Abstract
We have expressed the proline-rich antigen (PRA) from Coccidioides immitis in Escherichia coli and evaluated its potential as a vaccine candidate. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of the recombinant protein (rPRA) revealed two bands, which exhibited virtually identical primary amino acid sequences. T cells from rPRA-immunized BALB/c mice showed a significant in vitro proliferative response to rPRA. A small but statistically significant proliferative response was also induced by rPRA in T cells from mice immunized with whole-cell coccidioidal vaccines. BALB/c mice immunized with rPRA and challenged intraperitoneally with virulent C. immitis had a greatly reduced fungal burden in their lungs and spleens compared to unvaccinated mice. The number of organisms in the lungs was reduced 500-fold, and similar reductions were observed in the spleens of immunized mice. These studies support the continued development of rPRA as a candidate vaccine for prevention of coccidioidomycosis.
Coccidioides immitis is a fungus found in the soil of the deserts in the southwestern United States, northern Mexico, and other parts of the western hemisphere. The organism infects human beings by inhalation and causes disease in immunologically normal as well as immunocompromised hosts (10). In immunocompetent people, approximately 60% of infections are asymptomatic (10). Symptomatic pneumonia occurs in about 30% of the cases and can last for weeks to months (10). About 5% of immunocompetent people diagnosed with coccidioidomycosis develop disseminated disease (23, 24). Though some long-term residents in the area of endemicity have been infected and acquired immunity, the epidemic seen in 1990 to 1993 illustrates that many people in the area of endemicity have not been infected and would benefit from a vaccine (8, 19).
Natural infection with C. immitis results in lifelong immunity to the organism (23). In experimental infections, protection requires CD4 T cells (2). Experimental animals have been successfully vaccinated with formalin-killed spherules, which indicates that a nonviable vaccine strategy is feasible (16, 20).
The proline-rich antigen (PRA) is a protein which was identified independently by two laboratories and is also known as antigen 2 (5, 7, 28). PRA is a heavily glycosylated protein which is located in the spherule cell wall (11). In order to biochemically purify this protein, a spherule extract was chemically deglycosylated, which removes most O-linked carbohydrate (17). We found that people with coccidioidomycosis make both T-cell and antibody responses to deglycosylated PRA (6, 11). To further evaluate the antigenicity of this protein, we have expressed it in Escherichia coli. In this work, we evaluate the T-cell-mediated immune response to PRA in mice and test its ability to protect mice from infectious challenge.
MATERIALS AND METHODS
Expression of the PRA gene.
Total RNA was extracted from 48-h spherules and reverse transcribed with Superscript II and oligo(dT) (Gibco/BRL, Bethesda, Md.) as previously described (7). The resulting cDNA was used as a template in a PCR catalyzed by Pfu polymerase for 35 cycles as suggested by the manufacturer (Stratagene, La Jolla, Calif.). The PCR primers were no. 167 (5′CCGGATCCATGCAGTTCTCTCACGCTC) and no. 13 (5′CCGAATTCCAGTGAAATCAGGTGTGTT), each of which includes a restriction site to facilitate subcloning. The resulting 970-bp product was gel purified, digested with BamHI and EcoRI, and ligated into the BamHI/EcoRI sites of pBluescript SK+ (Stratagene) by standard techniques (22). The orientation, frame, and sequence of the cDNA insert were confirmed by automated DNA sequencing on an ABI 377 sequencer at the Arizona Research Laboratories, University of Arizona. The insert was excised from pBluescript with BamHI and EcoRI and ligated into pET32a as suggested by the supplier (Novagen, Madison, Wis.) to produce pPRA.B15. E. coli BL21(DE3)SlyD− (21) (which was given to us by Ry Young, Texas A&M University) was transformed with this construct by a CaCl2 technique (22). This strain was chosen because it lacks the FK-506 binding protein, which copurified with recombinant PRA (rPRA) (our unpublished observations).
Fifty milliliters of Luria-Bertani (LB) broth (22) containing 50 μg of carbenicillin per ml and 12.5 μg of tetracycline per ml was inoculated with a single colony and grown overnight at 37°C with continuous shaking at 225 rpm. The following morning, the bacteria were centrifuged for 15 min at 2,000 × g and resuspended in 50 ml of fresh LB broth, and 5 ml was used to inoculate each of eight 2-liter flasks containing 500 ml of LB broth containing carbenicillin and tetracycline. Cells were grown at 37°C with shaking at 225 rpm, until the absorbance at 600 nM was 0.6 (about 3 to 4 h), at which time expression of rPRA was induced with 1 mM isopropylthiogalactoside (Gibco/BRL), and the cells were incubated for another 4 h at 37°C.
Bacteria were harvested by centrifugation as described above, and the pellets were weighed. Cells were homogenized in column buffer (8 M urea, 100 mM Na2HPO4, 10 mM Tris, pH 8.0), 10 ml per g (wet weight). The homogenates were stirred for an hour at room temperature and then centrifuged at 10,000 × g for 15 min. Histidine-tagged recombinant protein was purified from the supernatant by batch metal affinity chromatography under denaturing conditions on Ni-nitrilotriacetic acid (NTA) agarose (Qiagen, Chatsworth, Calif.), by elution with a pH step gradient as recommended by the manufacturer. Thirty-two milliliters of packed, equilibrated Ni-NTA was incubated with 75 ml of supernatant for an hour at 25°C. The agarose matrix was then washed four times with 75 ml of column buffer, pH 8.0, and three times with 75 ml of column buffer, pH 6.3. The protein was eluted with three 32-ml aliquots of column buffer, pH 4.5. The eluate was dialyzed for 24 h against renaturation buffer (150 mM NaCl, 4 mM reduced glutathione, 40 nM oxidized glutathione, 20 mM Tris, pH 9.0) containing 3.5 M urea, followed by renaturation buffer containing 1 M urea, and finally, two 1-liter changes of 150 mM NaCl. The thioredoxin fusion peptide and histidine tag were removed from the purified His-tagged protein by proteolysis with biotinylated thrombin, and the thrombin was removed with a small aliquot of streptavidin-agarose as suggested by the supplier (Novagen). The cleaved protein was dialyzed into denaturing column buffer (pH 8.0) and passed over a 4-ml Ni-NTA agarose column to remove the histidine tag. After dialysis against renaturation buffers, as described above, the resulting purified protein (rPRA) was concentrated by centrifugal ultrafiltration. Protein content was determined by bicinchoninic acid with bovine serum albumin as a standard (Pierce, Rockford, Ill.), and a yield of 6 mg of culture of highly purified rPRA per liter was obtained. The level of endotoxin contamination was determined with a Limulus amebocyte lysate QCL-1000 kit (Biowhittaker, Walkersville, Md.). The PRA used in this study had 70 IU of endotoxin per μg of protein.
Protein analysis and sequencing.
Recombinant protein was electrophoresed on a sodium dodecyl sulfate (SDS)–12.5% polyacrylamide gel electrophoresis (PAGE) gel and stained with Coomassie blue or electroblotted to nitrocellulose or polyvinylidene difluoride membranes (Schleicher and Schuell, Keene, N.H.). Nitrocellulose blots were probed with a goat antiserum to spherule-derived PRA, which has previously been described (6). The N-terminal sequences of enterokinase-treated recombinant proteins were determined on the two bands seen with SDS-PAGE (see Results). Sequencing from the polyvinylidene difluoride membrane was done by automated Edman degradation at the Biotechnology Division of Arizona Research Laboratories, University of Arizona. The C-terminal amino acid sequence of one recombinant band (see Results) was determined at the Macromolecular Structure Facility, Michigan State University.
Animals.
All T-cell proliferation assays and infection experiments were conducted with 10-week-old female BALB/c mice supplied by the National Cancer Institute (Bethesda, Md.).
C. immitis antigens.
Formalin-killed spherules (13) and the mycelial filtrate-plus-lysate fraction (F+L) (4) were prepared as reported elsewhere. The concentration of mycelial F+L was determined with total dry weight.
T-cell proliferation assays.
Mice were immunized for these experiments in several ways. To test the antigenicity of rPRA in vivo, groups of 10 female BALB/c mice were immunized with 5 μg of PRA in 0.1 ml of incomplete Freund’s adjuvant (IFA) subcutaneously. This immunization was repeated 2 weeks later, and the final 5 μg of PRA was given in 0.1 ml of complete Freund’s adjuvant (CFA) at the base of the tail and in each hind footpad. The T-cell proliferation assay was done exactly as previously described (13).
The response of C. immitis-immune mice to rPRA was tested by two methods of immunization. Female BALB/c mice were immunized with 105 strain 95-291 arthroconidia subcutaneously (12). These organisms are auxotrophic and temperature sensitive; they convert to spherules but do not cause disease in mice (26). One month later, the mice were immunized with formalin-killed spherules in CFA as previously described (14). Alternatively, the mice were immunized three times with formalin-killed spherules as previously described (14). Control mice were given a single immunization in the footpads and the base of the tail with CFA. In all cases, T-cell proliferation assays were done 10 days after the last immunization.
Infectious challenge experiments.
Groups of 10 female BALB/c mice were immunized twice (2 weeks apart) with 5 μg of rPRA in IFA followed by 5 μg of PRA in CFA in the hind footpads and the base of the tail. Negative controls received IFA and CFA alone on the same schedule. Positive controls were given 106 formalin-killed spherules subcutaneously on days 1 and 14. Two weeks later, they were immunized with 1.5 × 106 formalin-killed spherules in 0.1 ml of CFA in the hind footpads and the base of the tail. Fourteen days after the last injection, the mice were infected with 50 arthroconidia (R.S. strain) intraperitoneally. Two weeks after infection, the mice were sacrificed and the numbers of organisms in their spleens and lungs were determined by quantitative culture as described elsewhere (13).
Statistical analysis.
T-cell proliferation assay results were compared with Student’s t test. The numbers of CFU per lung were expressed on a log scale. Because these values did not fall into a normal distribution, the Mann-Whitney U test was used to compare medians in all cases.
RESULTS
Protein expression.
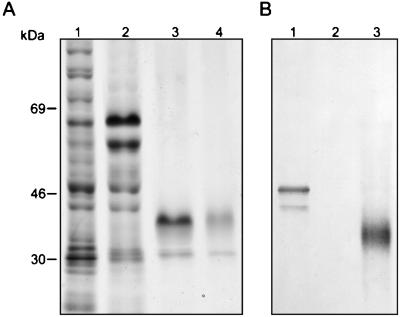
rPRA was analyzed by SDS-PAGE followed by staining with Coomassie blue. The protein resolved into two bands with apparent molecular masses of 47 and 43 kDa (Fig. 1A), both of which were recognized by monospecific antibody on an immunoblot (Fig. 1B). Following thrombin cleavage to remove the fusion partner, the two proteins migrated at about 36 and 27 kDa (Fig. 1A). Spherule-derived deglycosylated PRA has a molecular mass of 33 kDa (11), and the apparent mass of the upper band is consistent with the value expected, taking into account the residual vector-encoded amino acids. The N-terminal amino acid sequences were determined for both bands following removal of the fusion partner by enterokinase. They were identical and matched the predicted amino acid sequence for 10 amino acids following the initial methionine of PRA. The C-terminal amino acid sequence of the lower band was C′-A-A-L, indicating the loss of the two C-terminal amino acids. We concluded that the proteins represented by the two bands were essentially identical, despite the difference in migration on SDS-PAGE gels. Thrombin-cleaved rPRA was used as an immunogen and in the T-cell proliferation studies.
FIG. 1.
SDS-PAGE and immunoblot analysis of rPRA. Samples of rPRA were electrophoresed on 12.5% polyacrylamide gels and stained with Coomassie blue (A), or samples were transferred to nitrocellulose and proteins were detected with monospecific goat antiserum and alkaline phosphatase-conjugated secondary antibody (B). (A) Coomassie blue-stained gel. Lane 1, isopropylthiogalactoside-induced bacterial cell lysate; lane 2, eluate from Ni-NTA column; lane 3, thrombin-cleaved rPRA; lane 4, Ni-NTA-purified thrombin-cleaved rPRA. (B) Representative immunoblot. Lane 1, isopropylthiogalactoside-induced bacterial cell lysate; lane 2, isopropylthiogalactoside-induced bacterial cell lysate from empty vector; lane 3, biochemically purified spherule-derived PRA.
T-cell proliferation experiments.
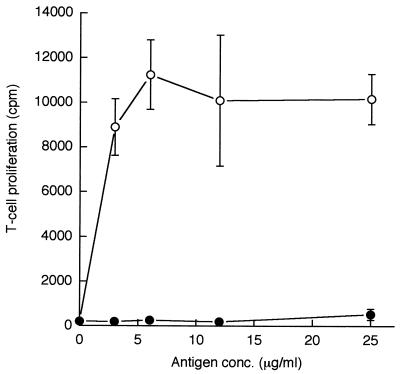
To find out whether rPRA immunization elicited a T-cell proliferative response to rPRA, mice were immunized and the T-cell proliferation response was measured 10 days later. The results are shown in Fig. 2. rPRA elicited a large proliferative response, indicating that the mice were immune to rPRA. In contrast, rPRA immunization did not prime cells to proliferate in response to mycelial F+L. This result is consistent with previous studies which found that C. immitis mycelia contain very little PRA (11).
FIG. 2.
Proliferative response of lymph node T cells from mice immunized with PRA plotted versus antigen concentration in vitro. The means and standard deviations of three determinations are shown. Open symbols represent PRA; filled symbols represent mycelial F+L.
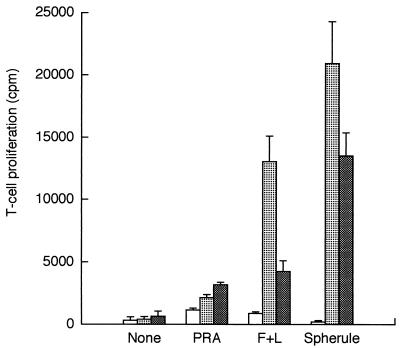
The response of C. immitis-immune T cells to several antigens, including rPRA, is shown in Fig. 3. The responses to a range of concentrations of antigen were measured; the maximal responses are shown. rPRA elicits a small, but significant, T-cell proliferative response in T cells from mice immunized against C. immitis. The maximal response to rPRA in mice immunized with the live attenuated mutant of C. immitis was 3,184 cpm compared to 661 cpm in the absence of antigen (P < 0.005). rPRA also elicits a very small response in T cells from control mice (1,154 cpm compared to 321 cpm in the absence of antigen [P < 0.02]). The response of mice immunized with C. immitis by either method is significantly greater than the response of control mice (P < 0.02). Immunization with the live attenuated mutant and immunization with formalin-killed spherules elicited similar responses to rPRA. The response to rPRA was substantially less than the response to mycelial F+L and much less than the response to formalin-killed spherules. Similar results were seen in three other T-cell proliferation experiments. Therefore, C. immitis-immune T cells make a very small but statistically significant T-cell proliferative response to rPRA in vitro.
FIG. 3.
Proliferative response of lymph node T cells from mice immunized with CFA alone (open bars), formalin-killed spherules (stippled bars), or live, attenuated C. immitis followed by formalin-killed spherules (solid bars). The maximal response for each antigen is shown. The concentration of PRA and mycelial F+L was 25 μg/ml. The formalin-killed spherules were at a concentration of 105/ml. Means and standard deviations of three determinations are shown.
Vaccination experiments.
The results of one vaccination experiment are shown in Fig. 4. The rPRA-immune mice had much lower numbers of organisms in the lungs than did the nonimmune controls; the median in rPRA-immune mice was 3.3 (log10) CFU (range, 0.7 to 4.47), compared to control mice with a median of 6 (log10) CFU (range, 4.3 to 6.89). This difference was statistically significant (P = 0.0001 by the Mann-Whitney U test). A similar difference was seen in the spleen; the median in rPRA-immune mice was 3.37 (log10) CFU (range, 0.7 to 5.06), compared to control mice with a median of 6 (log10) CFU (range, 3.3 to 6.89). The difference was also statistically significant (P = 0.003 by Mann-Whitney U test). Mice immunized with formalin-killed spherules had no viable organisms in their lungs or spleens. Similar data were seen in a second experiment with rPRA immunization in BALB/c mice. The median CFU in the control mice were 4.14 in the lung and 4.09 in the spleen. The medians in the PRA-immune mice were 1.5 in the lung and 1.6 in the spleen. The Mann-Whitney U test yielded P values of 0.0008 and 0.003, respectively, for the second experiment.
FIG. 4.
A box plot representation of the numbers of CFU found in the lungs and spleens of immune or control mice 14 days after infection with C. immitis. Control mice were immunized with CFA alone. The box indicates the 25th and 75th percentiles of the data; the line represents the 50th percentile. The capped bars represent the 10th and 90th percentiles. Symbols above and below the box indicate individual values above the 90th or below the 10th percentile.
DISCUSSION
One of the major reasons for studying antigenic proteins of C. immitis is to determine their potential as vaccines against coccidioidomycosis. Deglycosylated PRA biochemically purified from spherules is recognized by antibody from patients with coccidioidomycosis and by T cells from spherulin and coccidioidin skin test-positive people (6, 11). A recombinant PRA fusion protein has been shown to elicit delayed-type hypersensitivity in immunized mice and is also recognized by sera from patients with coccidioidomycosis (29). PRA has a predicted glycosylphosphatidylinositol attachment site (28). If the protein is in fact glycosylphosphatidylinositol linked, it should be extracellular and particularly accessible to the host immune system. All these observations suggested that PRA would be a good candidate for a C. immitis vaccine.
As we analyzed the rPRA by SDS-PAGE, we observed that it migrated as two bands with apparent molecular masses of 27 and 36 kDa. These two bands were virtually indistinguishable by amino acid sequence, amino acid content, or immunoreactivity on immunoblot. The abundance of proline almost certainly accounts for the anomalous migration on SDS-PAGE gels of both the recombinant and the spherule-derived proteins. Several reports have described proline-rich proteins expressed in E. coli that do not migrate in SDS-PAGE at the expected molecular weight or as a single band (15, 18, 25). The high proline content presumably increases the rigidity of the protein, which makes the protein migrate slower in SDS-PAGE than globular proteins of the same molecular weight. The appearance of two bands may be the result of conformational differences arising from cis-trans isomerizations in the central proline-rich domain.
There is abundant evidence that T-cell-mediated immunity is required for resistance to coccidioidomycosis. Athymic mice are more susceptible to infection (1), and CD4 T cells can adoptively transfer protective immunity in mice (2). The strongest clinical evidence is the propensity of AIDS patients and immunosuppressed transplant patients to develop disseminated disease (3, 9). The T-cell proliferation assay is an in vitro measure of T-cell-mediated immunity. For these reasons, we have used a T-cell proliferation assay as a screening test for identifying promising vaccine candidates.
Immunization of BALB/c mice with rPRA elicits a strong T-cell proliferative response to rPRA. Therefore, as expected, PRA is immunogenic. rPRA is a highly protective antigen in our mouse model system, but it elicits a very small amount of T-cell proliferation in C. immitis-immune mice. The magnitude of the proliferative response may not be a good predictor of the activity of an antigen as a vaccine.
The most important finding from this work is that rPRA is immunoprotective. rPRA elicits more impressive protection than we have seen with one other recombinant protein. The decrease in CFU of about 500-fold is at least 10-fold better than what we obtained with T-cell-reactive protein (TCRP) immunization (13). However, rPRA-immune mice did not achieve sterile immunity, as is seen in immunization with killed spherules. Therefore, whole formalin-killed spherules probably contain protective antigens other than PRA. Protection with rPRA was obtained with CFA, a very strong adjuvant which is too toxic for human use. Finally, we have done this experiment with intraperitoneal challenge rather than intranasal challenge, which is a more stringent test of protective immunity. Nevertheless, these data indicate that rPRA can be a highly protective antigen in this mouse model.
rPRA has no closely related homologs in the DNA database and, so far, appears to be unique to C. immitis. This is in contrast to TCRP, which is 50% identical to a mouse homolog (27). Furthermore, PRA is found in the spherule wall (11) and is presumably more accessible to the immune response than is TCRP, which is a cytoplasmic enzyme (13). These differences may contribute to the superior immunoprotection conferred by immunization with PRA compared to that with TCRP. Hopefully, the combination of two or three immunoprotective antigens will provide high-level immunoprotection in this model system. This would be a major step towards a human vaccine for coccidioidomycosis.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grants PO1 AI37232 and AI19149, the Research Service of the Department of Veterans Affairs, and the California Health Care Foundation.
REFERENCES
- 1.Beaman L, Pappagianis D, Benjamini E. Significance of T cells in resistance to experimental murine coccidioidomycosis. Infect Immun. 1977;17:580–585. doi: 10.1128/iai.17.3.580-585.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beaman L, Pappagianis D, Benjamini E. Mechanisms of resistance to infection with Coccidioides immitis in mice. Infect Immun. 1979;23:681–685. doi: 10.1128/iai.23.3.681-685.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen I M, Galgiani J N, Potter D, Ogden D A. Coccidioidomycosis in renal replacement therapy. Arch Intern Med. 1982;142:489–494. [PubMed] [Google Scholar]
- 4.Cole G T, Kruse D, Seshan K R. Antigen complex of Coccidioides immitis which elicits a precipitin antibody response in patients. Infect Immun. 1991;59:2434–2446. doi: 10.1128/iai.59.7.2434-2446.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox R A, Dolan M J, Magee D M, Galgiani J N. Production of a murine monoclonal antibody that recognizes an epitope specific to Coccidioides immitis antigen 2. Infect Immun. 1993;61:1895–1899. doi: 10.1128/iai.61.5.1895-1899.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dugger K O, Galgiani J N, Ampel N M, Sun S H, Magee D M, Harrison J, Law J. An immunoreactive apoglycoprotein purified from Coccidioides immitis. Infect Immun. 1991;59:2245–2251. doi: 10.1128/iai.59.7.2245-2251.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dugger K O, Villareal K M, Ngyuen A, Zimmermann C R, Law J H, Galgiani J N. Cloning and sequence analysis of the cDNA for a protein from Coccidioides immitis with immunogenic potential. Biochem Biophys Res Commun. 1996;218:485–489. doi: 10.1006/bbrc.1996.0086. [DOI] [PubMed] [Google Scholar]
- 8.Einstein H E, Johnson R H. Coccidioidomycosis: new aspects of epidemiology and therapy. Clin Infect Dis. 1993;16:349–354. doi: 10.1093/clind/16.3.349. [DOI] [PubMed] [Google Scholar]
- 9.Fish D G, Ampel N M, Galgiani J N, Dols C L, Kelly P C, Johnson C H, Pappagianis D, Edwards J E, Wasserman R B, Clark R J, Antoniskis D, Larsen R A, Englender S J, Petersen E A. Coccidioidomycosis during human immunodeficiency virus infection—a review of 77 patients. Medicine. 1990;69:384–391. doi: 10.1097/00005792-199011000-00006. [DOI] [PubMed] [Google Scholar]
- 10.Galgiani J N. Coccidioidomycosis. West J Med. 1993;159:153–171. [PMC free article] [PubMed] [Google Scholar]
- 11.Galgiani J N, Sun S H, Dugger K O, Ampel N M, Grace G C, Harrison J, Wieden M A. An arthroconidial-spherule antigen of Coccidioides immitis: differential expression during in vitro fungal development and evidence for humoral response in humans after infection or vaccination. Infect Immun. 1992;60:2627–2635. doi: 10.1128/iai.60.7.2627-2635.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirkland T N, Fierer J. Inbred mouse strains differ in resistance to lethal Coccidioides immitis infection. Infect Immun. 1983;40:912–917. doi: 10.1128/iai.40.3.912-916.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirkland T N, Thomas P W, Finley F, Cole G T. Immunogenicity of a 48-kilodalton recombinant T-cell reactive protein of Coccidioides immitis. Infect Immun. 1998;66:424–431. doi: 10.1128/iai.66.2.424-431.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kirkland T N, Zhu S W, Cruse D, Hsu L L, Seshan K R, Cole G T. Coccidioides immitis fractions which are antigenic for immune T lymphocytes. Infect Immun. 1991;59:3952–3961. doi: 10.1128/iai.59.11.3952-3961.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laqueyrerie A, Miltzer P, Romain F, Eiglmeier K, Cole S, Marchal G. Cloning, sequencing and expression of the apa gene coding for the Mycobacterium tuberculosis 45/47-kilodalton secreted antigen complex. Infect Immun. 1995;63:4003–4010. doi: 10.1128/iai.63.10.4003-4010.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lecara G, Cox R A, Simpson R B. Coccidioides immitis vaccine: potential of an alkali-soluble, water-soluble cell wall antigen. Infect Immun. 1983;39:473–475. doi: 10.1128/iai.39.1.473-475.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mort A J, Lamport D T. Anhydrous hydrogen fluoride deglycosylates glycoproteins. Anal Biochem. 1977;82:289–309. doi: 10.1016/0003-2697(77)90165-8. [DOI] [PubMed] [Google Scholar]
- 18.Ozaki L S, Svec P, Nussenzweig R S, Nussenzweig V, Godson G N. Structure of the Plasmodium knowlesi gene coding for the circumsporozoite protein. Cell. 1983;34:815–822. doi: 10.1016/0092-8674(83)90538-x. [DOI] [PubMed] [Google Scholar]
- 19.Pappagianis D. Marked increase in cases of coccidioidomycosis in California: 1991, 1992, and 1993. Clin Infect Dis. 1994;19:S14–S18. doi: 10.1093/clinids/19.supplement_1.14. [DOI] [PubMed] [Google Scholar]
- 20.Pappagianis D, Hector R, Levine H B, Collins M S. Immunization of mice against coccidioidomycosis with a subcellular vaccine. Infect Immun. 1979;25:440–445. doi: 10.1128/iai.25.1.440-445.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roof W S, Horne M, Young K D, Young R. slyD, a host gene required for φX174 lysis, is related to the FK506-binding protein family of peptidyl-prolyl cis-transisomerases. J Biol Chem. 1994;269:2902–2910. [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 23.Smith C E. Epidemiology of acute coccidioidomycosis with erythema nodosum (“San Joaquin” or “Valley Fever”) Am J Public Health. 1940;30:600–611. doi: 10.2105/ajph.30.6.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith C E, Saito M T, Simons S A. Pattern of 39,500 serologic tests in coccidioidomycosis. JAMA. 1956;160:546–552. doi: 10.1001/jama.1956.02960420026008. [DOI] [PubMed] [Google Scholar]
- 25.Staab J F, Ferrer C A, Sundstrom P. Developmental expression of a tandemly repeated proline- and glutamine-rich amino acid motif on hyphal surfaces of Candida albicans. J Biol Chem. 1996;271:6298–6305. doi: 10.1074/jbc.271.11.6298. [DOI] [PubMed] [Google Scholar]
- 26.Walch H A, Kalvoda A. Immunization of mice with induced mutants of Coccidioides immitis. I. Characterization of mutants and preliminary studies of their use as viable vaccines. Sabouraudia. 1971;9:173–184. [PubMed] [Google Scholar]
- 27.Wyckoff E E, Pishko E J, Kirkland T N, Cole G T. Cloning and expression of a gene encoding a T-cell reactive protein from Coccidioides immitis: homology to 4-hydroxyphenylpyruvate dioxygenase and the mammalian F antigen. Gene. 1995;161:107–111. doi: 10.1016/0378-1119(95)00250-a. [DOI] [PubMed] [Google Scholar]
- 28.Zhu Y, Chunmu Y, Magee M, Cox R A. Coccidioides immitis antigen 2: analysis of gene and protein. Gene. 1996;181:121–125. doi: 10.1016/s0378-1119(96)00486-6. [DOI] [PubMed] [Google Scholar]
- 29.Zhu Y, Yang C, Magee D M, Cox R A. Molecular cloning and characterization of Coccidioides immitis antigen 2 cDNA. Infect Immun. 1996;64:2695–2699. doi: 10.1128/iai.64.7.2695-2699.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]