Abstract
We provide evidence that Prp4p kinase activity is required for pre-mRNA splicing in vivo and show that loss of activity impairs G1–S and G2–M progression in the cell cycle. Prp4p interacts genetically with the non-SR (serine/arginine) splicing factors Prp1p and Prp5p. Bacterially produced Prp1p is phosphorylated by Prp4p in vitro. Prp4p and Prp1p also interact in the yeast two-hybrid system. In vivo labelling studies using a strain with a mutant allele of the prp4 gene in the genetic background indicate a change in phosphorylation of the Prp1p protein. These results are consistent with the notion that Prp4p kinase is involved in the control of the formation of active spliceosomes, targeting non-SR splicing factors.
INTRODUCTION
The recognition and removal of introns in eukaryotes is mediated by a highly complex and dynamic machinery called the spliceosome (Will and Lührmann, 1997). The basic RNA and protein components of this machinery have been well conserved throughout evolution; however, we found distinct differences when we compared the splicing factors of fission yeast Schizosaccharomyces pombe with those of Homo sapiens and the budding yeast Saccharomyces cerevisiae (Käufer and Potashkin, 2000).
The modification of some of the proteins by phosphorylation appears to play an important role in the splicing process. It has been shown that kinase activity in mammals is essential for pre-mRNA splicing in vitro (Tazi et al., 1993; Mermoud et al., 1994). In mammals, several protein kinases have been identified that specifically phosphorylate the RS domain of the SR-splicing factors in vitro (Gui et al., 1994; Colwill et al., 1996; Rossi et al., 1996). SR proteins consisting of one or two RNA-binding domains (RBDs) followed by an RS domain are involved in constitutive and differential pre-mRNA splicing (Manley and Tacke, 1996). The homologue of human SR protein kinase 1 (SRPK1) in fission yeast, Dsk1p, also phosphorylates SR proteins including Srp1p and Srp2p of fission yeast, the first typical SR proteins discovered in a unicellular organism (Tang et al., 1998, 2000; Groß et al., 1998; Lützelberger et al., 1999).
In fission yeast and mammals, we identified the protein kinase Prp4p (Groß et al., 1997). Budding yeast does not contain a kinase with sequence similarity to Prp4p (Käufer and Potashkin, 2000). Prp4p kinase is essential for growth, and the temperature sensitive allele prp4-73 shows a defect in pre-mRNA splicing. A strain containing this allele stops growing at the restrictive temperature, which correlates with the accumulation of pre-mRNA and the disappearance of mRNA (Rosenberg et al., 1991; Alahari et al., 1993; Groß et al., 1997). An extragenic suppressor, Spp42p, of this defect encodes a homologue of the mammalian and budding yeast splicing factor p220/Prp8p (Schmidt et al., 1999). Prp8p is a unique and highly conserved protein associated with U5 snRNP and is thought to be a major player in the switch from inactive to active spliceosome (Murray and Jarrell, 1999; Käufer and Potashkin, 2000).
We show in this report that Prp4p kinase activity is required for splicing in vivo whereas loss of activity leads not only, as expected, to the accumulation of pre-mRNA, but also to a cell cycle arrest phenotype, suggesting that kinase activity is essential for the transition of G1 to S and of G2 to M phase. Furthermore, prp4-73 is synthetically lethal with prp1-4 and prp5-1, indicating a functional relationship of Prp4p kinase with two additional highly conserved splicing factors. Bacterially produced Prp1p is phosphorylated by Prp4p in vitro. Prp1p and Prp4p also interact in the yeast two-hybrid system. We also provide evidence that the mutant allele prp4-73 causes a change in the phosphorylation of Prp1p in vivo. These data are consistent with our suggestion that Prp4p may play the role of a signalling kinase, controlling the formation of active spliceosomes by activating non-SR components.
RESULTS AND DISCUSSION
Genetic interactions of Prp4p kinase with splicing factors
All fourteen temperature sensitive strains prp1-prp14 (pre-mRNA processing) accumulate pre-mRNA at the restrictive temperature (Potashkin et al., 1989; Rosenberg et al., 1991; Urushiyama et al., 1996). In addition, the strains containing the temperature-sensitive alleles prp2-1, prp5-1, prp6-1 and prp8-1/cdc28–1 arrest at the restrictive temperature with a 2C DNA content (Lundgren et al., 1996; Potashkin et al., 1998; Beales et al., 2000).
In order to screen for genetic interactions with prp4-73, we crossed prp4-73 with each of the prp strains and performed tetrad analyses. Spores were grown at the permissive temperature of 25°C. This analysis revealed that the double mutants prp1-4 prp4-73 and prp5-1 prp4-73 are not viable at the permissive temperature of 25°C (Figure 1). Whenever the prp4-73 allele segregates with one of the indicated mutant alleles the spores do not grow out. This phenotype, called synthetic lethality, indicates a functional interaction between Prp4p protein kinase and these components.
Fig. 1. Synthetic lethality of prp4-73 with non-SR splicing components. Tetrad analyses of crosses of the indicated mutant strains, at 25°C. Four growing colonies, parental ditype, PD; two growing colonies, nonparental ditype, NPD; three growing colonies, tetratype,TT.
Prp1p, a tetratricopeptide protein containing 19 repeats of a 34 amino acid motif (TPR) with an Mr of ∼100 kDa, has been identified and characterized by Urushiyama et al. (1997). Prp1 shows a polymorphic phenotype at the restrictive temperature. The investigation of several alleles, such as prp1-1, prp1-4 and zer1, indicates that Prp1p is involved in pre-mRNA splicing, poly(A)+ transport and cell cycle progression (Urushiyama et al., 1997). The budding yeast homologue, Prp6p, was found to associate with snRNP U4/U6–U5 (Galisson and Legrain, 1993). Recently, the human homologue of Prp1p/Prp6p has been identified, tightly associated with the U5 snRNP (Makarov et al., 2000). It is interesting to note here that the fission yeast Prp1p and human Prp1p share 66% similar amino acids, whereas fission yeast Prp1p and budding yeast Prp6p share 46% similar amino acids (Käufer and Potashkin, 2000).
Prp5p is a protein containing seven WD motifs. Mass spectrometry of proteins isolated from in vitro assembled spliceosomes using HeLa cell extracts identified the Prp5p homologue as a spliceosomal protein (Neubauer et al., 1998). There are some hints that Prp5p might also be a U5 snRNP protein. The protein is highly conserved between mammals, fission yeast and budding yeast (Käufer and Potashkin, 2000).
Prp4p kinase and Prp1p mutations arrest the cell cycle in G1 and G2
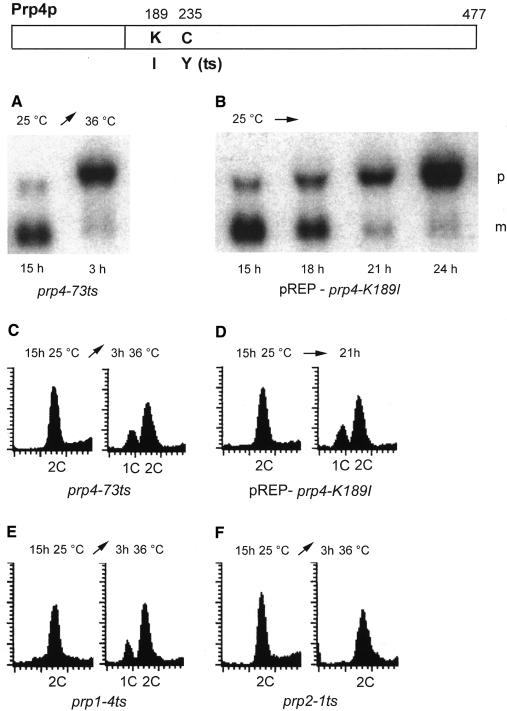
Next, we investigated in vivo the consequences of a loss-of-function mutation in the kinase domain of Prp4p when overexpressed in a prp4 background. For this purpose a strain containing the mutant allele prp4-K189I was constructed. We have replaced lysine in kinase subdomain II at position 189 with isoleucine (Figure 2). This lysine is involved in positioning ATP near the active site, to mediate proper catalysis, and has been found in many kinases invariable for function in vivo and in vitro. The prp4-73 allele harbours a mutation in kinase subdomain IV. Instead of a cysteine, a tyrosine residue is found at position 235. This subdomain is also involved in ATP binding (Taylor and Radzio-Andzelm, 1994). A strain containing prp4-73 ceases growing at the restrictive temperature within 3 h, which correlates with the accumulation of pre-mRNA and the disappearance of mRNA (Figure 2A). The mutant allele prp4-K189I, driven by the thiamine-repressible nmt1 promoter, was transformed in a prp4 background. The transformed strain grows on plates supplemented with thiamine, but does not grow on plates without thiamine (results not shown). For further analysis, we inoculated the strain pREP-prp4-K189I in medium without thiamine. Cells overexpressing prp4-K189I accumulate pre-mRNA at 25°C (Figure 2B). The accumulation of pre-mRNA also coincides with ceasing growth, which comes to a halt after 21 h (results not shown). In this approach we used the constitutively expressed tfIId gene as a probe; however, all other intron-containing genes tested also accumulate pre-mRNA.
Fig. 2. Response of cells overexpressing mutant Prp4p K189I. Northern analysis: (A) prp4-73ts strain 15 h after growth at the permissive temperature (25°C) and after a shift to the restrictive temperature (36°C) for 3 h; (B) the strain overexpressing the mutant allele after the indicated incubation time at 25°C in medium inducing the expression of pREP prp4-K189I. p, pre-mRNA; m, mRNA. The radiolabelled probe used is the complete tfIId gene, which encodes the TATA-binding protein, containing three introns. Each lane contains 15 µg total RNA. FACS analysis: cells were stained with propidium iodide and analysed by flow cytometry. (C) prp4-73ts strain after 15 h at 25°C and 3 h at 36°C; (D) strain expressing pREP prp4-K189I after the indicated time period at 25°C; (E) prp1-4ts strain after 15 h at 25°C and 3 h at 36°C; (F) prp2-1ts strain after 15 h at 25°C and 3 h at 36°C. The peaks labelled 1C and 2C represent cells with DNA contents characteristic of those in the G1 and G2 phase of the cell cycle, respectively.
We further analysed prp4-73 at the restrictive temperature and the strain overexpressing prp4-K189I by flow cytometry. In both strains we observed the same arrest phenotype. In correlation with ceasing growth and the accumulation of pre-mRNA, the cultures show a significant cell population with a 1C DNA content, and another with a 2C DNA content (Figure 2C and D). Whereas this FACS phenotype is seen when the mutations are in the C-terminal kinase domain, most likely causing a decrease of kinase activity, we found previously that expression of mutations in the N-terminal domain of Prp4p leads to the impairment of mitosis (Groß et al., 1997).
We also analysed the prp1-4 allele, which is synthetically lethal with prp4-73, by flow cytometry and found that some cells arrest in G1, and others in G2 at the restrictive temperature (Figure 2E). Remarkably, all the other prp mutants display a 2C DNA content at the restrictive temperature in this analysis. As an example and control we used prp2-1 (Figure 2F). These results indicate that Prp4p, as well as Prp1p activity, is essential for G1–S and for G2–M transitions.
Prp4p and Prp1p interact in the two-hybrid system
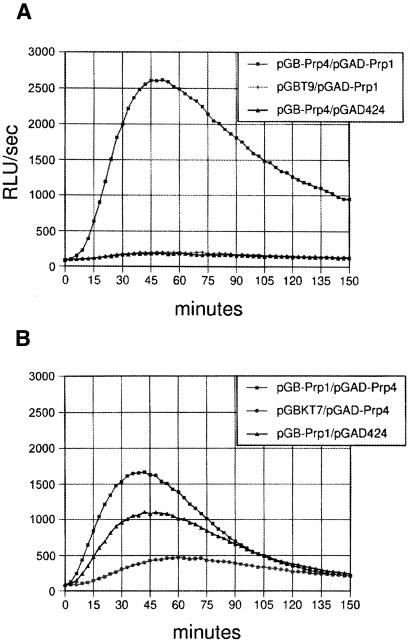
We investigated the interaction of Prp4p and Prp1p in the yeast two-hybrid system. In this approach we fused full-length Prp4p cDNA to the Gal4p DNA-binding domain, generating the plasmid pGB-Prp4, and fused the prp1 coding region to the activation domain in a plasmid called pGAD-Prp1, and vice versa (Figure 3). Several colonies containing both plasmids were analysed for the stimulation of the reporter gene β-galactosidase using a luminiscence-photometer. In both cases the interaction between Prp4p and Prp1p appears to be specific (Figure 3), although Prp1p fused with the DNA-binding domain generated a background activation of the reporter gene by itself (Figure 3B) and the activity measured is relatively low when compared with the activity induced by the interaction of p53 and SV40 large T-antigen, which were used as a positive control.
Fig. 3. Two-hybrid interaction between Prp4p and Prp1p. The yeast cells (Y190) containing the plasmids in the indicated combinations were cultured in SD media lacking leucine and uracil to mid log phase. Then, samples were mixed with an equal volume of Gal-Screen™ (Tropix, Inc.). The β-galactosidase activity in each mixture was then measured using a Microplate Luminometer LB96V (EG & G. Berthold).
Prp4p kinase phosphorylates Prp1p in vitro
We constructed a Prp1p protein containing a His6 tag at the N-terminus. The recombinant protein complements the prp1-4 allele when expressed in S. pombe (results not shown). We produced His6-Prp1p in bacteria and isolated it from the bacterial extract using the His-tag system. Then, HA-tagged Prp4p kinase was immunoprecipitated from a S. pombe extract using anti-HA antibodies. HA-tagged Prp4 kinase also complements the prp4-73 allele. We performed in vitro kinase assays with the immunoprecipitate, using [γ-32P]ATP. Adding increasing amounts of bacterially produced His6-Prp1p to the assay leads to the phosphorylation of a protein with a Mr of ∼100 kDa (Figure 4B). The protein is recognized on a western blot by anti-His antibodies (Figure 4A). The phosphorylated protein at ∼62 kDa is recognized by anti-HA antibodies, suggesting that Prp4p protein kinase strongly autophosporylates in vitro (Figure 4B and C). In contrast, the mutant protein HA-Prp4K189I, which was immunoprecipitated and used in the kinase assay, does not autophosphorylate, nor do we detect phosphorylation activity towards recombinant His6-Prp1p (Figure 4B). We also fused glutathione S-transferase (GST) to Prp4p, produced the recombinant GST–Prp4p molecules in bacteria and isolated them using glutathione–Sepharose. The fusion protein, GST–Prp4p, and Prp4p after the removal of GST show kinase activity towards His6-Prp1p (Figure 4D). These results provide clear evidence that Prp4p kinase is capable of phosphorylating Prp1p in vitro, and unambiguously show that the mutation of lysine 189 to isoleucine in Prp4p leads to the abolition of kinase activity in vitro.
Fig. 4. Phosphorylation of bacterially produced His6-Prp1p by immunoprecipitated HA-Prp4p and bacterially produced GST–Prp4p. Radiolabelled ATP was used in the kinase assay. SDS–PAGE electrophoresis. (A) Western blot analysis using anti-His antibodies as probe. (B) Autoradiography. (C) Western blot analysis using anti-HA antibodies as probe: lane 1, His6-Prp1p; lane 2, HA-Prp4p; lane 3, His6-Prp1p (0.05 µg) + HA-Prp4p; lane 4, His6-Prp1p (0.3 µg) + HA-Prp4p; lane 5, His6-Prp1p (0.6 µg) + HA-Prp4p; lane 6, HA-Prp4p K189I; lane 7, His6-Prp1p (0.3 µg) + HA-Prp4p K189I. (D) Autoradiography: lane 1, GST–Prp4p (0,6 µg); lane 2, His6-Prp1p (2 µg) + GST–Prp4p (0,6 µg); lane 3, PreScission Protease™ + GST–Prp4p (0,6 µg); lane 4, PreScission Protease™ + GST–Prp4p (0,6 µg) + His6-Prp1p (2 µg).
In vivo phosphorylation of Prp1p
We introduced an HA-tagged prp1+ gene into the genome of a strain containing the prp4-73 allele. The strain grows normally at 25°C and stops dividing 3 h after the shift to the restrictive temperature. This strain was labelled in vivo with [32P]ortho-phosphate for 4 h at the permissive temperature of 25°C and at the restrictive temperature of 36°C. Total protein was extracted and separated by SDS–PAGE. In the protein extract of the strain labelled at 25°C, we detect a signal in the range of Mr 100 kDa, which is much less intense in the protein extract of the cells labelled at 36°C (Figure 5A, arrow). A western blot analysis using HA-antibodies as probe indicates that the signal identified at 25°C is phosphorylated HA-Prp1p, which is not detected at 36°C (Figure 5B, arrow). These results indicate that in vivo phosphorylation of Prp1p appears to be dependent on Prp4p activity. Several labelling experiments in this series revealed that the amount of phosphorylated Prp1p must be very low in vivo.
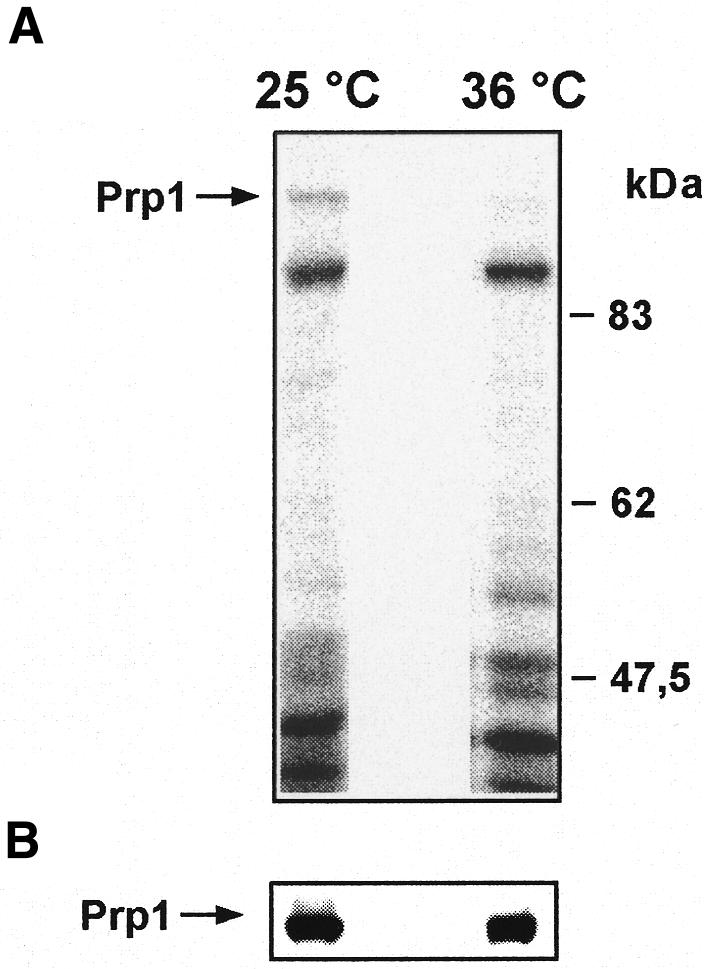
Fig. 5. In vivo labelling of HA-tagged Prp1p. [32P]ortho-phosphate was incubated for 4 h with a strain containing an HA-tagged prp1+ and the prp4-73 allele at 25 and 36°C. Total protein extract was subjected to SDS–PAGE. (A) Autoradiography. (B) Western blot analysis using HA antibodies as probe; the arrow indicates the labelled Prp1p protein.
This observation is consistent with the finding that we cannot co-immunoprecipitate Prp4p and Prp1p unless we overproduce both proteins on expression vectors (results not shown).
CONCLUSION
Collectively, the genetic and biochemical data presented above are consistent with the idea that Prp4p kinase activity is required for pre-mRNA splicing in vivo. The genetic and biochemical interactions between Prp4p and Prp1p, and the Prp4p kinase-dependent phosphorylation of Prp1p in vivo, suggest that Prp1p is a physiological substrate of Prp4p. In addition, the genetic interaction of prp4 with several genes encoding splicing components is consistent with our proposal that Prp4p is a kinase involved in the formation of active spliceosomes, targeting non SR-splicing factor(s). It remains to be determined whether Prp4p kinase functions as a signalling kinase, for example, labelling pleiotropic factors, such as Prp1p, as splicing factors. The human homologue of Prp1p has been discussed as functioning as a bridge between snRNPU5 and snRNPU4/U6 (Makarov et al., 2000). Therefore, it is conceivable that phosphorylation of Prp1p is part of a control mechanism to assemble a proper tri-snRNPU4/U6–U5 as a prerequisite for an active spliceosome. This is the first report showing that a kinase is involved in splicing regulation via phosphorylation of a non-SR splicing factor.
METHODS
Plasmid constructs and mutagenesis. The N-terminal HA-tagged expression vector pREP42 HA was used to insert prp4 cDNA using BamHI as cloning site as described (Craven et al., 1998; Lützelberger et al., 1999). To produce Prp1p in bacteria, a Sal1 fragment containing the ORF of prp1 was cloned into the vector pQE31 containing an N-terminal His-tag (Qiagen). All constructs were sequenced. The His-tag was used to isolate bacterially produced His6-Prp1p via Ni-NTA–agarose and to detect His6-Prp1p with the monoclonal anti-His antibodies. To construct the mutant allele prp4-K189I, site-specific mutagenesis was performed as described previously (Groß et al., 1997; Lützelberger et al., 1999).
In vitro phosphorylation assay. Protein extracts were made exactly as described (Groß et al., 1997). For immunoprecipitation, 400 µl of protein extract were used incubated with anti-HA antibodies for 2 h at 4°C. Twenty-five microlitres of protein A–Sepharose were added and incubated for a further 2 h at room temperature. The immunoprecipitate was first washed with 10 mM Tris pH 7.5, 0.4 M NaCl, 0.5 mM dithiothreitol (DTT), followed by 10 mM Tris pH 7.5, 1 M NaCl, 0.5 mM DTT, and then with kinase buffer (20 mM HEPES, 3 mM MgCl2, 0.5 mM DTT, 5% glycerol). Kinase assays were performed in a 20 µl volume containing kinase buffer, 5 µCi [γ-32P]ATP, 100 µM ATP and 5 µl immunoprecipitate. Bacterially produced Prp1p was added in the range of 0.05–0.6 µg protein. The samples were incubated at 37°C for 30 min.
In vivo labelling. Cultures were labelled as described by Moreno et al. (1991). Briefly, cells were grown in minimal medium (EMM) plus supplements at 25°C to mid log phase. Growing cells were resuspended in minimal low phosphate medium (EMMP). Five millilitres of the culture (5 × 106/ml) were kept at 25°C; another 5 ml of culture were shifted to 36°C. To each culture 0.5 mCi [32P]ortho-phosphate was added and incubated for 4 h. After 4 h labelling time, the cells were diluted with 15 ml of cultures kept under the same conditions without [32P]ortho-phosphate. Protein was extracted as described previously (Groß et al., 1997).
Acknowledgments
ACKNOWLEDGEMENTS
We thank J. Bode for the permission to use the FACSscan at the German Centre of Biotechnology (GBF, Braunschweig), and thank particularly Maria Höxter for her technical assistance with the FACSscan. We also appreciate the technical assistance of Susanne Zock-Emmenthal and the permanent support of Henning Schmidt. The prp2-1 strain was a gift from Judy Potashkin. Financial support for this study was provided by a grant from the Deutsche Forschungsgemeinschaft to N.F.K.
REFERENCES
- Alahari S.K., Schmidt, H. and Käufer, N.F. (1993) The fission yeast prp4+ gene involved in pre-mRNA splicing codes for a predicted serine/threonine kinase and is essential for growth. Nucleic Acids Res., 21, 4079–4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beales M., Flay, N., McKinney, R., Habara, Y., Ohshima, Y., Tani, T. and Potashkin, J. (2000) Mutations in the large subunit of U2AF disrupt pre-mRNA splicing, cell cycle progression and nuclear structure. Yeast, 16, 1001–1013. [DOI] [PubMed] [Google Scholar]
- Colwill K., Pawson, T. Andrews, B., Prasad, J., Manley, J., Bell, J.C. and Duncan, P.I. (1996) The Clk/Sty protein kinase phosphorylates SR-splicing factors and regulates their intracellular distributuion. EMBO J., 15, 265–275. [PMC free article] [PubMed] [Google Scholar]
- Craven R.A., Griffith, D.J. F., Sheldrick, K.S., Randall, R.E., Hagan, I.A. and Carr, A.M. (1998) Vectors for the expression of tagged proteins in Schizosaccharomyces pombe. Gene, 221, 59–68. [DOI] [PubMed] [Google Scholar]
- Galisson F. and Legrain, P. (1993) The biochemical defects of prp4-1 and prp6-1 yeast splicing mutants reveal that the PRP6 protein is required for the accumulation of the [U4/U6.U5] tri-snRNP. Nucleic Acids Res., 21, 1555–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groß T., Lützelberger, M., Wiegman, H., Klingenhoff, A., Shenoy, S. and Käufer, N.F. (1997) Functional analysis of the fission yeast Prp4 protein kinase involved in pre-mRNA splicing and isolation of a putative mammalian hommologue. Nucleic Acids Res., 25, 1028–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groß T., Richert, K., Mierke, C., Lützelberger, M. and Käufer, N.F. (1998) Identification and characterization of srp1, a gene of fission yeast encoding a RNA binding domain and a RS domain typical of SR splicing factors. Nucleic Acids Res., 26, 505–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui J.F., Lane, W.S. and Fu, X.-D. (1994) A serine kinase regulates intracellular localization of splicing factors in the cell cycle. Nature, 369, 678–682. [DOI] [PubMed] [Google Scholar]
- Käufer N.F. and Potashkin, J. (2000) Analysis of the splicing machinery in fission yeast: a comparison with budding yeast and mammals. Nucleic Acids Res., 28, 3003–3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundgren K., Allan, S., Urushiyama, S., Tani, T., Ohshima, Y., Frendewey, D. and Beach, D. (1996) A connection between pre-mRNA splicing and the cell cycle in fission yeast: cdc28+ is allelic with prp8+ and encodes an RNA-dependent ATPase/Helicase. Mol. Biol. Cell, 7, 1083–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lützelberger M., Groß, T. and Käufer, N.F. (1999) Srp2, a SR protein family member of fission: in vivo characterization of its modular domains. Nucleic Acids Res., 27, 2618–2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarov E.M., Makarova, O.V., Achsel, T. and Lührmann, R. (2000) The human homologue of the yeast splicing factor Prp6p contains multiple TPR elements and is stably associated with the U5 snRNP via protein–protein interactions. J. Mol. Biol., 298, 567–575. [DOI] [PubMed] [Google Scholar]
- Manley J.L. and Tacke, R. (1996) SR proteins and splicing control. Genes Dev., 10, 1569–1579. [DOI] [PubMed] [Google Scholar]
- Mermoud M.D., Cohen, P.T.W. and Lamond, A.I. (1994) Regulation of mammalian spliceosome assembly by a protein phosphorylation mechanism. EMBO J., 13, 5679–5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray H.L. and Jarrell, K.A. (1999) Flipping the switch to an active spliceosome. Cell, 96, 599–602. [DOI] [PubMed] [Google Scholar]
- Moreno S., Klar, A. and Nurse, P. (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol., 194, 795–823. [DOI] [PubMed] [Google Scholar]
- Neubauer G., King, A., Rappsilber, J., Calvio, C., Watson, M., Ajuh, P., Sleeman, J., Lamond, A. and Mann, M. (1998) Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nature Genet., 20, 46–50. [DOI] [PubMed] [Google Scholar]
- Potashkin J., Li, R. and Frendewey, D. (1989) Pre-mRNA splicing mutants of Schizosaccharomyces pombe. EMBO J., 8, 551–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potashkin J., Kim, D., Fons, M., Humphrey, T. and Frendewey, D. (1998) Cell-division-cycle defects associated with fission yeast pre-mRNA splicing mutants. Curr. Genet., 34, 153–163. [DOI] [PubMed] [Google Scholar]
- Rosenberg G.H., Alahari, S.K. and Käufer, N.F. (1991) Prp4 from Schizosaccharomyces pombe, isolated using genes containing artificial introns. Mol. Gen. Genet., 226, 305–309. [DOI] [PubMed] [Google Scholar]
- Rossi F., Labourier, E., Forne, T., Divita, G., Derancourt, J., Riou, J.F., Antoine, E., Cathala G., Brunel, C. and Tazi, J. (1996) Specific phosphorylation of SR proteins by mammalian DNA topoisomerase 1. Nature, 381, 80–82. [DOI] [PubMed] [Google Scholar]
- Schmidt H., Richert, K., Drakas, R.A. and Käufer, N.F. (1999) spp42, identified as a classical suppressor of prp4-73, which encodes a kinase involved in pre-mRNA splicing in fission yeast, is a homologue of the splicing factor Prp8p. Genetics, 153, 1183–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z., Yanagida, M. and Lin, R.-J. (1998) Fission yeast mitotic regulator Dsk1 is an SR protein-specific kinase. J. Biol. Chem., 273, 5963–5969. [DOI] [PubMed] [Google Scholar]
- Tang Z., Kuo, T., Shen, J. and Lin, R.-J. (2000) Biochemical and genetic conservation of fission yeast Dsk1 and human SR protein-specific kinase 1. Mol. Cell. Biol., 20, 816–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S. and Radzio-Andzelm, E. (1994) Cyclic AMP-dependent protein kinase. In Woodgett, J.R. (ed.), Protein Kinases. IRL Press, Oxford, UK, pp. 1–29.
- Tazi J., Kornstädt, U., Rossi, F., Jeanteur, P., Cathala, G., Brunel, C. and Lührmann, R. (1993).Thiophosphorylation of U1-70K protein inhibits pre-mRNA splicing. Nature, 363, 283–286. [DOI] [PubMed] [Google Scholar]
- Urushiyama S., Tani, T. and Ohshima, Y. (1996) Isolation of novel pre-mRNA splicing mutants of Schizosaccharomyces pombe. Mol. Gen. Genet., 253, 118–127. [DOI] [PubMed] [Google Scholar]
- Urushiyama S., Tani, T. and Ohshima, Y. (1997) The prp1+ gene required for pre-mRNA splicing in Schizosaccharomyces pombe encodes a protein that contains TPR motifs and is similar to Prp6p of budding yeast. Genetics, 147, 101–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will C. and Lührmann, R. (1997) Protein functions in pre-mRNA splicing. Curr. Opin. Cell Biol., 9, 320–328. [DOI] [PubMed] [Google Scholar]