Abstract
Introduction: The purpose of this study was to compare the transcriptomes of poorly cohesive carcinoma (PCC; diffuse-type) and well-differentiated tubular adenocarcinoma (WD; intestinal-type) using gastric cancer (GC) tissues and cell lines and to evaluate the prognostic role of HIV-1 Tat Interactive Protein 2 (HTATIP2). Materials and Methods: We performed next-generation sequencing with 8 GC surgical samples (5 WD and 3 PCC) and 3 GC cell lines (1 WD: MKN74, and 2 PCC: KATOIII and SNU601). Immunohistochemistry was used to validate HTATIP2 expression. We performed functional analysis by HTATIP2 overexpression (OE). Kaplan-Meier survival plots and the PrognoScan database were used for survival analysis. Results: The genes with significantly reduced expression in PCC versus WD (in both tissues and cell lines) were HTATIP2, ESRP1, GRHL2, ARHGEF16, CKAP2L, and ZNF724. According to immunohistochemical staining, the HTATIP2-OE group had significantly higher number of patients with early GC (EGC) (T1) (P = .024), less lymph node (LN) metastasis (P = .008), and low TNMA stage (P = .017) than HTATIP2 underexpression (UE) group. Better survival rates were confirmed in the HTATIP2 OE group by Kaplan-Meir survival and PrognoScan analysis. In vitro, HTATIP2-OE in KATO III cells caused a significant decrease in cancer cell migration and invasion. Decreased Snail and Slug expression in HTATIP2 OE cells suggested that epithelial-mesenchymal transition is involved in this process. Conclusion: HTATIP2 might be a good prognostic marker and a candidate target for GC treatment.
Keywords: stomach neoplasm, NGS, immunohistochemistry, biomarker, prognosis
Introduction
Gastric cancer (GC) is the sixth most common cancer (5.6% of total cancer cases) worldwide and remains the fourth leading cause of death (7.7% of total cancer deaths) according to Global Cancer Statistics 2020. 1 Despite the development of therapeutic modalities through numerous clinical and translational studies, this disease has a serious impact on patients and care providers.
The 2010 World Health Organization (WHO) classification system for gastric adenocarcinoma was revised based on the morphological features and includes 5 main categories: papillary, tubular, mucinous, poorly cohesive (PCC, diffuse-type, including signet ring cell carcinoma), and uncommon histological variants. 2 PCC consists of tiny clusters of small, uniform signet ring cells, is poorly differentiated, and lacks glands. It tends to spread submucosally, with early metastatic spread via transmural extension and lymphatic invasion, resulting in a poorer prognosis than other types of GC. 3 Despite the clinical severity of PCC, there are few studies comparing PCC and tubular adenocarcinoma to identify genes for prognosis prediction and therapeutic development.
The authors identified 30 cancer-promoting gene candidates through next-generation sequencing (NGS) analysis of GC tissues. 4 In this study, additional genes were discovered through comparing the transcriptomes of PCC and well-differentiated (WD; intestinal-type) GC tissues as well as GC cell lines; from these experiments, HIV-1 Tat Interactive Protein 2 (HTATIP2) was identified as a prognostic marker candidate.
HTATIP2 (also known as TIP30 or CC3) is a tumor-suppressor gene initially reported in 1997. The CC3 gene was first identified from a differential display analysis of messenger RNA (mRNA) from highly metastatic human variant small-cell lung carcinoma versus less metastatic classic small-cell lung carcinoma cell lines. 5 HTATIP2 is frequently downregulated in some cancer cells, including melanoma, breast cancer, neuroblastoma, glioblastoma, colon cancer, and hepatocellular carcinoma5-7. In one study using GC tissue, a decrease in HTATIP2 was reported to be related to tumor growth and metastasis. 8 However, the relevance of HTATIP2 function to cancer development in PCC-type GC remains unclear and warrants further research.
The purpose of this study was to compare the transcriptomes of PCC and WD tubular adenocarcinoma using GC tissues and cell lines and to evaluate the prognostic role of HTATIP2 identified here.
Materials and Methods
Western Blotting
The cells were lysed using RIPA (Radioimmunoprecipitation assay) buffer (Thermo Fisher Scientific, Inc. #89901) supplemented with protease inhibitors (Sigma, P8340) and phosphatase inhibitors (Thermo Fisher Scientific, Inc., 78420). The proteins (20 μg) were separated by 10% sodium dodecyl-sulfate polyacrylamide gel electrophoresis gel and then transferred onto PVDF membranes (iBlot 2PVDF Regular Stacks, Invitrogen, Thermo Fisher Scientific, Inc.). The membranes were blocked with 5% skim milk for 1 h at room temperature and then incubated overnight at 4°C with primary antibodies as follows: anti-E-cadherin (Cell Signaling #14472), anti-HTATIP2 (PA5-82247), anti-Snail (Abcam ab216347), anti-Slug (Abcam ab51772), and anti-Actin (sc-8432, Santa Cruz Biotechnology, Inc.). The membranes were then washed 3 times with TBST/Tween 20 buffer and incubated with a secondary antibody for 1 h at room temperature. The antibody-bound proteins were detected using the ECL system (Bio-Rad Laboratories, Inc.).
RNA Extraction and Next-Generation Sequencing
Previous studies on NGS using GC tissue have been referenced in a previous publication. 4 In brief, a total of 8 surgical samples were collected, including 5 samples of WD-GC and 3 samples of poorly differentiated signet ring cell GC (SRC-GC). Additionally, the tubular cell lines MKN74-1 and MKN74-2, as well as 2 PCC cell lines, KATO3 and SNU601, were obtained. RNA extraction was performed using a Total Purification Kit (Norgene Bioteck Corp) following the manufacturer's instructions. The mRNA sequencing was conducted using NGS by Macrogen, Inc., and we compared the expression profiles between clinically comparable samples using transcriptome resequencing data. Hierarchical clustering using Log2(FPKM + 1) values for each sample was conducted to group similar expression levels, and multidimensional scaling plots were generated in a binomial space using the 2 components that best explained the variance of the overall data for analysis of the degree of similarity between samples. Significant changes of 2-fold or greater with P < .05 were indicated in comparisons between WD and SRC, and commonly upregulated or downregulated genes between groups of tissues and cells were identified with a Venn diagram.
Ethics Statement
These studies were designed and conducted in accordance with the principles of the Helsinki Declaration (as revised in 2013). The studies were approved by the Institutional Review Board (GNUHIRB 2009-54). Consent was obtained from all patients prior to the experiment.
Validation of HTATIP2 by Tissue Microarray Analysis With Immunohistochemistry
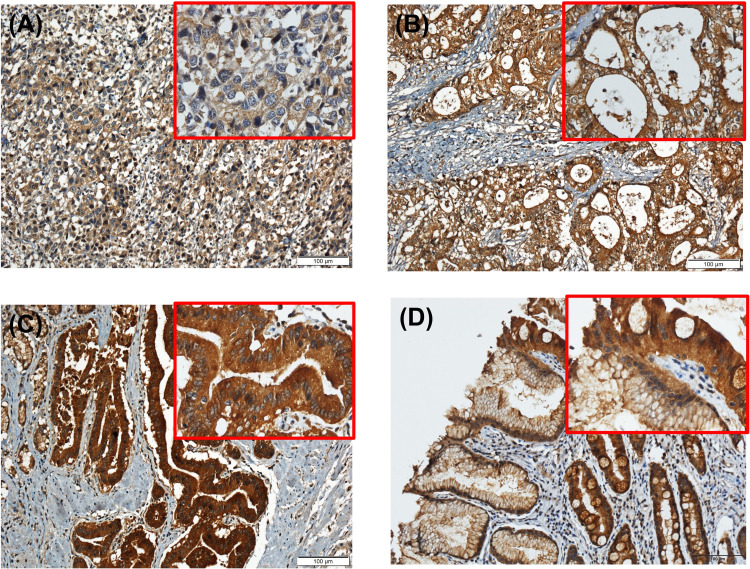
The quantification of Immunohistochemistry (IHC)-stained and immunostained cells was carried out as described in a previous study. 1 Specimens were fixed with 4% paraformaldehyde (F8775, Sigma), embedded, and sectioned to 5 μm thickness. The sections were incubated with a polyclonal anti-rabbit antibody against HTATIP2 (PA5-82247), followed by incubation with a secondary antibody (Dako Real Envision/HRP, K5007) for 15 min at room temperature. The intensity of immunochemical staining was divided into 3 categories: weak staining, moderate staining, and strong staining (see Figure 1). Based on the degree of immunohistochemical staining, we divided the samples into 2 groups. The criterion for the HTATIP2 overexpression (OE) was a sum of strong and moderate staining of 50% or more or a strong staining rate of 25% or more. Other samples were included in the HTATIP2 underexpression (UE) group.
Figure 1.
HTATIP2 expression in gastric carcinoma. The gastric carcinomas enrolled in this study showed (A) weak, (B) moderate, or (C) strong cytoplasmic staining on HTATIP2 immunohistochemical staining (×200) with magnification (×400). Cytoplasmic staining of HTATIP2 in gastric normal tissue also can be seen in (D) with magnification. Abbreviation: HTATIP2, HIV-1 Tat Interactive Protein 2.
Cell Lines and Transfection
Human gastric cell lines (MKN74-1, MKN74-2, SNU601, and KATO III) were obtained from the Cell Line Bank. GES-1 cells were obtained from the laboratory of Professor Nam Ki-Tae. The cells were cultured in RPMI 1640 medium (Gibco, Thermo Fisher Scientific, Inc.) supplemented with 10% heat-inactivated fetal bovine serum (FBS, Gibco, Thermo Fisher Scientific, Inc.) and penicillin (100 U/ml, Thermo Fisher Scientific, Inc.) and were incubated at 37°C in a humidified chamber containing 5% CO2. The cells were transfected with 2 μg of the pCMV6 control vector (CAT#: PS100001) and the pCMV6-HTATIP2-FLAG-DDK-tagged expression construct vector (CAT#: RC212332) using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), following the manufacturer's instructions. Refer to the previous study materials and method for detailed experimental methods of western blotting, proliferation assay, Transwell migration, invasion assay, and assay of the epithelial-mesenchymal transition markers. 9
Transwell Cell Migration and Invasion Assay
Refer to the previous study for detailed experimental methods of western blotting, proliferation assay, Transwell migration, invasion assay, and assay of epithelial-mesenchymal transition markers. 8 Transwell migration and invasion assay chambers with 8μm pores were purchased from Corning. The cells were harvested and resuspended in serum-free RPMI medium at a concentration of 1 × 105 cells/mL for MKN74 and KATO III cells and then seeded into the upper chamber of a 24-well plate. The lower chambers were filled with RPMI containing 10% FBS. The cells were then incubated for 2 days. At the end of the experiment, the cells that had migrated to the underside of the Transwell membrane were fixed with a 4% formalin solution, stained with DAPI solution, and counted under an inverted light microscope. Matrigel was used in the invasion assay, and Transwell chambers with 8μm pores and Transwell membranes precoated with Matrigel (2.2 mg/mL, cat. no. 356234; BD Bioscience) were purchased from BD Bioscience. Microscope images of 3 marked locations for migration and invasion cells were counted at the indicated distances from the wound edge using NIH ImageJ software. The data are presented as the mean ± standard deviation (SD) for 3 independent experiments (original magnification, ×40).
Statistical Analysis
Statistical analysis was performed using IBM® SPSS® Statistics version 27 software (IBM Corp.). Data are presented as the mean ± SD. The significance of the differences was determined using the chi-square test. Student's t-test and Kaplan-Meier's (KM) method were used to analyze patient outcomes and overall survival (OS). Statistical tests were 2-sided or 1-sided. P < .05 was considered to indicate a statistically significant difference. GRAPHPAD Prism 7.0 software (GraphPad Software, Inc.) was used for in vitro analysis.
Kaplan-Meier Survival Curve Analysis
A KM OS analysis (KM plot, https://kmplot.com/analysis/index.php?p=service& cancer=gastric) was performed on the larger KM plot GC dataset using the HTATIP2 gene probe set (209448_at) on the available Affymetrix arrays. The line color in the figure indicates the gene expression (red: high, black: low). KM survival plots with the number indicated hazard ratio (HR) with 95% confidence intervals (CI) and log-rank P value were obtained on the webpage. P < .05 was considered statistically significant.
PrognoScan
A database named “PrognoScan” has been developed to make the most of public resources. A large collection of published cancer microarray datasets with clinical annotations is a very useful tool for evaluating the biological relationship between gene expression and prognosis. PrognoScan uses a minimal P value approach to group patients for survival analysis. It finds optimal cut points for continuous gene expression measurements without prior biological knowledge or assumptions and, as a result, enables systematic meta-analysis on multiple datasets, making it useful to several researchers. 10 By using the online platform PrognoScan (http://www.prognoscan.org), the correlation between gene expression and the prognosis of patients with cancer was investigated. The correlation between HTATIP2 expression and survival in various carcinomas was analyzed by PrognoScan. The minimum P value and HR with 95% CI were calculated automatically according to the mRNA level (high or low expression).
Results
Comparison of the Transcriptome Between PCC-GC and WD-GC
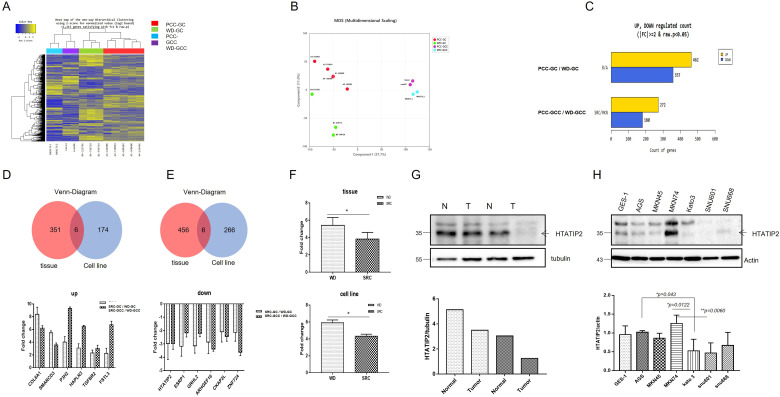
We analyzed the NGS data (3 PCC-GC and 5 WD-GC samples) obtained in a previous study with data for the newly analyzed GC cell lines PCC-GCC (KATO III, SNU601) and WD-GCC (MKN 74-1, 74-2). The genes of the 4 groups of PCC-GCC (blue), WD-GCC (purple), PCC-GC (red), and WD-GC (green) showed clear differences in the 2-dimensional heatmap for hierarchical clustering (Figure 2A). Four groupings were confirmed in the multidimensional scaling analysis, suggesting that the genetic characteristics of cancer tissues and cancer cell lines are different (Figure 2B).
Figure 2.
Comparison of the transcriptome between PCC-GC and WD-GC tissues and cell lines. (A) Heatmap for hierarchical clustering. (B) Multidimensional scaling plot. (C) Number of significantly upregulated and downregulated genes according to fold change and P value. Venn diagram showing the number of upregulated (D) and downregulated (E) genes common to tissues and cell lines. (F) Expression levels of HTATIP2 in tissues and cell lines. (G) Western blot of HTATIP2 in normal gastric tissue and cancer tissue. (H) Western blot of HTATIP2 in cell lines (normal gastric; GES1 & WD; AGS, MKN45, MKN74 vs PCC; Kato III, SNU601, SNU668). Abbreviations: GC, gastric cancer; HTATIP2, HIV-1 Tat Interactive Protein 2; PCC, poorly cohesive carcinoma; WD, well-differentiated.
In the comparison of GC tissues, 462 genes showed 2-fold increases in expression in WD-GC compared to PCC-GC, and 357 genes showed 2-fold increases in expression in PCC-GC versus WD-GC. In the comparison of GC cell lines, 272 genes showed 2-fold increases in expression in in WD-GCC compared to PCC-GCC, and 180 genes showed 2-fold increases in expression in PCC-GC versus WD-GC (Figure 2C).
In PCC, the number of genes that with a fold change greater than 2 in WD was 357 in tissue and 180 in cell lines, and 6 genes (COL6A1, SMARCD3, P3H2, HAPLN3, TGFBR2, and FSTL3) were commonly observed (Figure 2D). In addition, there were 462 genes in tissues and 272 in cell lines that decreased more than 2-fold in PCC versus WD, and 6 genes (HTATIP2, ESRP1, GRHL2, ARHGEF16, CKAP2L, and ZNF724) were commonly observed (Figure 2E).
HTATIP2 expression was significantly reduced in PCC compared to WD in GC tissues (P = .047) and showed a significant decrease in the same pattern in cell lines (P = .043) (Figure 2F). Western blot analysis confirmed that the expression of HTATIP2 was lower in GC tissue than in normal gastric tissue in same patients (Figure 2G). In cell line, the expression of HTATIP2 was lower in PCC GC cell lines (KATO III, SNU-601, and SNU-668) with poor differentiation than in normal gastric cell line (GES-1) and GC cell lines with differentiation (AGS, MKN-45, and MKN-74) (Figure 2H).
HTATIP2 Expression is Reduced in PCC Compared to WD and This Phenotype Indicates a Poor Prognosis in Patients
The results of the immunohistochemical analyses of TMAs from the HTATIP2-UE (HTATIP2-UE, score 1) and HTATIP2-OE (HTATIP2-OE, score 2&3) groups of patients were compared (Table 1). In the HTATIP2-OE group, the proportion of patients with early GC (T1) was 76.8%, which was significantly higher than that of patients with AGC (64.6%) (P = .024); the OE group also had a significantly lower rate of lymph node (LN) metastasis (P = .008) and a significantly lower proportion of TNM (Tumour, Lympn node, Metastasis) stage patients (P = .017). However, there was no difference between the patient groups according to age, sex, Lauren classification, and WHO classification, and the number of cancer-related deaths was low in the HTATIP2-OE group, but there was no significant difference between groups; there was no difference in the recurrence rate (Table 1).
Table 1.
Comparison of the Clinicopathological Features of the HTATIP2-UE and HTATIP2-OE Groups According to Immunohistochemistry of Tissues.
| Level of HTATIP2 expression | Total | P value | ||
|---|---|---|---|---|
| HTATIP2-UE (0) | HTATIP2-OE (1&2) | |||
| Age | 63.0 ± 11.7 | 62.92 ± 10.8 | .938 | |
| Sex | .639 | |||
| Male | 55 (61.1%) | 137 (64.9%) | 192 | |
| Female | 35 (38.9%) | 74 (35.1%) | 109 | |
| Lauren classification | .080 | |||
| Intestinal type | 53 (58.9%) | 110 (52.1%) | 163 | |
| Diffuse type | 17 (18.9%) | 41 (19.4%) | 58 | |
| Mixed type | 6 (6.7%) | 5 (2.4%) | 11 | |
| Unknown | 14 (15.6%) | 55 (26.1%) | 69 | |
| WHO classification | .050 | |||
| Differentiated | 41 (24.6%) | 126 (75.4%) | 167 | |
| Undifferentiated | 44 (35.8%) | 79 (64.2%) | 123 | |
| Tumor invasion | .024 | |||
| EGC (T1) | 33 (23.2%) | 109 (76.8%) | 142 | |
| AGC (T2-T4) | 57 (35.4%) | 104 (64.6%) | 161 | |
| LN metastasis | .008 | |||
| No | 42 (23.7%) | 135 (76.3%) | 177 | |
| Yes (≥ 1) | 48 (38.1%) | 78 (61.9%) | 126 | |
| TNM stage | .017 | |||
| I | 37 (23.1%) | 123 (76.9%) | 160 | |
| II | 18 (32.1%) | 38 (67.9%) | 56 | |
| III-IV | 35 (40.2%) | 52 (59.8%) | 87 | |
| Chemotherapy | .151 | |||
| No | 27 (24.3%) | 84 (75.7%) | 131 | |
| Yes | 63 (32.8%) | 129 (67.2%) | 192 | |
| Cancer-related death | .089 | |||
| No | 65 (27.2%) | 174 (72.8%) | 239 | |
| Yes | 25 (39.1%) | 39 (60.9%) | 64 | |
| Recurrence | .118 | |||
| No | 61 (27.2%) | 163 (72.8%) | 224 | |
| Yes | 29 (36.7%) | 50 (63.3%) | 79 | |
Abbreviations: AGC, advanced gastric cancer; EGC, early gastric cancer; HTATIP2, HIV-1 Tat Interactive Protein 2; LN, lymph node; TNM, Tumour, Lympn node, Metastasis; OE, overexpressed; UE, underexpressed; WHO, World Health Organization.
The significance of the differences was determined using the chi-square test.
The High HTATIP2 Expression Group Showed Better Overall Survival According to Kaplan-Meier Plotter Data
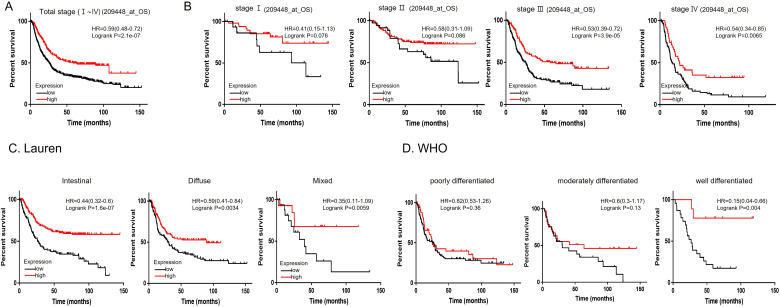
The OS analysis of the KM plotter dataset (209448) including data on patients with GC demonstrated that patients with high HTATIP2 gene expression had a higher OS rate than those with low expression when patients were not grouped by TNM stage (HR 0.59, P = 2.1e-07) (Figure 3A). There were no significant differences in OS between the low and high gene expression groups for the patients with TNM stages I and II. However, for patients with TNM stages III and IV, the higher the expression level of HTATIP2 was, the higher the survival rate was (***P < .001, P = 3.9e-05, **P < .01, P = .0065, Figure 3B).
Figure 3.
Kaplan-Meier survival curves of (A) OS in all patients and patients in the following groups: (B) stage Ⅰ∼Ⅳ, (C) intestinal, diffuse, and mixed according to Lauren classification, and (D) poorly, moderately and well-differentiated according to WHO classification. Abbreviations: OS, overall survival; WHO, World Health Organization.
According the Lauren classification (intestinal, diffuse, and mixed type), it was confirmed that the higher the expression level of HTATIP2 was, the higher the survival rate was (intestinal, HR = 0.44, CI 0.32-0.6, I = 1.6e-07; diffuse, HR = 0.59, CI 0.41-0.84, P = .0034; mixed, HR = 0.39, CI 0.11-1.09, P = .0059) (Figure 3C). According to the WHO classification, the well-differentiated group showed a higher expression level of HTATIP2 and a higher OS rate than the low-differentiated group (HR = 0.15, CI 0.04-0.66, P = .0004), but OS rate was not significantly different in the poorly differentiated (P = .36) and moderately differentiated (P = .13) subgroups (Figure 3D).
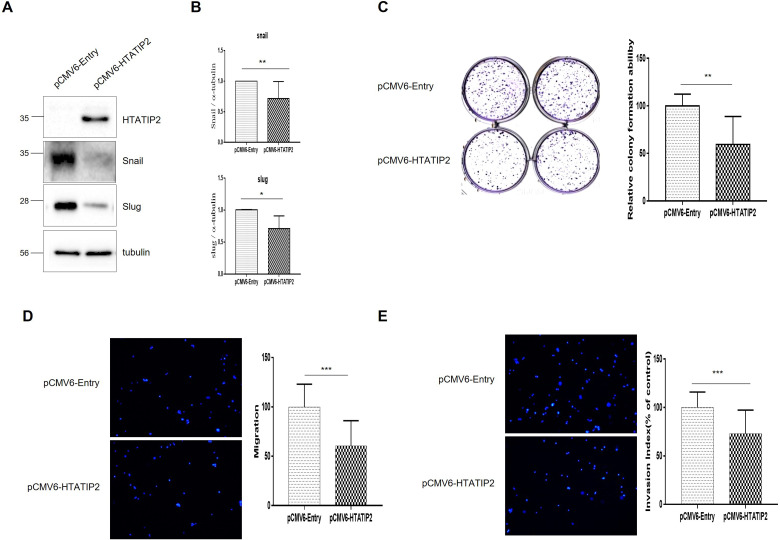
HTATIP2-Overexpressing KATOII Cell Lines Showed Decreased Migration, Invasion, and EMT
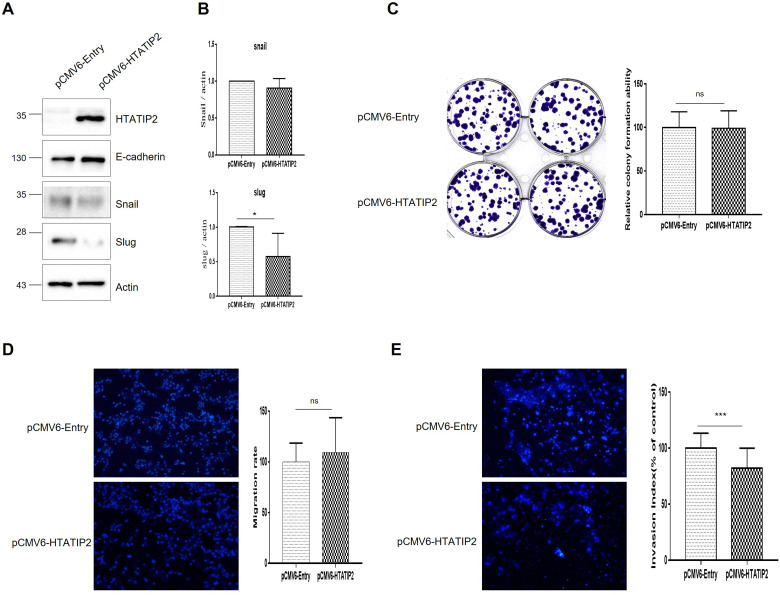
HTATIP2 was overexpressed in KATO III (HTATIP2-OE GC cell line), which was identified in pCMV6-HTATIP2 GCC compared to pCMV6-Entry GCC (Figure 4A). The epithelial-mesenchymal transition (EMT) markers snail (P = .0249) and slug (P = .0403) were significantly downregulated after OE (Figure 4B). In the colony formation assay, pCMV6-HTATIP2 GCC showed a significant decrease compared to pCMV6-Entry GCC (Figure 4C, P = .0015). In the Transwell migration assay, pCMV6-HTATIP2 GCC was significantly decreased compared to pCMV6-Entry GCC (Figure 4D, P < .0001) and was also significantly decreased in the invasion assay (Figure 4E, P < .0001).
Figure 4.
Functional study of HTATIP2-Overexpressing KATOIII. (A) Observation of changes in snail and slug, markers of EMT in HTATIP2-overexpressing Kato III cells using western blotting, (B) Data graphs of snail and slug, which are markers of EMT, (C) Colony formation assay (CFU), (D) Transwell migration assay, (E) Transwell invasion assay. Abbreviations: EMT, epithelial-mesenchymal transition; HTATIP2, HIV-1 Tat Interactive Protein 2.
HTATIP2-Overexpressing MKN74 Cell Lines Showed Decreased Invasion and Slug Expression
The expression of HTATIP2 was increased in the MKN74 cell line, which had a relatively high expression of HTATIP2 compared to the KATOIII cell line. HTATIP2-OE was confirmed in pCMV6-HTATIP2 GCC compared to pCMV6-Entry GCC using western blotting (Figure 5A). After OE, the EMT marker slug was significantly downregulated (P = .0135) but snail was not (P = .1428) (Figure 5B). In the colony formation assay, pCMV6-HTATIP2 GCC showed no significant difference compared to pCMV6-Entry GCC (Figure 5C). There was no significant difference in migration rate between pCMV6-HTATIP2 GCC and pCMV6-Entry GCC (Figure 5D), but the invasion rate was significantly decreased in pCMV6-HTATIP2 GCC versus pCMV6-Entry GCC (Figure 5E, P < .0001).
Figure 5.
Functional study of HTATIP2-Overexpressing MKN-74 (A) Observation of changes in snail and slug, markers of EMT, using western blot after overexpression of HTATIP2 in MKN74 cells, (B) Data graphs of snail and slug, which are markers of EMT among the data in (A). (C) Colony formation assay, (D) Transwell migration assay, (E) Transwell invasion assay. Abbreviations: EMT, epithelial-mesenchymal transition;HTATIP2, HIV-1 Tat Interactive Protein 2.
The HTATIP2 Overexpression Group had Improved Survival Rates in Various Carcinomas
The authors analyzed the expression of HTATIP2 in various carcinomas and the survival rate of patients in different expression groups using PrognoScan (Supplemental Figure 1 and Table 1). In bladder cancer, the HTATIP2-overexpressing group showed a significantly higher OS rate than the low HTATIP2 expression group (corrected P value, cor P = .041366, Supplemental Figure 1A). In acute myeloid leukemia, the HTATIP2-overexpressing group showed a significantly higher relapse-free survival rate than the low HTATIP2 group (cor P = .029298, Supplemental Figure 1B). In breast cancer, the HTATIP2-overexpressing group showed significantly higher relapse-free survival (cor P = .048505, Figure 6C-1,2) and distant metastasis-free survival (cor P = .047994, 0.007453, Supplement Figure 1C-3, 4) than the low HTATIP2 group. In colorectal cancer, the HTATIP2-overexpressing group showed significantly higher disease-free survival (cor P = .01244, Supplemental Figure 1D-1) and OS rates (cor P = .023681, .045703, Supplemental Figure 1D-2, 3) than the low HTATIP2 group. In ovarian cancer, the HTATIP2-overexpressing group showed a significantly higher OS rate than the low HTATIP2 group, and there was also a significant difference in the Cox proportional hazard model (P = .000229, .001941, Supplemental Figure 1E, F).
Discussion
This study aimed to facilitate the development of target agents and stomach cancer biomarkers by discovering genes that are upregulated and downregulated in both cancer tissues and cell lines. Among the several markers discovered, HTATIP2 was confirmed to be significantly downregulated in the signet ring cell subtype compared to the differentiated subtype in GC tissue and cell lines. Through IHC analysis and functional study of GC tissue, patients with HTATIP2-OE showed a good prognosis and a good survival rate. In addition, in a study of an HTATIP2-overexpressing GC cell line conducted to confirm the mechanism, EMT-related features such as migration and invasion were reduced. Therefore, HTATIP2 might be a good prognostic marker and a candidate target for GC treatment.
In a previous study, the authors conducted a study comparing WD-GC and PCC-GC subtypes using GC tissue. 4 Through GO analysis, it was confirmed that the genes constituting the adhesion, vascular development, and cell-to-cell function components showed a very large difference between the 2 subtypes. In addition, 30 cancer driver gene candidates were discovered through cancer variant analysis, and the NUDC-positive group showed significantly better survival than the NUDC-negative group via variant analysis. This study is a follow-up study that includes not only GC tissue data but also GC cell line NGS data. We used WD-GC-derived MKN74 cells and PCC-GC-derived KATO III and SNU601 cells to identify genes that increased and decreased more than 2-fold in the PCC-GC subtype compared to the WD-GC subtype in tissue and cell lines. We confirmed that 6 genes (HTATIP2, ESRP1, GRHL2, ARHGEF16, CKAP2L, and ZNF724) are commonly decreased in PCC. Epithelial splicing regulatory protein (ESPR1) is an epithelial cell-type-specific splicing regulator that regulates EMT-related isoforms and regulates the splicing of CD44, CTNND1, and ENAH. 11 Grainyhead-like transcription factor 2 (GRHL2) encodes a protein that is a cancer suppressor transcription factor that can act as a homodimer or as a heterodimer with either GRHL1 or GRHL3. Recently, Liang et al 12 found that miR-1290 in extracellular vesicles secreted from GC cells contributed to immune escape through the Grhl2/ZEB1/PD-L1 axis in a mouse model.
Human HTATIP2 is a gene that is ubiquitous in normal tissue, and its biological cellular functions are unknown (2014 T). HTATIP2 is thought to be related to cell death, growth, metastasis, angiogenesis, DNA repair, and regulation of tumor cell metabolism of HTATIP2. 9 It is known to play a role as a tumor suppressor in cancer. Significant downregulation of HTATIP2 was reported in lung cancer, and this phenotype was associated with poor differentiation, poor prognosis, and a high rate of metastatic progression. 13 In one study, HTATIP2-mediated EGFR augmentation was reported as a mechanism of the occurrence of lung adenomas and adenocarcinoma. 14 It has also been reported that HTATIP2-UE is associated with axillary LN metastasis and vascular invasion. In addition, it was reported that the deletion of the HTATIP2 gene in mice caused ductal hyperplasia and extensive mammary hyperplasia in mouse mammary glands and eventually caused rapid neoplastic transformation. 15 It was reported that hepatocellular carcinoma occurred naturally in a higher proportion of HTATIP2-/-mice than HTATIP2 wild-type mice, and hepatocellular carcinoma generation also increased in nude mice upon HTATIP2 deletion. 16 In addition, it is known that OE of HTATIP2 interferes with the growth and lung metastasis of hepatocellular carcinoma in nude mice. 17
In one study using gastric carcinoma tissue, a decrease in HTATIP2 was reported to be related to tumor growth and metastasis. 8 They reported that HTATIP2-mediated cell cycle modification by downregulating cyclin D1, Bcl-2, and Bcl-xL by OE of HTATIP2 and upregulation of p27, Bax, p53, caspase 3 and caspase 9. 8
In this study, the PCC-GC subtype had low HTATIP2 expression, which explains the high rates of LN metastasis and hematogenic metastasis in the WD-GC subtype. When HTATIP2 was overexpressed in KATO III cells, the number of CFUs decreased, the migration rate decreased, and EMT markers such as snail and slug were downregulated. However, in MKN74 cells, when HTATIP2 was overexpressed, the invasion rate was decreased, but the migration rate was unaffected; slug expression decreased, but snail expression was not affected. The difference in functional results for these cell lines may be due to the difference in the expression level of HTATIP2 between the 2 cell lines. Since HTATIP2 expression is lower in KATO III cells, this difference may explained by the tumor-suppressive effect of increased HTATIP2 expression. However, since MKN74 has high expression of HTATIP2, it is expected that increased HTATIP2 expression played a limited role.
In the analysis of patients with GC based on HTATIP2 expression in tissues, it was shown that GC showed decreasing HTATIP2 expression from the early stage to the advanced stage. This phenomenon may be related to the mechanism by which HTATIP2 increases LN metastasis and hematogenous metastasis, considering its role as a tumor suppressor. There was no difference in the diffuse and intestinal subtypes in the Lauren classification, and the decrease in HTATIP2 was reduced in the diffuse subtype, but the diffuse subtype does not necessarily mean that HTATIP2 is reduced. In addition, this study revealed that an increase in HTATIP2 expression suppresses EMT. However, although there was a difference in this TMA cohort, it was not significant. This was due to the limited number of samples with HTATIP2 expression data. We found differences in OS in the analysis using a large-scale cohort using KM plotter, a public database. In addition, using the results of PrognoScan data analysis, it was confirmed that the survival rate was good when HTATIP2 expression was increased in various cancers, such as bladder, blood, breast, color, and ovarian cancers.
The main limitation of this study is the small number of tissues (8 samples) and cancer cell lines (3 samples). We could not analyze the effects of gene knockdown. However, this study is the first to study the transcriptomes of PCC-GCs and WD-GCs simultaneously in tissue and cancer cell lines and the first to conduct a functional study of HTATIP2 based on phenotypic changes in GC.
In conclusion, HTATIP2 might be a good prognostic marker and a candidate therapeutic target for GC. Further research needs to be performed to develop a targeting agent.
Supplemental Material
Supplemental material, sj-docx-1-tct-10.1177_15330338231187254 for HTATIP2 Overexpression was Associated With a Good Prognosis in Gastric Cancer by Sun Yi Park, PhD, Ji-Ho Park, MD, Jung Wook Yang, MD, Eun-Jung Jung, MD, Young-Tae Ju, MD, Chi-Young Jeong, MD, Ju-Yeon Kim, MD, Taejin Park, MD, Miyeong Park, MD, Young-Joon Lee, MD, and Sang-Ho Jeong, MD in Technology in Cancer Research & Treatment
Abbreviations
- AGC
advanced gastric cancer
- EGC
early gastric cancer
- EMT
epithelial-mesenchymal transition
- ESPR1
epithelial splicing regulatory protein
- FBS
fetal bovine serum
- GC
gastric cancer
- GES-1
gastric cell line
- GRHL2
Grainyhead-like transcription factor 2
- HR
hazard ratio
- HTATIP2
HIV-1 Tat Interactive Protein 2
- LN
lymph node
- MDS
multidimensional scaling
- NGS
next-generation sequencing
- OE
overexpression
- OS
overall survival
- PCC
poorly cohesive carcinoma
- SRC-GC
signet ring cell gastric cancer
- UE
underexpression
- WD
well-differentiated.
Footnotes
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethics Approval: The study was approved by the Institutional Review Board of Gyeongsang National University Hospital (GNUHIRB 2009-54).
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by National Research Foundation of Korea (NRF) grant funded by the Republic of Korean government (2020R1F1A1074077).
ORCID iDs: Chi-Young Jeong https://orcid.org/0000-0001-9061-6236
Sang-Ho Jeong https://orcid.org/0000-0001-9061-6236
Supplemental Material: Supplemental material for this article is available online.
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209-249. doi: 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2.Bosman FT, Carneiro F, Hruban RH, Theise ND. WHO classification of tumours of the digestive system. World Health Organization; 2010. [Google Scholar]
- 3.Voron T, Messager M, Duhamel A, et al. Is signet-ring cell carcinoma a specific entity among gastric cancers? Gastric Cancer. 2016;19(4):1027-1040. doi: 10.1007/s10120-015-0564-2 [DOI] [PubMed] [Google Scholar]
- 4.Jeong SH, Park M, Park SY, et al. Transcriptome analysis and the prognostic role of NUDC in diffuse and intestinal gastric cancer. Technol Cancer Res Treat. 2021;20:15330338211019501. doi: 10.1177/15330338211019501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shtivelman E. A link between metastasis and resistance to apoptosis of variant small cell lung carcinoma. Oncogene. 1997;14(18):2167-2173. doi: 10.1038/sj.onc.1201059 [DOI] [PubMed] [Google Scholar]
- 6.Ito M, Jiang C, Krumm K, et al. TIP30 deficiency increases susceptibility to tumorigenesis. Cancer Res. 2003;63(24):8763-8767. [PubMed] [Google Scholar]
- 7.NicAmhlaoibh R, Shtivelman E. Metastasis suppressor CC3 inhibits angiogenic properties of tumor cells in vitro. Oncogene. 2001;20(2):270-275. doi: 10.1038/sj.onc.1204075 [DOI] [PubMed] [Google Scholar]
- 8.Li X, Zhang Y, Cao S, et al. Reduction of TIP30 correlates with poor prognosis of gastric cancer patients and its restoration drastically inhibits tumor growth and metastasis. Int J Cancer. 2009;124(3):713-721. doi: 10.1002/ijc.23967 [DOI] [PubMed] [Google Scholar]
- 9.Park SY, Lee YJ, Park J, et al. PRDX4 overexpression is associated with poor prognosis in gastric cancer. Oncol Lett. 2020;19(5):3522-3530. doi: 10.3892/ol.2020.11468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mizuno H, Kitada K, Nakai K, Sarai A. Prognoscan: a new database for meta-analysis of the prognostic value of genes. BMC Med Genomics. 2009;2:18. doi: 10.1186/1755-8794-2-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell. 2009;33(5):591-601. doi: 10.1016/j.molcel.2009.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang Y, Liu Y, Zhang Q, Zhang H, Du J. Tumor-derived extracellular vesicles containing microRNA-1290 promote immune escape of cancer cells through the Grhl2/ZEB1/PD-L1 axis in gastric cancer. Transl Res. 2021;231:102-112. doi: 10.1016/j.trsl.2020.12.003 [DOI] [PubMed] [Google Scholar]
- 13.Tong X, Li K, Luo Z, et al. Decreased TIP30 expression promotes tumor metastasis in lung cancer. Am J Pathol. 2009;174(5):1931-1939. doi: 10.2353/ajpath.2009.080846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359(13):1367-1380. doi: 10.1056/NEJMra0802714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pecha J, Ankrapp D, Jiang C, et al. Deletion of Tip30 leads to rapid immortalization of murine mammary epithelial cells and ductal hyperplasia in the mammary gland. Oncogene. 2007;26(53):7423-7431. doi: 10.1038/sj.onc.1210548 [DOI] [PubMed] [Google Scholar]
- 16.Jiang C, Pecha J, Hoshino I, Ankrapp D, Xiao H. TIP30 Mutant derived from hepatocellular carcinoma specimens promotes growth of HepG2 cells through up-regulation of N-cadherin. Cancer Res. 2007;67(8):3574-3582. doi: 10.1158/0008-5472.Can-06-0831 [DOI] [PubMed] [Google Scholar]
- 17.Zhao J, Lu B, Xu H, et al. Thirty-kilodalton Tat-interacting protein suppresses tumor metastasis by inhibition of osteopontin transcription in human hepatocellular carcinoma. Hepatology. 2008;48(1):265-275. doi: 10.1002/hep.22280 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-tct-10.1177_15330338231187254 for HTATIP2 Overexpression was Associated With a Good Prognosis in Gastric Cancer by Sun Yi Park, PhD, Ji-Ho Park, MD, Jung Wook Yang, MD, Eun-Jung Jung, MD, Young-Tae Ju, MD, Chi-Young Jeong, MD, Ju-Yeon Kim, MD, Taejin Park, MD, Miyeong Park, MD, Young-Joon Lee, MD, and Sang-Ho Jeong, MD in Technology in Cancer Research & Treatment