP are not present in GfcR. This is because the catalytic loop of OMPS is structured very differently in GfcR (i.e., the right-hand side of the binding site as viewed in Figure S4). For this reason, GfcR would not be expected to bind PRPP and this may explain why GfcR does not exhibit orotate phosphoribosyltransferase activity (Bitan-Banin et al., 2003).
2.9 |. Molecular docking of GfcR ligands
The four molecules that bind GfcR are all sugar phosphates (or sugar alcohol phosphates in the case of glycerol phosphate) and are therefore expected to bind to the region of GfcR that is homologous to the PRPP-binding site of OMPS. To examine potential interactions made by the effector molecules, we manually docked KDPG, FBP, KDPGal and G3P into the GfcR model using PRPP bound to OMPS as a guide. Emphasis was placed on positioning the 6′ phosphate of KDPG, FBP, KDPGal and 3′ phosphate of G3P into the pocket occupied by the 5′ phosphate of PRPP. Each model was then subjected to energy minimization and 5 ns of molecular dynamics simulations.
The models show there is ample space in the putative binding site for all four molecules and that most of the contacts involve the 6′/3′ phosphate forming hydrogen bonds with residues 157–163 of GfcR. For most of the intermediates, there are also hydrogen bonds with Asp155. The exception is FBP, where the presence of the furanose ring has produced a slightly different mode of binding. As noted above, residues 155–162 are highly conserved in the PRT superfamily. By contrast, there are relatively few contacts involving residues at the right side of the binding site (as viewed in Figure 8), consistent with the variability of effector molecules occupying these regions. Only Lys124 is consistently involved in binding via electrostatic contact with the negatively charged carboxylate or phosphate groups of the intermediates. Consistent with the 6′/3′ phosphate being present in all effectors, residues around this group exhibit relatively minimal differences in position after the MD simulations, whereas those on the right-hand side of the binding site vary more in structure. Together, these models provide a plausible framework to explain how the regulation of four different glycolytic pathways can be achieved by the binding of the four specific effector molecules to the same transcriptional regulator, GfcR.
FIGURE 8.

Molecular docking of effector molecules into the homology model of Haloferax volcanii GfcR. Only the region of GfcR corresponding to the PRPP-binding site of OMP synthase is shown where the effector molecules are predicted to bind. The panels are as follows: (a) Fructose-1,6-bisphosphate (FBP), (b) 2-keto-3-deoxy-6-phosphogluconate (KDPG), (c) 2-keto-3-deoxy-6-phosphogalactonate (KDPGal) and (d) Glycerol-3-phosphate (G3P). Potential hydrogen or electrostatic bonds are shown as dashed lines. Effector molecules are colored in yellow bonds and the GfcR residues in the binding site are gray.
3 |. DISCUSSION
In this study, we report the identification and functional characterization of GfcR as an activator of four different catabolic pathways in the haloarchaeon Haloferax volcanii; namely, the degradation pathways for the sugars D-glucose, D-fructose and D-galactose, and of the non-sugar substrate glycerol. Regulation is achieved through activation of specific steps in the upper part of the pathways for glucose and fructose, and also through activation of the common enzymes of the lower part of all four pathways. EMSAs demonstrate direct binding of GfcR to promoters and thermal denaturation experiments suggest that the mechanism of activation is by direct binding of intermediates of the four catabolic pathways to GfcR. Overall, these data reveal a new transcriptional regulator of archaeal sugar metabolism.
The degradation pathways of the three hexoses regulated by GfcR each comprise an upper half that is specific for the hexose and a common lower pathway commencing with GAP (Figure 6). Through activation of both GAP dehydrogenase and pyruvate kinase in the lower half of the pathways, GfcR regulates the catabolism of all three hexoses and also of glycerol. GfcR also regulates specific steps in two of the upper pathways. In the modified EM pathway for D-fructose, these are the transport of D-fructose into cell catalyzed by PTS (via upregulation of ptfC) and the subsequent cleavage of FBP (via upregulation of FBA). In the spED pathway, GfcR regulates the dehydration of gluconate into KDG catalyzed by gluconate dehydratase. By contrast, GfcR does not apparently regulate any steps in the DeLey-Doudoroff pathway for D-galactose or steps in the conversion of glycerol to DHAP. This might be due to the fact that these pathways are controlled by different regulators, GacR for galactose and GlpR for glycerol (Martin et al., 2018; Tästensen et al., 2020). Overall, it appears that GfcR is both a global regulator of hexose catabolism and a local regulator of specific steps in the pathways for D-glucose and D-fructose.
Having established GfcR as an inducer of the hexose catabolic pathways in H. volcanii, we then investigated the molecular mechanism of regulation. Differential scanning fluorimetry of purified GfcR against a panel of intermediates from the four pathways revealed four molecules that bind the protein: 2-keto-3-deoxy-6-phosphoglu conate (KDPG), fructose-1,6-bisphosphate (FBP), 2-keto-3-deoxy-6-phosphogalactonate (KDPGal) and glycerol-3-phosphate (Table 5). Notably, this corresponds to one intermediate from each of the four pathways regulated by GfcR, suggesting coordinated control of all four catabolic pathways. The strongest binding effect was observed with intermediates of glucose and galactose degradation, KDPG and KDPGal, which are C-4 epimers of each other. These experiments also revealed that GfcR is activated by binding of intermediates in the pathways under its regulatory control, rather than by an indirect mechanism. Furthermore, the underlying mechanism is one of feed-forward stimulation, where intermediates in the regulated pathways increase expression of enzymes or transporters within the pathways. In this context, the activation by GfcR of the fructose transport component ptfC on glucose can be explained by binding of KDPG, an intermediate in the spED pathway, although this activation has no physiological relevance for growth on glucose.
Another element of regulation in the GfcR system is the cotranscription of gfcR and gdh. This is not unique to H. volcanii because we also identified GfcR homologs in other haloarchaea, e.g., in Haloferax mediterranei, Halorubrum lacusprofundi and Haloterrigena turkmenica, whose encoding genes are also adjacent to the gene encoding glucose dehydrogenase (Figure S5). In Haloarcula species, GfcR homologs were also found in genomic vicinity to their respective glucose dehydrogenase genes but separated by nine genes (including four genes encoding components of a putative ABC transporter) (Figure S5). Thus, we propose that these GfcR homologs show a similar regulatory function in these haloarchaea in activating the catabolism of D-glucose and D-fructose, and of D-galactose and glycerol.
Sequence analysis of GfcR suggests that the protein belongs to the phosphoribosyl transferase (PRT) superfamily. This is a group of enzymes with well-known functions in purine, pyrimidine, and amino acid biosynthesis. A common feature of these enzymes is a central 5-stranded beta sheet surrounded by 5 alpha helices and many bind phosphoribosyl pyrophosphate (PRPP) as a donor of phosphoribosyl. PRTs are homologous to bacterial PurR regulators present in a few Gram-positive bacteria, i.e., Bacillus subtilis and Lactobacillus lactis (Hove-Jensen et al., 2017; Kilstrup & Martinussen, 1998; Sinha et al., 2003). Each of the four intermediates shown to increase the Tm of GfcR could be plausibly docked into an AlphaFold model of the protein. These models suggest that recognition of the 6′ phosphate of FBP, KDPG, and KDPGal or the 3′-phosphate of glycerol-3-phosphate by a conserved loop of mostly serines and threonines (residues 128–131 in GfcR) is the key for binding. On the other hand, the lack of homology in the region of the binding site that contacts the pyrophosphate moiety of PRPP in OMP synthase may explain why GfcR shows no activity with PRPP (Bitan-Banin et al., 2003).
It has been speculated that the bacterial PurR regulators originated from catalytically active enzymes of the PRT family by gene duplication followed by addition at the N-terminal of a DNA-binding domain containing a helix-turn-helix motif (Sinha et al., 2003; Sinha & Smith, 2001). This enables PurR to function as a transcriptional regulator of purine biosynthesis genes. Consistent with this, a putative HTH domain is observed at the N-terminal region of GfcR, and constructs lacking this motif no longer complement a GfcR deletion mutant, supporting the idea that GfcR binds DNA. This was subsequently confirmed using EMSAs, where we observed that GfcR binds to the promoters of gdh-gfcR, gad and ptfC. By analogy to PurR, we therefore speculate that GfcR originated from an archaeal PRT enzyme by fusing an HTH domain with a catalytically inactive PRT domain to form a transcription factor for the regulation of sugar catabolism.
GfcR of H. volcanii, formerly named PyrE1, is homologous to PyrE2. PyrE2 is an enzyme with orotate phosphoribosyl transferase activity that is involved in pyrimidine biosynthesis (Bitan-Banin et al., 2003). H. volcanii and many other haloarchaea, e.g., H. lacusprofundi and H. turkmenica, contain both GfcR and a PyrE2 homolog, and the same is true in a few other archaea, e.g., in methanogens and Thermoplasmatales. However, most archaea contain only the PyrE2 enzyme in accordance with its universal involvement in pyrimidine biosynthesis and lack a detectable GfcR.
In order to analyze the phylogenetic relationship of GfcR regulators with catalytically active PyrE enzymes (orotate phosphoribosyl transferases), we constructed a phylogeny of GfcR and PyrE sequences from bacteria and archaea (Figure 9). The phylogenetic tree also includes transcriptional regulators for purine (PurR) and for pyrimidine (PyrR) biosynthesis of selected Bacillus/Firmicutes species. The sequences clustered into four distinct groups. Cluster 1 (orange) comprises GfcR from haloarchaea and its homologs from methanogens and Thermoplasmatales. In Thermoplasmatales, the homologs might be involved in regulation of sugar degradation since in these archaea, sugar degradation has been reported via a non-phosphorylative ED pathway involving glucose dehydrogenase as the first enzyme (Angelov et al., 2005; Reher et al., 2010). By contrast, in methanogens, which do not grow on sugars, a role in regulation of sugar catabolism is not likely. Cluster 2 (light green) and Cluster 3 (dark green) includes bacterial PyrR regulators that regulate pyrimidine synthesis via attenuation, and bacterial PurR regulators that regulate purine biosynthesis genes by acting as DNA-binding regulators (Hove-Jensen et al., 2017). Cluster 4 (blue) comprises sequences of the PyrE enzymes (orotate phosphoribosyl transferases) from archaea and few bacteria. Most of these archaea, with the exception of Pyrobaculum aerophilum and Desulfurococcus amylolyticus, also contain a GfcR homolog that is present in cluster 1. Together, the tree topology indicates that PyrE enzymes and the transcriptional regulators, GfcR, PurR and PyrR and their homologs, form distinct clusters that reflect different evolutionary lines according to their distinct function.
FIGURE 9.

Phylogenetic relationship of GfcR of Haloferax volcanii and archaeal homologs, PurR and PyrR regulators from bacteria and PyrE enzymes from archaea and bacteria. UniProt and PDB entries as follows: GfcR archaea (orange, Cluster 1): Methanocaldococcus jannaschii, Q59040; Methanosarcina mazei, Q8PVD0; Haloterrigena turkmenica, D2RW31; Haloarcula marismortui, Q5V3K1; Halorubrum lacusprofundi, B9LUS0; Haloferax volcanii, Q8NKQ2; Haloterrigena turkmenica, D2S0M3; Thermoplasma acidophilum, Q9HJ11; Picrophilus torridus, Q6L1V2. PyrR bacteria (light green, Cluster 2): B. subtilis, pdb 1A3C; B. caldolyticus, pdb 1N0N; L. lactis, P0A3Z9. PurR bacteria (dark green, Cluster 3): B. subtilis, pdb 1O57; Lactococcus lactis, P0A3Z9. PyrE archaea and bacteria (blue, Cluster 4): B. subtilis, P25972; Salmonella typhimurium, pdb 1LH0; P. torridus, Q6L2K9; T. acidophilum, Q9HM15; M. mazei, Q8Q0J4; M. jannaschii, Q58509; H. turkmenica, D2RXD2; H. marismortui, Q5UZH9; H. volcanii, D4GZW2; H. lacusprofundi, B9LTQ2; Desulfurococcus amylolyticus, I3XQY8; Pyrobaculum aerophilum, Q8ZTG3.
GfcR is not the first transcriptional regulator identified for the spED and modified EM pathways in H. volcanii. The DeoR-type regulator, GlpR, has been identified as repressor of enzymes involved in glucose and fructose catabolism, including phosphofructokinase, FBA and KDGK (Martin et al., 2018; Rawls et al., 2010). Among genes whose expression is altered by GlpR is ptfC. Similar to GfcR, GlpR activates ptfC in the modified EM pathway caused by derepression during growth on fructose. However, for the spED pathway at least, GfcR appears to be the major transcriptional regulator because it elicits higher levels of activity compared to GlpR. Finally, we note that GfcR differs from the recently characterized transcriptional activators of sugar catabolism in haloarchaea, i.e., XacR for pentoses, RhcR for L-rhamnose and GacR for D-galactose (Johnsen et al., 2015; Reinhardt et al., 2019; Tästensen et al., 2020). All these haloarchaeal regulators belong to the bacterial-type IclR family that have likely been acquired by Haloferax volcanii from bacteria via a lateral gene transfer event.
To conclude, we have identified a novel type of transcriptional regulator that functions as an activator of sugar catabolic pathways in haloarchaea by binding an intermediate from each pathway under its regulatory control. GfcR likely originated in the archaeal domain by fusion of an HTH motif containing a DNA-binding domain and an archaeal PRT domain. With the identification and characterization of GfcR as a novel type of regulator, we expand our knowledge of transcriptional regulation of central metabolic pathways in archaea.
4 |. EXPERIMENTAL PROCEDURES
4.1 |. Growth of H. volcanii
Haloferax volcanii H555 and the ΔgfcR mutant were grown aerobically at 42°C on synthetic medium, as described (Sutter et al., 2016), containing either D-glucose, D-fructose, D-galactose (each at 25 mM), glycerol (20 mM) or 1% casamino acids. This medium was supplemented with uracil (50 μg/mL); for growth of the ΔgfcR mutant complemented with gfcR, the medium was supplemented with 100 μM L-tryptophan and growth was performed in the absence of uracil for selection of the plasmid. Growth was monitored by measuring optical density at 600 nm.
4.2 |. Reporter assays
Haloferax volcanii H555 transformed with promoter-reporter gene constructs were grown on synthetic medium with 0.2% casamino acids (except for the fba promoter analysis, 5 mM D-glucose was used) up to an optical density at 600 nm of 1.0. Then, either D-glucose, D-fructose, D-galactose or glycerol (each 5 mM) was added, followed by further incubation. β-Galactosidase activity was analyzed before and up to 150 h after the addition of the sugar or glycerol. For analysis of GfcR-dependent transcriptional regulation of gad, the same growth condition was used and RNA was isolated before (0 h) and 2 h after the addition of D-glucose (5 mM).
4.3 |. Construction of deletion mutants
Deletions of gfcR and glpR were introduced in H. volcanii strain H555 using the pop-in/pop-out strategy (Allers & Mevarech, 2005; Bitan-Banin et al., 2003). The upstream and downstream regions of gfcR or glpR were amplified and the 0.5 kb-fragments were fused by PCR. The fused fragments were each ligated into the vector pTA131 and multiplied in E. coli XL1 Blue MRF′. After transformation of H. volcanii H555 with the pTA131-ΔgfcR or pTA131-ΔglpR plasmids (Table S2), pop-in clones were selected in uracil-free medium containing casamino acids (1%) and passaged in liquid medium. For pop-out selection, a pop-in culture was streaked on agar plates containing uracil (50 μg/mL) and 5-fluoroorotic acid (50 μg/mL) followed by several passages in liquid medium. Clones carrying the deletion were identified by PCR and the deletion was verified by Southern hybridisation using digested genomic DNA (EcoRV and SmaI for ΔgfcR; PciI and EcoRV for ΔglpR). Probes were synthesized with the PCR DIG Probe Synthesis Kit (Roche Diagnostic). The double deletion mutant ΔgfcRΔglpR was generated by transforming ΔgfcR with the pTA131-ΔglpR plasmid following the procedure described above.
4.4 |. RNA isolation and transcriptional analyses
For gdh-gfcR transcript analysis, RNA was prepared from exponentially grown H. volcanii cells on D-glucose or casamino acids (OD600 ~ 1.3). For gad transcript analysis, RNA was obtained from stationary grown cells at the indicated times (see growth section). The cells were centrifuged for 2 min at 10,000 g and suspended in 1 mL TRI reagent (Sigma-Aldrich). RNA was prepared as described previously by Johnsen et al. (2013). Northern blot analysis was performed with 4 to 8 μg RNA as described by Pickl et al. (2012). The probe for gdh-gfcR transcript analysis was prepared using the PCR DIG Probe Synthesis Kit (Roche Diagnostic).
The transcript of gad was analyzed by RT-PCR with DNaseI-treated RNA using the GoScript Reverse Transcription System (Promega) as described by the manufacturer. First, 100 ng of DNA-free RNA was transcribed into cDNA using DNA hexamers of random sequence (C1181, Promega) and the following PCR steps used GoTaq G2 Flexi DNA polymerase (Promega) and a gad specific primer. Ribosomal protein L10 gene (RibL) was used as a marker for constitutive expression (Reinhardt et al., 2019).
4.5 |. Construction of promoter-reporter-gene fusions
Promoter fragments were amplified and ligated into the vector pTA919, as described by Johnsen et al. (2015). By this cloning strategy, the reporter gene bgaH (Holmes & Dyall-Smith, 2000) was controlled by the respective promoter. The resulting plasmids (Table S2) were multiplied in Escherichia coli XL1-Blue MRF′ and used for transformation of wild-type H. volcanii H555 or of the ΔgfcR, ΔgacR, ΔglpR and ΔgfcRΔglpRmutants.
4.6 |. β-galactosidase assay
β-galactosidase activity was analyzed in cell lysates of transformed H. volcanii H555 stains at 30°C using the o-Nitrophenyl-β-D-galactopyranoside (ONPG) assay and a molar extinction coefficient of 3.3 mM/cm as described Johnsen et al. (2015). The reaction mixture contained 50–200 μL cell suspension, 1 μL β-mercaptoethanol, 0.1% SDS in 1 mL of 50 mM Tris–HCl (pH 7.2) containing 2.5 M NaCl and 10 μM MnCl2. The reaction was initiated by addition of 8 mM ONPG and the increasing absorbance was followed at 405 nm. One unit of enzyme activity is defined as 1 μmol product formed per minute. For calculation of specific activities (U/mg), the protein content of cells was estimated from the optical density of these cultures. An optical density at 600 nm of 1.0 corresponds to a protein concentration of 0.28 mg/mL.
4.7 |. Overexpression and purification of GfcR
GfcR was cloned into the plasmid pT963 (Table S2), a shuttle vector for E. coli and H. volcanii. The plasmid was multiplied in E. coli XL1-Blue MRF′ and used for transformation of the expression strain H. volcanii H1209. For expression of GfcR, cells were grown in Hv-YPC medium (Allers et al., 2010) up to an optical density at 600 nm of 0.5 and expression was started by the addition of 2 mM L-tryptophan. After 12 h of further growth cells were harvested by centrifugation. For purification, cells were suspended in 50 mM Tris–HCl, pH 8.2, containing 5 mM imidazole and 2 M KCl, and disrupted by passaging a French pressure cell followed by a centrifugation step. The supernatant was applied on a Ni-NTA Superflow cartridge (Qiagen) and bound protein was eluted by increasing the concentration of imidazole up to 250 mM. This protein solution was applied on a Superdex 200 HiLoad 16/60 column (Cytiva) and protein was eluted by an isocratic step in 50 mM Tris–HCl, pH 7.5, containing 2 M KCl. Purity of protein and estimation of protein concentration was checked by SDS-PAGE and the method of Bradford, respectively (Figure S2).
4.8 |. Site-directed mutagenesis
The plasmids pTA963-gfcRΔα1 and pTA963-gfcRΔα1Δα2 were generated using the Q5 Site-directed Mutagenesis Kit (New England Biolabs) according to the manufacturer’s instructions. Primers were designed using the NEBaseChanger tool and pTA963-gfcR was used as template (Table S2). These plasmids were used for complementation experiments of H. volcanii H555 ΔgfcR.
4.9 |. Thermal shift assay
The thermal shift assay was performed as described (Martin et al., 2018; Reinhardt et al., 2019). The assay mixture contained 1–5 μM recombinant GfcR, 5 mM ligand, SYPRO Orange (Sigma, S5692) and 2 M NaCl in 50 mM Tris–HCl, pH 8.0. Melting curves were measured between 25 and 95°C (scan rate of 1°C per minute) using a Real-Time PCR System (Applied Biosystems). Melting temperatures were calculated using the Boltzman equation with Origin 2018 software (OriginLab Corporation).
4.10 |. Electrophoretic mobility shift assay (EMSA)
For EMSAs GfcR was fused with StrepII at its C-terminal end (GfcR-StrepII). To make this construct, HVO_1082 was amplified by PCR from H. volcanii genomic DNA using the primers HVO_1082-NdeI (5′-tagcatATGAA GAA CGT CGA CGA CCT CATCGCC) and HVO_1082-KpnI (5′-aaggtaccCTGCT CGC CGA CGC GGAC) and cloned into pJAM809, yielding pJAM3945 (Table S2). A plasmid constructed using E. coli TOP10 (Invitrogen) was transferred to E. coli GM2163 (New England Biolabs) prior to H. volcanii H26 transformation. GfcR-StrepII was then expressed in the transformed H. volcanii strain. A detailed description of the expression and the purification procedure of GfcR-StrepII is given in Appendix S1.
EMSAs were performed as described previously by Mondragon et al. (2022) with the following modifications: GfcR-StrepII protein was exchanged into binding buffer (50 mM HEPES, pH 7.5, 2 M NaCl, 10% sorbitol, 15 mM MgCl2, 2 mM DTT, 1 mM EDTA) immediately prior to incubation with the 5′-biotinylated dsDNA probes. Reactions (10 μL) included GfcR, 2 nM biotinylated DNA probe (Pgdh-gfcR, Pgad, PpftC or Prad3b, Figure S6), 50 mM HEPES, pH 7.5, 2 M NaCl, 10% sorbitol, 15 mM MgCl2, 2 mM DTT, 1 mM EDTA, ultrapure sheared salmon sperm DNA at 0.1 mg/mL (Ambion AM9680), and molecular biology-grade BSA at 0.25 mg/mL (New England Biolabs). Reactions were incubated for 30 min at room temperature. The effect of KDPG was examined by supplementing the reactions with 2–keto–3–deoxy-6-phosphogluconic acid lithium salt (KDPG, Sigma 79156). After incubation, samples were applied without loading dye to 10% PAGE gels in 0.5 × TBE buffer pH 8.3 (110 mM Tris, 90 mM borate, 2.5 mM EDTA) preequilibrated with 0.5 × TBE buffer pH 8.3 at 20 mA for 50 min (4°C). Samples were separated by electrophoresis at 4°C for 50 min at 60 V followed by 2 h at 120 V. After electrophoresis, DNA and nucleoprotein complexes were transferred to a nylon membrane (BrightStar Plus, Ambion) equilibrated in 0.5 × TBE pH 8.3 using the trans blot semidry system (Bio-Rad) at 20 V for 20 min. DNA and nucleoprotein complexes were cross-linked to the membrane via UV radiation using the autocrosslink mode (UV Stratalinker 2400, Stratagene). Signal was detected using a Chemiluminescent Nucleic Acid Detection Kit (Thermo Scientific) with visualization using an iBright FL1500 Imaging System (Invitrogen).
4.11 |. Molecular modeling
A model for GfcR was obtained from the AlphaFold Protein Structure Database (https://alphafold.com) and superimposed onto the structure of Salmonella typhimurium OMP synthase in complex with PRPP using the COOT program (Emsley et al., 2010). The position of the PRPP 5-O-phosphonate was used to model fructose-1,6-bisphosphate (FBP), 2-keto-3-deoxy-6-phosphogluconate (KDPG), 2-keto-3-deoxy-6-phosphogalactonate (KDPGal), and glycerol-3-phosphate (G3P) into the binding site of GfcR. Each resulting ligand-GfcR complex was subject to molecular dynamics (MD) simulations using the Desmond Molecular Dynamics System (D. E. Shaw Research) in the Maestro environment (Schrödinger Release 2021-1 Schrödinger) as follows: Models were prepared by charge-neutralization at physiologic pH by addition of Na+ ions, solvation using the SPC water model, and application of the OPLS4 forcefield (Lu et al., 2021). After energy minimization, each structure was then subject to 5 ns molecular dynamics simulation, maintained at 300 K and 1 bar pressure using the NPT ensemble.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant of the Deutsche Forschungsgemeinschaft (SCHO 316/12-1). This work is also supported by JMF grants of the U.S. Department of Energy, Office of Basic Energy Sciences, Division of Chemical Sciences, Geosciences and Biosciences, Physical Biosciences Program (DOE DE-FG02-05ER15650) in archaeal bioenergy and National Institutes of Health (NIH R01 GM057498) in archaeal evolution. Open Access funding enabled and organized by Projekt DEAL.
Funding information
Deutsche Forschungsgemeinschaft, Grant/Award Number: SCHO 316/12-1; National Institutes of Health, Grant/Award Number: R01 GM057498; U.S. Department of Energy, Grant/Award Number: DE-FG02-05ER15650; Office of Basic Energy Sciences; Division of Chemical Sciences, Geosciences and Biosciences; Physical Biosciences Program
Footnotes
ETHICS STATEMENT SECTION
Authors declare that no human or animal subjects were used in this study.
SUPPORTING INFORMATION
Additional supporting information can be found online in the Supporting Information section at the end of this article.
DATA AVAIL ABILIT Y STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.e made available upon request.
REFERENCES
- Allers T, Barak S, Liddell S, Wardell K & Mevarech M (2010) Improved strains and plasmid vectors for conditional overexpression of his-tagged proteins in Haloferax volcanii. Applied and Environmental Microbiology, 76, 1759–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allers T & Mevarech M (2005) Archaeal genetics – the third way. Nature Reviews. Genetics, 6, 58–73. [DOI] [PubMed] [Google Scholar]
- Angelov A, Fütterer O, Valerius O, Braus GH & Liebl W (2005) Properties of the recombinant glucose/galactose dehydrogenase from the extreme thermoacidophile, Picrophilus torridus. The FEBS Journal, 272, 1054–1062. [DOI] [PubMed] [Google Scholar]
- Bitan-Banin G, Ortenberg R & Mevarech M (2003) Development of a gene knockout system for the halophilic archaeon Haloferax volcanii by use of the pyrE gene. Journal of Bacteriology, 185, 772–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräsen C, Esser D, Rauch B & Siebers B (2014) Carbohydrate metabolism in archaea: current insights into unusual enzymes and pathways and their regulation. Microbiology and Molecular Biology Reviews, 78, 89–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan DWA & Jones DT (2019) The PSIPRED protein analysis work-bench: 20 years on. Nucleic Acids Research, 47, W402–W407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG & Cowtan K (2010) Features and development of Coot. Acta Crystallographica. Section D, Biological Crystallography, 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubmeyer C, Hansen MR, Fedorov AA & Almo SC (2012) Structure of Salmonella typhimurium OMP synthase in a complete substrate complex. Biochemistry, 51, 4397–4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes ML & Dyall-Smith ML (2000) Sequence and expression of a halobacterial β-galactosidase gene. Molecular Microbiology, 36, 114–122. [DOI] [PubMed] [Google Scholar]
- Hove-Jensen B, Andersen KR, Kilstrup M, Martinussen J, Switzer RL & Willemoes M (2017) Phosphoribosyl diphosphate (PRPP): biosynthesis, enzymology, utilization, and metabolic significance. Microbiology and Molecular Biology Reviews: MMBR, 81, e00040–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen U, Dambeck M, Zaiss H, Fuhrer T, Soppa J, Sauer U et al. (2009) D-xylose degradation pathway in the halophilic archaeon Haloferax volcanii. The Journal of Biological Chemistry, 284, 27290–27303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen U, Reinhardt A, Landan G, Tria FDK, Turner JM, Davies C et al. (2019) New views on an old enzyme: allosteric regulation and evolution of archaeal pyruvate kinases. The FEBS Journal, 286, 2471–2489. [DOI] [PubMed] [Google Scholar]
- Johnsen U, Sutter JM, Reinhardt A, Pickl A, Wang R, Xiang H et al. (2020) D-Ribose catabolism in archaea: discovery of a novel oxidative pathway in Haloarcula species. Journal of Bacteriology, 202, e00608–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen U, Sutter JM, Schulz AC, Tästensen JB & Schönheit P (2015) XacR - a novel transcriptional regulator of D-xylose and L-arabinose catabolism in the haloarchaeon Haloferax volcanii. Environmental Microbiology, 17, 1663–1676. [DOI] [PubMed] [Google Scholar]
- Johnsen U, Sutter JM, Zaiss H & Schönheit P (2013) L-Arabinose degradation pathway in the haloarchaeon Haloferax volcanii involves a novel type of L-arabinose dehydrogenase. Extremophiles, 17, 897–909. [DOI] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O et al. (2021) Highly accurate protein structure prediction with AlphaFold. Nature, 596, 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai T, Akerboom J, Takedomi S, van de Werken HJ, Blombach F, van der Oost J et al. (2007) A global transcriptional regulator in Thermococcus kodakaraensis controls the expression levels of both glycolytic and gluconeogenic enzyme-encoding genes. The Journal of Biological Chemistry, 282, 33659–33670. [DOI] [PubMed] [Google Scholar]
- Kilstrup M & Martinussen J (1998) A transcriptional activator, homologous to the Bacillus subtilis PurR repressor, is required for expression of purine biosynthetic genes in Lactococcus lactis. Journal of Bacteriology, 180, 3907–3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Surma M, Hausner W, Thomm M & Boos W (2008) The role of TrmB and TrmB-like transcriptional regulators for sugar transport and metabolism in the hyperthermophilic archaeon Pyrococcus furiosus. Archives of Microbiology, 190, 247–256. [DOI] [PubMed] [Google Scholar]
- Lu C, Wu C, Ghoreishi D, Chen W, Wang L, Damm W et al. (2021) OPLS4: improving force accuracy on challenging regimes of chemical space. Journal of Chemical Theory and Computation, 17, 4291–4300. [DOI] [PubMed] [Google Scholar]
- Martin JH, Sherwood Rawls K, Chan JC, Hwang S, Martinez-Pastor M, McMillan LJ et al. (2018) GlpR is a direct transcriptional repressor of fructose metabolic genes in Haloferax volcanii. Journal of Bacteriology, 200, e00244–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondragon P, Hwang S, Kasirajan L, Oyetoro R, Nasthas A, Winters E et al. (2022) TrmB family transcription factor as a thiol-based regulator of oxidative stress response. mBio, 13, e0063322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patenge N, Haase A, Bolhuis H & Oesterhelt D (2000) The gene for a halophilic beta-galactosidase (bgaH) of Haloferax alicantei as a reporter gene for promoter analyses in Halobacterium salinarum. Molecular Microbiology, 36, 105–113. [DOI] [PubMed] [Google Scholar]
- Pickl A, Johnsen U, Archer RM & Schönheit P (2014) Identification and characterization of 2-keto-3-deoxygluconate kinase and 2-keto-3-deoxygalactonate kinase in the haloarchaeon Haloferax volcanii. FEMS Microbiology Letters, 361, 76–83. [DOI] [PubMed] [Google Scholar]
- Pickl A, Johnsen U & Schönheit P (2012) Fructose degradation in the haloarchaeon Haloferax volcanii involves a bacterial type phosphoenolpyruvate-dependent phosphotransferase system, fructose-1-phosphate kinase, and class II fructose-1,6-bisphosphate aldolase. Journal of Bacteriology, 194, 3088–3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls KS, Martin JH & Maupin-Furlow JA (2011) Activity and transcriptional regulation of bacterial protein-like glycerol-3-phosphate dehydrogenase of the haloarchaea in Haloferax volcanii. Journal of Bacteriology, 193, 4469–4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls KS, Yacovone SK & Maupin-Furlow JA (2010) GlpR represses fructose and glucose metabolic enzymes at the level of transcription in the haloarchaeon Haloferax volcanii. Journal of Bacteriology, 192, 6251–6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reher M, Fuhrer T, Bott M & Schönheit P (2010) The non-phosphorylative Entner-Doudoroff pathway in the thermoacidophilic euryarchaeon Picrophilus torridus involves a novel 2-keto-3-deoxygluconate-specific aldolase. Journal of Bacteriology, 192, 964–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt A, Johnsen U & Schönheit P (2019) L-Rhamnose catabolism in archaea. Molecular Microbiology, 111, 1093–1108. [DOI] [PubMed] [Google Scholar]
- Robert X & Gouet P (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Research, 42, W320–W324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid AK, Reiss DJ, Pan M, Koide T & Baliga NS (2009) A single transcription factor regulates evolutionarily diverse but functionally linked metabolic pathways in response to nutrient availability. Molecular Systems Biology, 5, 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood KE, Cano DJ & Maupin-Furlow JA (2009) Glycerol-mediated repression of glucose metabolism and glycerol kinase as the sole route of glycerol catabolism in the haloarchaeon Haloferax volcanii. Journal of Bacteriology, 191, 4307–4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebers B & Schönheit P (2005) Unusual pathways and enzymes of central carbohydrate metabolism in archaea. Current Opinion in Microbiology, 8, 695–705. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W et al. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular Systems Biology, 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha SC, Krahn J, Shin BS, Tomchick DR, Zalkin H & Smith JL (2003) The purine repressor of Bacillus subtilis: a novel combination of domains adapted for transcription regulation. Journal of Bacteriology, 185, 4087–4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha SC & Smith JL (2001) The PRT protein family. Current Opinion in Structural Biology, 11, 733–739. [DOI] [PubMed] [Google Scholar]
- Sutter JM, Johnsen U & Schönheit P (2017) Characterization of a pentonolactonase involved in D-xylose and L-arabinose catabolism in the haloarchaeon Haloferax volcanii. FEMS Microbiology Letters, 364, fnx140. [DOI] [PubMed] [Google Scholar]
- Sutter JM, Tästensen JB, Johnsen U, Soppa J & Schönheit P (2016) Key enzymes of the semiphosphorylative Entner-Doudoroff pathway in the haloarchaeon Haloferax volcanii: characterization of glucose dehydrogenase, gluconate dehydratase, and 2-keto-3-d eoxy-6-phosphogluconate aldolase. Journal of Bacteriology, 198, 2251–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tästensen JB, Johnsen U, Reinhardt A, Ortjohann M & Schönheit P (2020) D-galactose catabolism in archaea: operation of the DeLey-Doudoroff pathway in Haloferax volcanii. FEMS Microbiology Letters, 367, fnaa029. [DOI] [PubMed] [Google Scholar]
- Tästensen JB & Schönheit P (2018) Two distinct glyceraldehyde-3-phosphate dehydrogenases in glycolysis and gluconeogenesis in the archaeon Haloferax volcanii. FEBS Letters, 592, 1524–1534. [DOI] [PubMed] [Google Scholar]
- Todor H, Dulmage K, Gillum N, Bain JR, Muehlbauer MJ & Schmid AK (2014) A transcription factor links growth rate and metabolism in the hypersaline adapted archaeon Halobacterium salinarum. Molecular Microbiology, 93, 1172–1182. [DOI] [PubMed] [Google Scholar]
- Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G et al. (2021) AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Research, 50, D439–D444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M, Wagner A, Ma X, Kort JC, Ghosh A, Rauch B et al. (2014) Investigation of the malE promoter and MalR, a positive regulator of the maltose regulon, for an improved expression system in Sulfolobus acidocaldarius. Applied and Environmental Microbiology, 80, 1072–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.e made available upon request.


