Abstract
The Tox21 Program has investigated thousands of chemicals with high-throughput screening assays using cell-based assays to link thousands of chemicals to individual molecular targets/pathways. However, these systems have been widely criticized for their suspected lack of ‘metabolic competence’ to bioactivate or detoxify chemical exposures. In this study, 9 cell line backgrounds used in Tox21 assays (i.e., HepG2, HEK293, Hela, HCT116, ME180, CHO-K1, GH3.TRE-Luc, C3H10T1/2 and MCF7) were evaluated via metabolite formation rates, along with metabolic clearance and metabolite profiling for HepG2, HEK293, and MCF-7aroERE, in comparison to pooled donor (50) suspensions of primary human hepatocytes (PHHs). Using prototype clinical drug substrates for CYP1A2, CYP2B6, and CYP3A4/5, extremely low-to-undetectable CYP450 metabolism was observed (24 hours), and consistent with their purported ‘lack’ of metabolic competence. However, for Phase II metabolizing enzymes and metabolic clearance, surprisingly proficient metabolism was observed for bisphenol AF, bisphenol S, and 7-hydroxycoumarin. Here, comparatively low glucuronidation relative to sulfation was observed in contrast to equivalent levels in PHHs. Overall, while a lack of CYP450 metabolism was confirmed in this benchmarking effort, Tox21 cell lines were not ‘incompetent’ for xenobiotic metabolism, and displayed surprisingly high proficiency for sulfation that rivaled PHHs. These findings have implications for the interpretation of Tox21 assay data, and establish a framework for evaluating of ‘metabolic competence’ with in vitro models.
Keywords: Tox21, xenobiotic metabolism, metabolic clearance, metabolic competence, CYP450, primary human hepatocytes, bisphenol
1. Introduction
The Toxicology in the 21st Century Program (Tox21) is a federal collaboration among the National Institute of Environmental Health Sciences, National Center for Advancing Translational Sciences, the U.S. Environmental Protection Agency, and the Food and Drug Administration (Collins et al., 2008). Tox21 researchers utilize alternative high-throughput screening methods to evaluate chemical-induced bioactivities (Kavlock et al., 2009; Tice et al., 2013). These approaches have been effective in characterizing thousands of chemicals with respect to individual molecular targets and pathways, but have often been criticized for their presumed lack of tissue-like differentiation and functionality that includes xenobiotic metabolism ‘competence’. Given the central role of liver in human drug metabolism, drug-drug interactions and toxicity, hepatic metabolism is often the focus of these critiques. While hepatic metabolism (i.e., metabolic clearance, metabolite formation rates/profiles, reactive metabolites) is not directly operative in extrahepatic tissues (e.g., endocrine-related), the notion of conferring ‘metabolism’ to Tox21 assay systems remains a primary focus of researchers working towards new approach methodologies (NAMs) in the toxicology field to confer bioactivation of parent chemicals to reactive or bioactive metabolites. However, to date ‘metabolic competence’ in Tox21 assay systems has not been directly evaluated in context with established benchmarks of hepatic metabolic competence. Previous studies of metabolism with immortalized cell lines have been limited, but a recent study evaluating of benzo(a)pyrene metabolism in ARE-bla, GR-bla, and AREc32 cells (i.e., engineered reporter cells in HepG2 and HEK293) reported fluorescent-based CYP1A1 enzymatic activities, which reflect fetal liver metabolism (Fischer et al., 2020).
The benchmark for modeling human liver response to xenobiotic exposures in vitro has centered around isolation and culture of primary liver cells (e.g., those isolated from living tissue). Suspensions of primary human hepatocytes (PHHs) are widely recognized as the ‘gold standard’ to quantitatively model human hepatic drug metabolism, as they retain in vivo-expressed enzymes (e.g., cytochromes P450, CYPs) in the initial minutes/hours post isolation from human liver tissue (Hewitt et al., 2007; Guengerich, 1997). Given the myriad of metabolizing enzymes and transporters involved in drug biotransformation and clearance, sentinel drug metabolizing enzyme activities such as CYP3A4/5, CYP1A2 and CYP2B6 are often evaluated to gauge the fidelity, differentiation status, and metabolic proficiency of in vitro liver models including isolated PHHs from donor tissues (Kafert-Kasting et al., 2006). These 3 major CYPs are involved in clearance of > 60% of marketed drugs (Gerin et al., 2013; Lee and Kim, 2013). PHHs are routinely used for drug development and environmental toxicology applications to quantitatively estimate metabolic clearance rates and metabolite formation profiles in humans (Wetmore et al., 2012). In addition, confluent cultures of PHHs faithfully reflecting the in vivo-like cobblestone networks of cell-cell interactions observed in vivo have been widely used to model a broad complement of chemical-biological interactions for drug regulatory studies to estimate the potential for drug-drug interactions (e.g., liver enzyme induction, biliary excretion, and time-dependent inhibition) (Kafert-Kasting et al., 2006; Mao et al., 2016).
In addition, CYP450-mediated xenobiotic metabolism, Phase II conjugation via sulfate, glucuronide, glutathione and other facilitated quenching/excretion products plays a key role in xenobiotic exposure dynamics. To evaluate Phase II glucuronidation and sulfation proficiency, reference substrates 7-hydroxycoumarin, bisphenol AF (BPAF), and bisphenol S (BPS) were selected for this study. BPAF and BPS represent 2 environmentally-relevant bisphenol A (BPA) alternatives with existing Tox21 assay data (Yamasaki et al., 2003) (Liao et al., 2012) (Song et al., 2012; Wang et al., 2015).. Interpretation of these relative potencies is largely presumed to be direct reflections of parent chemical exposures based on nominal parent chemical concentrations in the absence of xenobiotic metabolism. An improved understanding the dynamics of in vitro exposures over time that includes Phase II metabolism of Tox21 assays is essential to contextualize and translate bioactivity assay data to humans towards the emergence of next generation risk assessment frameworks.
In this study, we set out to benchmark the ‘metabolic competence’ of the most prevalent cell lines applied in the Tox21 Program (henceforth abbreviated as “Tox21 cell lines”): HepG2, HEK293, Hela, HCT116, ME180, CHO-K1, GH3.TRE-Luc, C3H10T1/2 and MCF-7aroERE (MCF7). Specific activities for major human drug metabolizing enzymes CYP3A4/5, CYP1A2, and CYP2B6 were evaluated using clinically-relevant probe substrates and contextualized to the benchmark of pooled-donor suspensions of PHHs (50-donor pool). Moreover, the extents of metabolic clearance (i.e., loss of parent compound) and capacity for formation of Phase II glucuronidation and sulfation metabolites were evaluated for a subset of Tox21 cell lines following in vitro exposures to 7-hydroxycoumarin (7HC), BPAF, and BPS. Given the purported lack of xenobiotic metabolism in cell lines used in Tox21 assays, our findings reveal both anticipated and surprising results with implications to the interpretation of Tox21 and in vitro toxicology bioactivity assay data. The study provides a benchmark to contextualize the proficiency of metabolic competence with in vitro model systems, and highlights important considerations for chemicals with direct Phase II sulfation and glucuronidation metabolism.
2. Material and methods
2.1. Chemicals and reagents
All metabolism substrates and assay reagents were of the highest grade/purity (e.g., HPLC grade). Phenacetin, bupropion, midazolam (MI), acetaminophen, hydroxybupropion, 1-hydroxy midazolam, aminobenzotriazole (ABT), omeprazole (OMP), rifampicin (RIF), 7-HC, 7hydroxycoumarin glucuronide (7HCG), 7-hydroxycoumarin sulfate (7HCS), BPAF and BPS were obtained from Sigma-Aldrich (Saint Louis, MO, USA). Acetonitrile, methanol and formic acid were purchased from Fisher Scientific (Waltham, MA, USA). Eagle’s MEM medium (catalog No. 30–2003) was purchased from ATCC (Manassas, VA, USA) and was used for HepG2 and Hela cells culture. DMEM medium (catalog No. 11995) was purchased from Invitrogen (Carlsbad, CA, USA) and was used for HEK293 cells culture. McCoy’s 5a medium (catalog No. 30–2007) was purchased from ATCC (Manassas, VA, USA) and was used for HCT116 and ME-180 cells culture. F-12K medium (catalog No. 30–2004) was purchased from ATCC (Manassas, VA, USA) and was used for CHO-K1 cells culture. DMEM/F12, GlutaMax™ supplement medium (catalog No. 10565) was purchased from Life Technologies (Carlsbad, CA, USA) and was used for GH3.TRE-Luc cells culture. Eagle’s Basal medium (catalog No. 21010) was purchased from Invitrogen (Carlsbad, CA, USA) and was used for C3H10T1/2 cells culture. MEM/EBSS medium (catalog No. SH 30024.01) was purchased from Hyclone (Seattle, WA, USA) and was used for MCF7 cells culture.
2.2. Cell lines, cell incubations and cell culture
2.2.1. Primary human hepatocytes
Cryopreserved pooled PHHs (from 50 individual donor preparations) were purchased from Life Technologies/Thermo (HUE50-D) and were stored in liquid nitrogen vapor until use. Hepatocytes were thawed using hepatocyte thawing medium (Lonza Biosciences Solutions; Morrisville, NC, USA). Cell viability and yield were determined using a Cellometer (Nexcelom Bioscience, Lawrence, MA, USA) with acridine orange/propidium iodide (AOPI) staining per the manufacturer’s protocol. Once thawed, hepatocyte maintenance medium (Lonza Biosciences Solutions; Morrisville, NC, USA) was added to attain a concentration of 1×106 cells/mL. 50 μL of cell suspension was added to a V-bottom 96-well plate (Corning, NY, USA) at 50,000 cells/well with 50 μL maintenance medium (control) or with 50 μL of substrate at 2X final desired concentration in maintenance medium. The plates were incubated at 37°C, 5% CO2 with individual metabolism substrates (specific activity assays) or parent chemicals for metabolic clearance and metabolite formation assays (7HC, BPAF, BPS). PHHs were incubated for no more than 240 minutes in suspension form due to their loss of viability after 4 hours.
2.2.2. Cell lines used for Tox21 chemicals screening (Tox21 cell lines)
Tox21 cell lines including HepG2, HEK293, Hela, HCT116, ME180, CHO-K1, GH3.TRE-Luc, C3H10T1/2 and MCF7 cells were obtained from National Center for Advancing Translational Sciences (NCATS, Rockville, MD) and cultured analogous to established cell culture conditions for each respective line. Evaluation of enzymatic specific activities, metabolic clearance, and metabolite formation was performed in Costar flat bottom 96-well cell culture plates (Corning, NY, USA) analogous to culture conditions (https://tripod.nih.gov/tox21/assays/) used for each respective Tox21 cell line (e.g., culture media and initial seeding densities). Prior to initiating metabolism incubations, cultures were allowed to reach approximately 80–90% confluence (Hsieh et al., 2017).
2.3. Cytochrome P450 specific activity assays
2.3.1. Metabolism incubations
PHHs and Tox21 cell lines (i.e., HepG2, HEK293, Hela, HCT116, ME180, CHO-K1, GH3.TRE-Luc, C3H10T1/2 and MCF7) were each incubated at 37°C, 5% CO2 with phenacetin, bupropion and midazolam (i.e., individually) with shaking (200 rpm) on an orbital shaker (model BT1502, Benchmark Scientific, Edison, NJ, USA). Phenacetin o-deethylase, bupropion hydroxylase, and midazolam 1-hydroxylation activities were assessed using in situ metabolism evaluations of metabolite formation in cell culture median as previously described metabolic competence evaluations (Jackson et al., 2016; Ramaiahgari et al., 2017). All samples were stored at −80ºC prior to analysis by Liquid Chromatography-Mass Spectrometry (LC-MS).
2.3.2. CYP450 specific activities
The formation of acetaminophen, hydroxybupropion and 1-hydroxymidazolam was quantified by LC-MS. The analytical method used was a modification of protocol described earlier, and a complete list of instrumentation and analysis parameters are described in Supplementary Table 1–3. (Ramaiahgari et al., 2017). Briefly, 50 μL of incubation medium samples from each well were transferred to a 96-well plate and 170 μL of 0.1% formic acid in acetonitrile was added along with 10 μL of isotopically labeled internal standard mixture (Supplementary Table 1, 250 ng/mL each in acetonitrile). Samples were mixed, centrifuged (4000 rpm for 5 min) and 100 μL of the supernatant was transferred to a Waters Acquity 96-well plate and mixed with 100 μL of methanol:water (1:1). Samples were analyzed using an API 5000 triple quadrupole mass spectrometer (SCIEX, Concord, Ontario, Canada) coupled to a Waters Acquity HPLC system (Waters, Milford, MA, USA). A Phenomenex Luna C18 (50 × 2 mm, 5μm) Column (Phenomenex, Torrance, CA, USA) with LC gradient method consisting of mobile phase A (0.05% formic acid in 5 mM ammonium formate) and mobile phase B (0.05% formic acid in acetonitrile:methanol (95:5)) at 2% mobile phase B increased linearly to 5% over 0.5 minute, followed by Mobile phase B increase to 71% over 2 minutes and then return to 2% over 0.5 minutes. The injection volume was 10 μL. LC flow rates were 0.7 mL/min for acetaminophen method and 0.5 mL/min for hydroxybupropion and hydroxymidazolam method. Standard curves ranged from 1 to 1000 ng/ml. Standards were prepared by mixing 50 μL of blank media with 10 μL of spiking solution for each compound at each respective concentration level and extracted as per standards. Accuracy of all standards were within 15% of nominal for all concentrations except at limit of quantitation (LOQ, 1 ng/ml). A quadratic equation with 1/x weighting was used to relate peak area response ratio of analyte to internal standard and concentration. Concentrations of specific metabolites were calculated by using the analyte peak area/internal standard peak area and using regression analysis in Analyst 1.6.2 (SCIEX, Concord, Ontario, Canada) to determine the ng/ml quantities. Metabolite formation rates were normalized to number of cells and incubation time, and expressed as pmol/min-million cells. MichaelisMenten curve fitting was performed and graphed with GraphPad Prism (GraphPad Software, San Diego, CA).
2.4. Glucuronidation and sulfation activity
2.4.1. Probe substrates exposures
7HC (50 μM), BPAF (50 μM), and BPS (50 μM) were used as probe substrates to evaluate glucuronidation and sulfation proficiency with 3 cell lines used in Tox21 assays: HepG2, HEK293, and MCF7. Assay plates were incubated at 37°C, 5% CO2 with these drug substrates individually with shaking (200 rpm) on an orbital shaker (model BT1502, Benchmark Scientific, Edison, NJ, USA). Reactions were stopped at 150 minutes (PHH & Tox21 cell lines) and 24 hours (Tox21 cell lines) by addition of 100 μL cold acetonitrile to each well, and plates were then centrifuged at 100 x g for 10 minutes. 100 μL aliquots of solvent-crashed lysates were subsequently were transferred to analytical plates and stored in −80°C until analysis.
2.4.2. Glucuronidation and sulfation LC-MS
Phase II metabolites for 7HC, 7HCS and 7HCG were analyzed using a targeted Selected Ion Monitoring (SIM) method, while metabolites of BPAF and BPS were analyzed using a semi-targeted method with data collected in a full scan MS experiment but using inclusion lists of mass-to-charge values corresponding to glucuronide and sulfate conjugates of BPAF and BPS (Supplementary Table 4–5) using a Q Exactive Plus (Thermo Fisher, Waltham, MA, USA) with a Vanquish Ultra performance Liquid Chromatography (UPLC) system (Thermo Fisher, Waltham, MA, USA). Both methods used a Hypersil Gold aq Column (100 × 2.1 mm, 1.9 μm) (Thermo Fisher, Waltham, MA, USA). LC conditions consisted of a gradient method where mobile phase A was 0.1 % formic acid in water and mobile phase B was 0.1% formic acid in acetonitrile. The gradient method started with a 2-minute equilibration at 10% mobile phase B. Mobile phase B was then increased to 100% over 3 minutes and then held at 100% for 2 minutes. In the last minute of the gradient, mobile phase B was decreased back to 10%. LC flow rate was 0.3 mL/min. The injection volume was 2 μL. The column temperature was held at ambient temperature whereas the autosampler tray temperature was set at 4°C. The mass spectrometer was calibrated with Thermo Fisher Scientific positive mode calibration solution. A complete list of MS analysis parameters is described in Supplementary Table 6–8.
2.5. Metabolic clearance assays
2.5.1. Chemicals exposure
Metabolic clearance assays following disappearance of parent compound over time were performed to determine respective rates of molecular transformation for 7HC, BSAF, and BPS in 50-donor pooled suspensions of PHH and 3 Tox21 cell lines (HepG2, HEK293, and MCF7) that produced detectable metabolite concentrations during specific activity evaluations. For these exposures, 1 μM 7HC, BPAF, or BPS prepared in cell culture medium. Assay plates were incubated at 37°C, 5% CO2 with individual substrates with shaking (200 rpm) on an orbital shaker (model BT1502, Benchmark Scientific, Edison, NJ, USA). Reactions were stopped at times 0, 15, 30, 60, 90, 120, 240, and 1440 minutes by adding 100 μL of cold acetonitrile to each well (i.e., whole well crash), and centrifuged at 100 x g for 10 minutes. 100 μL of each acetonitrile-crashed lysate were transferred to analytical plates and stored in −80°C for subsequent LC-MS analysis.
2.5.2. Metabolic clearance LC-MS
Parent compounds 7HC, BPAF, and BPS were detected using a single ion monitoring method with MS/MS used for analyte confirmation. All data were collected using a Q Exactive Plus (Thermo Fisher, Waltham, MA, USA) with a Vanquish UPLC system (Thermo Fisher, Waltham, MA, USA). All compounds were analyzed in negative mode following LC separation with a Hypersil Gold aq Column (100 × 2.1 mm, 1.9 μm) (Thermo Fisher, Waltham, MA, USA) using a gradient method. LC conditions consisted of a gradient method where mobile phase A was 0.1 % formic acid in water and mobile phase B was 0.1% formic acid in acetonitrile. The gradient method started with a 2-minute equilibration at 10% mobile phase B. Mobile phase B was then increased to 30% over 2 minutes and then to 100% over the next 3 minutes. Gradient was held at 100% mobile phase B for a minute before being decreased back to 10% over the last minute of the run. The mass spectrometer was calibrated with Thermo Fisher Scientific positive mode calibration solution. A complete list of MS analysis parameters is described in Supplementary Table 9. Data was processed using Xcalibur 4.1 (Thermo Fisher, Waltham, MA, USA). Parent substrate concentration versus time plots were created using GraphPad Prism and fitted using non-linear regression analysis to determine half-life.
2.6. CYP450-Glo bioluminescent assays
Luminescent screening assays were established with CYP450-Glo assays (Promega, Madison WI) for CYP1A2 and CYP3A4/5. These assays monitor the conversion by CYPs of inactive D-luciferin derivatives to an active form that produces light when firefly luciferase is added to the reaction mixture, with light intensity proportionally changing with enzymatic activity (Larson et al., 2011). Tox21 cell lines were cultured as a monolayer in 96-well white plate (Corning, NY, USA) at ~50,000 cells/well for 24 hours. Then, cells were pretreated with the vehicle 0.1% DMSO (Basal), or OMP (100 μM), a well known CYP1A2 inducer, or ABT (1 mM), a CYP1A2 inhibitor, for 24 hours. In other experiments, cells were pretreated with the vehicle 0.1% DMSO (Basal), or RIF (20 μM), well known CYP3A4 inducer, or MI (30 μM), a CYP3A4 inhibitor, for 24 hours. Assay background (BGCYP1A2 or BGCYP3A4) was measured in medium without cells. After treatment, the medium was replaced with fresh medium containing Luciferin-1A2 P450-Glo substrate (6 μM), or Luciferin-3A4 P450-Glo substrate (3 μM), cells were incubated for an additional 60 minutes. CYP450-Glo bioluminescent assays were performed by manufacturer’s protocol using a CLARIOSTAR luminometer (BMG LABTECH).
2.7. Statistical Analysis
All data represent the mean ± standard deviation (SD) of three or four independent experiments (n = 3 or 4), unless otherwise stated, using Microsoft Excel. Graphs were plotted using GraphPad Prism 7 (GraphPad software, San Diego, CA). Student’s t-test or ANOVA with subsequent Dunnett’s test were used as appropriate, with a p < 0.05 considered significant.
3. Results
3.1. Phase I metabolism evaluation
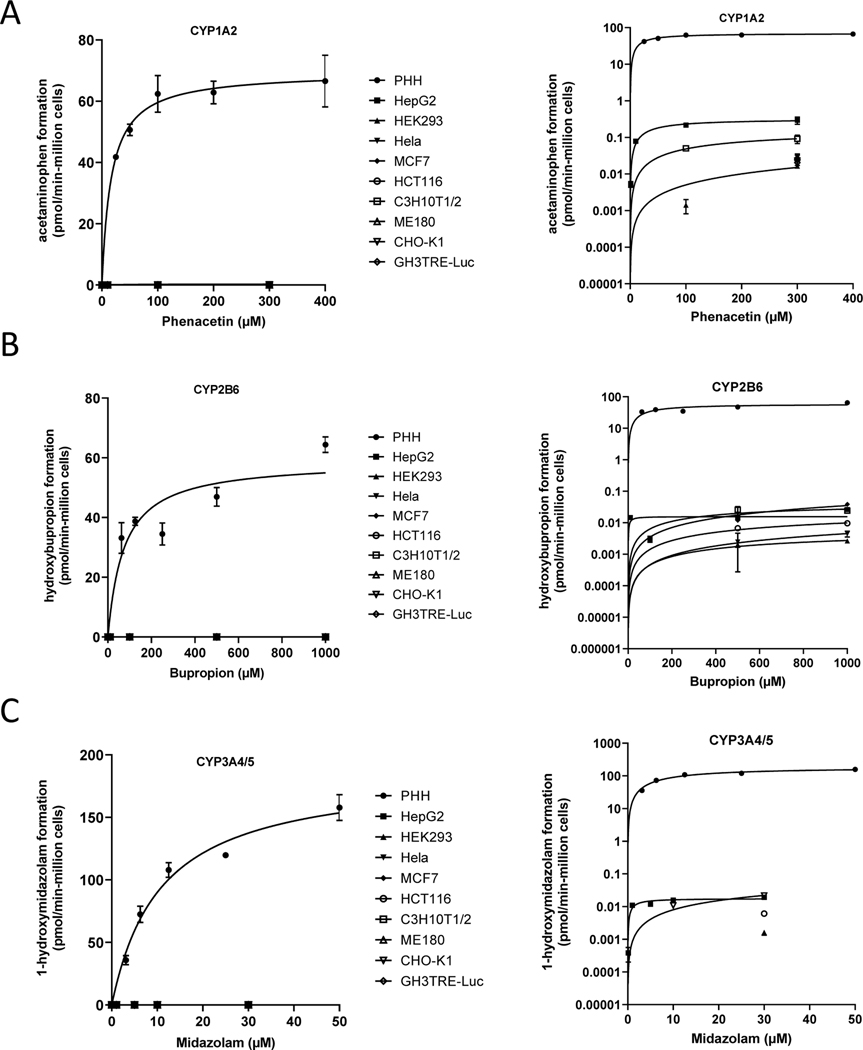
To characterize the perceived limitations for Tox21 cell lines to metabolize xenobiotics, HepG2, HEK293, Hela, HCT116, ME180, CHO-K1, GH3.TRE-Luc, C3H10T1/2 and MCF7 cultures were established and assayed in 96-well microplate formats for CYP specific activities to major human drug metabolizing enzymes CYP3A4/5, CYP1A2, and CYP2B6. Both substrate concentration and time course ranges were evaluated. As shown in Figure 1 and Table 1–3, robust metabolite formation for marker metabolites with clinical drug substrates phenacetin, bupropion, and midazolam was observed after 30 minutes incubations with suspensions of PHHs, which appeared to plateau with increasing substrate concentrations consistent with Michaelis-Menten kinetics. In contrast, Tox21 cell lines cultured over a much longer 24 hours incubation period produced profoundly lower specific activities requiring much longer incubation times to observe detectable metabolite formation. Determinable Vmax and Km values are shown in Table 4.
Figure 1. Metabolism comparison between PHHs and Tox21 cell lines.
Fifty-donor pool of PHHs and Tox21 cell lines at ~50,000 cells/well were treated with probe substrates phenacetin (PHH = 25, 50, 100, 200, 400, Tox21 = 0.1, 1, 10, 100, 300) bupropion (PHH = 62.5, 125, 250, 500, 1000, Tox21 = 1, 10, 100, 500, 1000), and midazolam (PHH = 3.125, 6.25, 12.5, 25, 50, Tox21 = 0.1, 1, 5, 10, 30). LC-MS assays were performed to quantify acetaminophen (A: CYP1A2), hydroxybupropion (B: CYP2B6), and 1-hydroxymidazolam (C: CYP3A4/5/7) plotted on both linear (left) and log (right) scales. Metabolite concentrations were converted to pmol of total metabolite per million cells plated over the respective incubation periods used for each model (pmol/min-106 cells).
Table 1.
Metabolism Comparison between PHHs and Tox21 Cell Lines for Acetaminophen Formation (pmol/min-106 cells); CYP1A2
| Phenacetin (μM) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cell Lines | Incubation Time | 0.1 | 1 | 10 | 25 | 50 | 100 | 200 | 300 | 400 |
|
| ||||||||||
| PHH | 30 min | - | - | - | 41.8 ± 0.353 | 50.7 ± 1.85 | 62.4 ± 6.00 | 62.8 ± 3.69 | - | 66.6 ± 8.43 |
| HepG2 | 24 h | 0.00 | 0.00527 ± 0.000925 | 0.0786 ± 0.00898 | - | - | 0.217 ± 0.0257 | - | 0.293 ± 0.0648 | - |
| HEK293 | 24 h | 0.00 | 0.00 | 0.00 | - | - | 0.00141 ± 0.000591 | - | 0.0163 ± 0.00150 | - |
| Hela | 24 h | 0.00 | 0.00 | 0.00 | - | - | 0.00 | - | 0.00 | - |
| MCF7 | 24 h | 0.00 | 0.00 | 0.00 | - | - | 0.00 | - | 0.00 | - |
| HCT116 | 24 h | 0.00 | 0.00 | 0.00 | - | - | 0.00 | - | 0.0248 ± 0.00770 | - |
| C3H10T1/2 | 24 h | 0.00 | 0.00 | 0.00 | - | - | 0.0500 ± 0.00414 | - | 0.0914 ± 0.0236 | - |
| ME180 | 24 h | 0.00 | 0.00 | 0.00 | - | - | 0.00 | - | 0.0294 ± 0.00589 | - |
| CHO-K1 | 24 h | 0.00 | 0.00 | 0.00 | - | - | 0.00 | - | 0.0233 ± 0.00102 | - |
| GH3TRE-Luc | 24 h | 0.00 | 0.00 | 0.00 | - | - | 0.00 | - | 0.0204 ± 0.00175 | - |
Table 3.
Metabolism Comparison between PHHs and Tox21 Cell Lines for 1-Hydroxymidazolam Formation (pmol/min-106 cells); CYP3A4/5
| Midazolam (μM) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cell Lines | Incubation Time | 0.1 | 1 | 3 | 5 | 10 | 12.5 | 25 | 30 | 50 |
|
| ||||||||||
| PHH | 30 min | - | - | 35.8 ± 3.65 | 72.4 ± 6.53 | - | 107.9 ± 5.87 | 119.80 ± 1.41 | - | 157.9 ± 10.3 |
| HepG2 | 24h | 0.00038 ± 0.00018 | 0.0109 ± 0.000694 | - | 0.0118 ± 0.000588 | 0.0157 ± 0.000372 | - | - | 0.0192 ± 0.000124 | - |
| HEK293 | 24h | 0.00 | - | 0.00 | 0.00 | - | - | 0.00167 ± 0.000283 | - | |
| Hela | 24h | 0.00 | - | 0.00 | 0.00 | - | - | 0.00 | - | |
| MCF7 | 24h | 0.00 | - | 0.00 | 0.00 | - | - | 0.00 | - | |
| HCT116 | 24h | 0.00 | - | 0.00 | 0.00 | - | - | 0.00607 ± 0.000435 | - | |
| C3H10T1/2 | 24h | 0.00 | - | 0.00 | 0.00 | - | - | 0.00 | - | |
| ME180 | 24h | 0.00 | - | 0.00 | 0.00 | - | - | 0.00 | - | |
| CHO-K1 | 24h | 0.00 | - | 0.00 | 0.0108 ± 0.00180 | - | - | 0.0217 ± 0.00212 | - | |
| GH3TRE-Luc | 24h | 0.00 | 0.00 | 0.00 | 0.00 | |||||
Table 4.
Specific Activity Maximum Enzymatic Velocity (Vmax) and Michelis-Menten Equilibrium Constant (Km) Estimates Summary
| Cell Type | Incubation Time | Vmax | Km | ||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| CYP1A2 | CYP2B6 | CYP3A4/5 | CYP1A2 | CYP2B6 | CYP3A4/5 | ||
| PHH | 30 min | 69.5 ± 2.12 | 59.0 ± 4.49 | 187 ± 8.35 | 16.7 ± 2.79 | 73.2 ± 23.3 | 10.7 ± 1.34 |
| HepG2 | 24 h | 0.320 ± 0.0234 | 0.0157 ± 0.00259 | 0.0177 ± 0.000912 | 38.4 ± 11.8 | 4.79 ± 5.48 | 0.969 ± 0.268 |
| HEK293 | 24 h | NA | 0.00616 ± 0.00221 | NA | NA | 1260 ± 734 | NA |
| Hela | 24 h | NA | NA | NA | NA | NA | NA |
| MCF7 | 24 h | NA | NA | NA | NA | NA | NA |
| HCT116 | 24 h | NA | 0.0286 ± 0.0110 | NA | NA | 1910 ± 1050 | NA |
| C3H10T1/2 | 24 h | 0.168 ± 0.0393 | 0.0422 ± 0.0119 | NA | 247 ± 111 | 566 ± 354 | NA |
| ME180 | 24 h | NA | NA | NA | NA | NA | NA |
| CHO-K1 | 24 h | NA | NA | 0.175 ± 0.300 | NA | NA | 209 ± 405 |
| GH3TRE-Luc | 24 h | NA | NA | NA | NA | NA | NA |
NA - Not applicable due to insufficient data for model fit
In general, hepatic lineage HepG2 cultures were the most proficient Tox21 cell line when summing CYP1A, CYP2B and CYP3A specific activities. The ranking for all Tox21 cell lines was HepG2 > C3H10T1/2 > CHO-K1 >> MCF7 ~ HCT116 ~ ME180 ~ HEK293 ~ GH3TRE-Luc > Hela. For CYP1A metabolism, seven cell lines produced detectable acetaminophen metabolite levels following phenacetin substrate exposures (24 hours). The relative ranking of activity was: HepG2 > C3H10T1/2 > ME180 > HCT116 > CHO-K1 > GH3TRE-Luc > HEK293 (Table 1).
For CYP2B activity, assessed by bupropion hydroxylation formation, 7 cell lines produced some detectable, but near the lower limit of quantitation activity after very long (i.e., 24-hour incubation periods) to accumulate detectable metabolite. The ranking of CYP2B activities observed was MCF7 > HepG2 > C3H10T1/2 > HCT116 > Hela > HEK293 at 1000 μM bupropion concentrations (Table 2). For CYP3A activity, only four cell line cultures produced detectable 1-hydroxymidazolam after 24 hours incubations with the following ranking CHO-K1 ~ HepG2 > HCT116 > HEK293 (Table 3). All Tox21 culture model activities, when benchmarked to PHH suspensions, produced very low CYP3A-type metabolism activities that are widely considered the most important enzymes for drug/xenobiotic metabolism in humans.
Table 2.
Metabolism Comparison between PHHs and Tox21 Cell Lines for Hydroxybupropion Formation (pmol/min-106 cells); CYP2B6
| Bupropion (μM) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cell Lines | Incubation Time | 0.1 | 1 | 10 | 62.5 | 100 | 125 | 250 | 500 | 1000 |
|
| ||||||||||
| PHH | 30 min | - | - | - | 33.1 ± 5.14 | - | 38.7 ± 1.34 | 34.5 ± 3.70 | 46.9 ± 3.10 | 64.4 ± 2.60 |
| HepG2 | 24 h | 0.00 | 0.00 | 0.0146 ± 0.000928 | - | 0.00305 ± 0.000668 | - | - | 0.0141 ± 0.000381 | 0.0262 ± 0.00206 |
| HEK293 | 24 h | 0.00 | 0.00 | - | 0.00 | - | - | 0.00203 ± 0.000380 | 0.00262 ± 0.000174 | |
| Hela | 24 h | 0.00 | 0.00 | - | 0.00 | - | - | 0.00245 ± 0.00218 | 0.00447 ± 0.000947 | |
| MCF7 | 24 h | 0.00 | 0.00 | - | 0.00 | - | - | 0.0119 ± 0.0206 | 0.0382 ± 0.00250 | |
| HCT116 | 24 h | 0.00 | 0.00 | - | 0.00 | - | - | 0.00667 ± 0.000451 | 0.00958 ± 0.000485 | |
| C3H10T1/2 | 24 h | 0.00 | 0.00 | - | 0.00 | - | - | 0.0257 ± 0.00337 | 0.0241 ± 0.000575 | |
| ME180 | 24 h | 0.00 | 0.00 | - | 0.00 | - | - | 0.00 | 0.00 | |
| CHO-K1 | 24 h | 0.00 | 0.00 | - | 0.00 | - | - | 0.00 | 0.00 | |
| GH3TRE-Luc | 24 h | 0.00 | 0.00 | - | 0.00 | - | - | 0.00 | 0.00 | |
Hepatic lineage HepG2 were only weakly effective in metabolizing phenacetin to acetaminophen (<1% of PHHs), bupropion to hydroxybupropion (<0.1% of PHHs) and midazolam to 1-hydroxymidazolam (< 0.1% of PHHs). These data benchmark and define the highly limited capacity of Tox21 cell lines to model major human xenobiotic metabolism functionality, and indicate extremely limited capacity for CYP1A-, CYP2B-, or CYP3A-dependent metabolic transformation or bioactivation in response to chemical exposures.
To further evaluate the extent of xenobiotic metabolism functionality with Tox21 cell lines, inhibitors of xenobiotic metabolism were applied. As shown in Supplemental Figure 1, HepG2, MCF7, CHO-K1, C3H10T1/2, and G3H3TRE-Luc were assayed using CYP1A2 luminescent substrate. Comparison of normalized luminescence data showed MCF7 > HepG2 > CHO-K1 >> C3H10T1/2 ~ G3H3TRE-Luc, and marginally inhibitable metabolite formation after exposure to the P450 inhibitor ABT only inhibited in CHO-K1 cells. These data were consistent with the very low enzymatic activities observed, and further confirm their deficiency for CYP450 metabolism.
While it is generally known that immortalized cell lines lack proficient signal transduction pathways related to drug metabolizing enzymes (e.g., PXR, CAR), we further evaluated Tox21 cell line exposures with prototype inducers of xenobiotic metabolism. AhR activator OMP only marginal increased luminescence ranking: MCF7 > CHO-K1 > HepG2 > C3H10T1/2 > G3H3TRE-Luc. Overall, CYP1A2/AhR functionality was highest in MCF7 among the Tox21 assays evaluated. For CYP3A/PXR functionality, HepG2 were superior in basal CYP3A luminescence with a ranking of HepG2 > MCF7 > HCT116 >> C3H10T1/2 ~ Hela (Supplemental Figure 2), which was appreciably inhibitable in all 5 cell lines with substrate-depleting high concentrations of MI with a ranking HepG2 > C3H10T1/2 > HCT116 > MCF7 >> Hela. As expected, no response to prototype PXR agonist rifampicin (RIF) was observed in any of the Tox21 cell lines evaluated, with the exception of a small increase in CYP3A4 activity in Hela cells.
3.2. Phase II metabolism evaluation
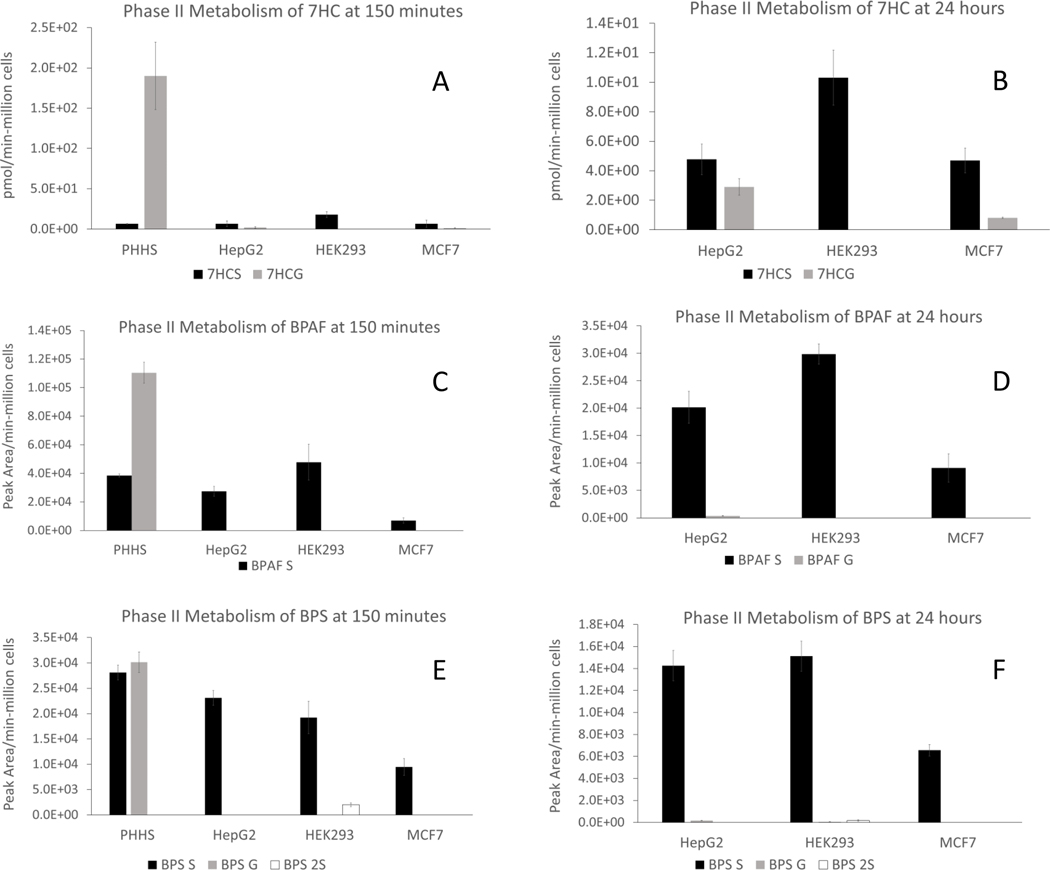
Phase II metabolism in Tox21 cell lines was evaluated with bisphenols BPAF and BPS, along with 7-HC, and metabolite formations were assayed using LC-MS. Direct overlay of existing Tox21 assay data for estrogen receptor (ER) activation revealed BPAF to be more potent than BPA or BPS (Supplemental Figure 3), thus investigation of their in vitro metabolism was explored (https://sandbox.ntp.niehs.nih.gov/tox21-curve-visualization (Huang et al., 2014). Preliminary experiments revealed detectable concentrations of 7HC-glucuronide (7HCG) and sulfate (7HCS) only in HepG2, HEK293 and MCF7 (data not shown). Based on these data, more extensive evaluation of glucuronide and sulfate metabolites of 7HC, BPAF, BPS (50 μM for each) was evaluated over 150 minutes and 24 hours incubations (i.e., 7HCG, 7HCS, BPAFG, BPAFC, BPSG, and BPSS). As shown in Figure 2 and Table 5, PHH suspensions produced far more 7HCG than 7HCS conjugate (Figure 2A) than Tox21 cell lines after 150 minutes. Since PHH suspensions lose cell viability after 2–4 hours in culture, it was not possible to evaluate the 24-hour time point. After 24 hours, HepG2 and MCF7 cultures achieved substantial amounts of 7HCG, whereas HEK293 produced no detectable 7HCG at either 150 minutes or 24 hours. However, 7HCS conjugates were detected in all 3 Tox21 cell lines (i.e., HepG2, HEK293 and MCF7) after both 150 minutes and 24 hours, demonstrating a greater proficiency for sulfation over glucuronidation.
Figure 2. Glucuronide and sulfate conjugates assay in PHH and three Tox21 cell lines.
HepG2, HEK293 and MCF7 cells were incubated at 50,000 cells/well for 24 hours individually in comparison to PHHs (50,000 cells/well) incubated with 50 μM 7HC (A, 150 minutes; B, 24 hours); 50 μM BPAF (C, 150 minutes; D, 24 hours), or 50 μM BPS (E, 150 minutes; F, 24 hours). LC-MS assays were performed to quantify 7HC-glucuronide and sulfate conjugates, semi-quantify BPAF-glucuronide, sulfate conjugates, BPS-glucuronide and sulfate conjugates. Metabolite concentrations were converted to pmol of total metabolite per million cells plated for 7HC conjugates (pmol/min-106 cells) and peak area of total metabolite per million cells plated (peak area/min-106 cells) for BPAF and BPS conjugates.
Table 5:
Phase II Metabolite Formation Rates
| 150 minutes | |||||||
|---|---|---|---|---|---|---|---|
| Metabolites | 7HCS | 7HCG | BPAF S | BPAF G | BPS S | BPS G | BPS 2S |
| Cell Lines | pmol/minute-million cells | Peak area/minute-million cells | Peak area/minute-million cells | ||||
| PHHs | 6.55 ± 0.365 | 190 ± 41.8 | 38500 ± 1110 | 110000 ± 7240 | 28100 ± 1440 | 30100 ± 2010 | -------------1 |
| HepG2 | 6.46 ± 3.63 | 1.96 ± 0.887 | 27400 ± 3450 | -------------1 | 23100 ± 1470 | -------------1 | -------------1 |
| HEK293 | 17.9 ± 3.56 | -------------1 | 47800 ± 12600 | -------------1 | 19200 ± 3210 | -------------1 | 1990 ± 378 |
| MCF7 | 6.63 ± 4.56 | 1.34 ± 0.206 | 7000 ± 1880 | -------------1 | 9460 ± 1680 | -------------1 | -------------1 |
| 24 hours | |||||||
| Metabolites | 7HCS | 7HCG | BPAF S | BPAF G | BPS S | BPS G | BPS 2S |
| Cell Lines | pmol/minute-million cells | Peak area/minute-million cells | Peak area/minute-million cells | ||||
| PHHs | -------------2 | -------------2 | -------------2 | -------------2 | -------------2 | -------------2 | -------------2 |
| HepG2 | 4.77 ± 1.04 | 2.89 ± 0.563 | 20200 ± 2930 | 375 ± 48.0 | 14300 ± 1380 | 170 ± 11.4 | -------------1 |
| HEK293 | 10.3 ± 1.86 | -------------1 | 29800 ± 1830 | -------------1 | 15100 ± 1360 | 53.8 ± 6.89 | 1990 ± 378 |
| MCF7 | 4.69 ± 0.848 | 0.809 ± 0.0457 | 9090 ± 2570 | -------------1 | 6560 ± 527 | -------------1 | -------------1 |
: Metabolite not detected
: PHH suspension cells no longer viable at this time point
BPAFG conjugate was readily observed in PHH suspensions after 150-minute incubations, with higher proportions of BPAFS vs. BPAFG (Figure 2C). By comparison, very low BPAFG was observed in these 3 Tox21 cell lines. Interestingly, BPAF sulfation was clearly observed in all 3 Tox21 cell lines after 150-minute incubations, with levels rivaling those produced in PHH suspensions. Only HepG2 cultures retained observable levels of both sulfate- and glucuronide-conjugated BPAF after 24 hours (Figure 2D), consistent with its hepatic lineage.
As shown in Figure 2E & 2F, BPS incubations produced robust BPSS and BPSG metabolites equivalent to 150-minute PHH incubations. In contrast, no detectable BPS G was observed in Tox21 cell lines after 150-minute incubations. BPSS was observed at comparable levels to PHH suspensions, consistent with results with 7HC and BPAF. After 24 hours incubations, BPSS concentrations further increased for all 3 Tox21 cell lines and was analogous to the profile observed for BPAFS with proportionally small amounts of BPSG. BPS2S conjugates (i.e., 2 sulfate conjugates) were also assayed and detectable in HEK293 cells after both 150 minutes and 24 hours (Figure 2E, 2F). Taken together, these data demonstrate that Tox21 cell lines HepG2, HEK293, and MCF7 do display robust Phase II sulfation capacity with all three substrates evaluated (7HC, BPAF, & BPS), yet minimally supported Phase II glucuronidation.
3.3. Metabolic clearance evaluation
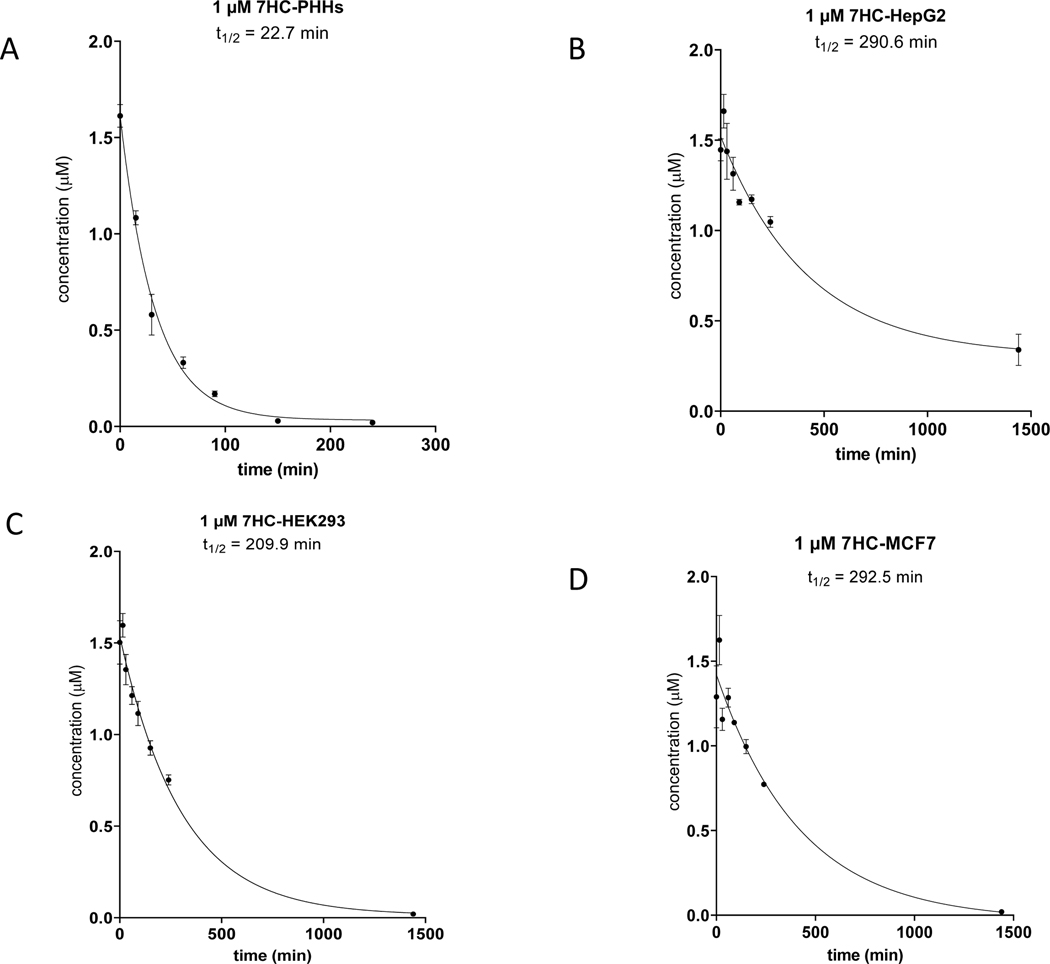
In vivo, the spectrum of adaptive and toxicological responses to xenobiotic exposure occurs as a result of the combined dynamic exposure to both parent chemical and downstream metabolites over time. Therefore, to better understand the impact of observed metabolite formations, the extent of metabolic clearance (i.e., loss of parent form of chemical to alternate forms) was further evaluated with HepG2, HEK293, and MCF7 cell lines. For this, 7HC, BPAF, and BPS were assayed over 24 hours (Table 6, Figures 3–5). Analogous to Phase II metabolite formation evaluations, 7HC was measured via LC-MS to monitor loss of 7HC (1 μM initial substrate concentration) and derive percent depletion and apparent half-life (T1/2) values (Table 6). 7HC was extensively ‘cleared’ (i.e., 99% conversion to other chemical structure forms) in PHH suspensions after 240-minute incubations, which revealed an estimated half-life of ~23 minutes (Table 6, Figure 3). Twenty-four-hour incubations with PHH suspensions were not possible due to their short-term viability (i.e., ~2–4 hours). In contrast, HepG2 cells revealed 27% depletion after 240 minutes with an estimated half-life of ~290 minutes. Interestingly, HEK293 had a faster loss of 7HC relative to hepatic lineage HepG2 with 50% depletion after 240 minutes, and 99% turnover after 24 hours with an overall T1/2 ~210 minutes. This observation was consistent with the elevated extent of 7HC sulfation observed in Figure 2. MCF7 produced a comparable extent of 7HC depletion with 40% depletion after 240 minutes with T1/2 ~ 290 minutes (Figure 3D).
Table 6.
Chemicals Clearance Comparison between PHHs and Tox21 Cell Lines
| Chemicals | Concentration (μM) | Cell Culture Model | Percent Depletion 240 min (% ±/- SD) | Percent Depletion 24 hour (% ±/- SD) | Mean Half-life (min)* | 95% Confidence Interval Half-life (min) | Fit Method Description |
|---|---|---|---|---|---|---|---|
| 7-HC | 1 | 50-donor pool PHHs | 99 ±/- 5.9 | Not determined | 22.7 | 19.8 – 26.1 | †One-phase exponential decay |
| 7-HC | 1 | HepG2 | 27 ±/−6.8 | 76 ±/- 11 | 290.6 | 196 – 512 | One-phase exponential decay |
| 7-HC | 1 | HEK293 | 50 ±/- 12 | †99 ±/- 12 | 209.9 | 173 – 259 | †One-phase exponential decay |
| 7-HC | 1 | MCF7 | 40 ±/- 18 | 99 ±/- 12 | 292.5 | 196 – 530 | †One-phase exponential decay |
| BPAF | 1 | 50-donor pool PHHs | 82 ±/- 2.9 | Not determined | 86.3 | 51.2 – 202 | One-phase exponential decay |
| BPAF | 1 | HepG2 | 36 ±/- 8.2 | 66 ±/- 6.6 | 31.2 (fast) 1,626 (slow) | very wide | Two-phase exponential decay |
| BPAF | 1 | HEK293 | 36 ±/- 5.4 | 60 ±/- 4.5 | 43.0 (fast) ~1,887 (slow) | very wide | Two-phase exponential decay |
| BPAF | 1 | MCF7 | 42 ±/- 7.7 | 55 ±/- 7.4 | 23.3 (fast) ~9,684 (slow) | very wide | Two-phase exponential decay |
| BPS | 1 | 50-donor pool PHHs | 59 ±/- 6.7 | Not determined | 205.2 | 73.5 – 268 | One-phase exponential decay |
| BPS | 1 | HepG2 | 17 ±/- 11 | 39 ±/- 7.4 | 326.1 | 164 – 3053 | One-phase exponential decay |
| BPS | 1 | HEK293 | 9.5 ±/- 11 | 36 ±/- 12 | NC | NC | One-phase exponential decay |
| BPS | 1 | MCF7 | 22 ±/- 7.0 | 26 ±/- 7.3 | 119.7 | 48.9 – 272 | One-phase exponential decay |
value from 150 minutes
value applies lower limit of quantification (LLOQ) where mean concentrations less than LLOQ
NC: Not calculated due to insufficient trend within dataset for fit model estimates
Figure 3. Clearance of 7HC by PHH and three Tox21 cell lines.
HepG2, HEK293 and MCF7 cells were incubated at 50,000 cells/well for 24 hours individually in comparison to PHHs (50,000 cells/well) incubated with 1.0 μM 7HC at 0,15, 30, 60, 90, 150 and 240 minutes. LC-MS analysis was performed to monitor the clearance of 7HC parent compound at 1.0 μM to derive percent depletion and apparent half-life (t1/2). 7HC clearance in PHHs (A); 7HC clearance in HepG2 cells (B); 7HC clearance in HEK293 cells (C); 7HC clearance in MCF7 cells (D).
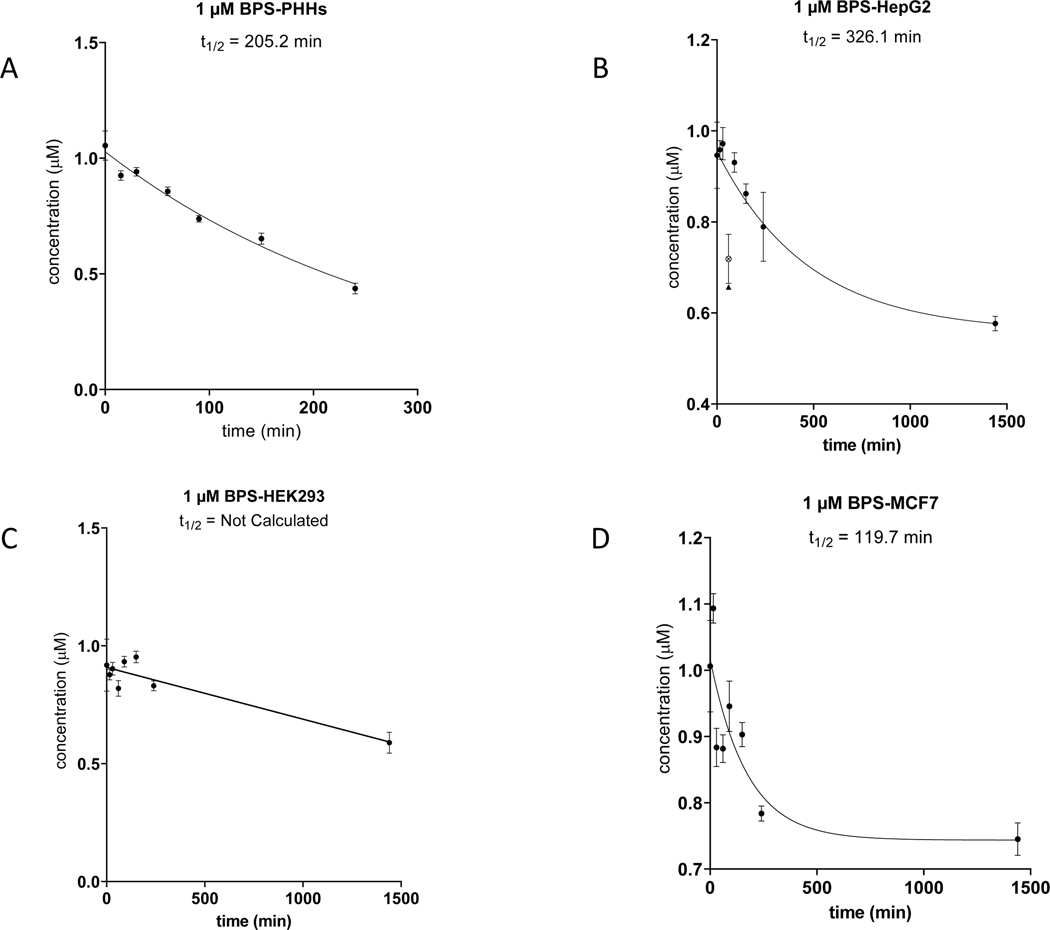
Figure 5. Clearance of BPS by PHH and three Tox21 cell lines.
HepG2, HEK293 and MCF7 cells were incubated at 50,000 cells/well for 24 hours individually in comparison to PHHs (50,000 cells/well) incubated with 1.0 μM BPS at 0, 15, 30, 60, 90, 150 and 240 minutes. LC-MS analysis was performed to monitor the clearance of BPS parent compound at 1.0 μM to derive apparent half-life (t1/2). BPS clearance in PHHs (A); BPS clearance in HepG2 cells (B); BPS clearance in HEK293 cells (C); BPS clearance in MCF7 cells (D).
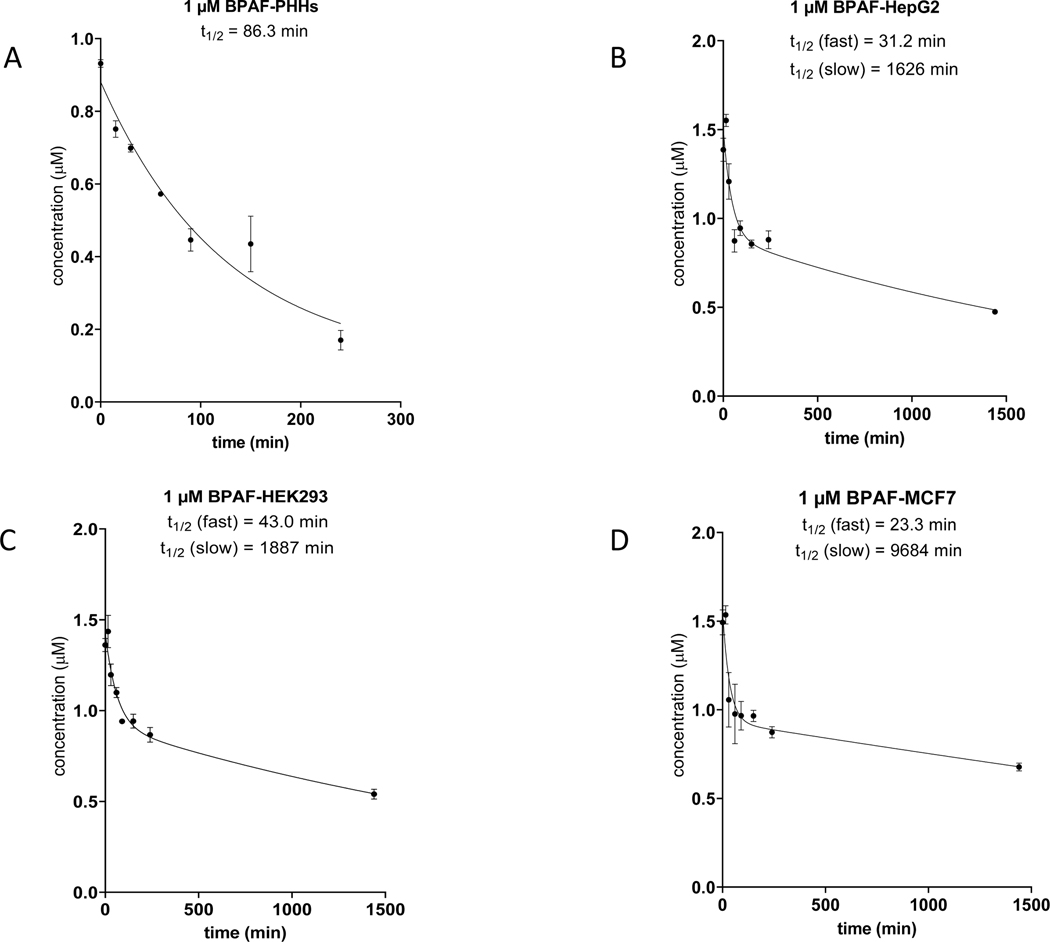
The metabolic clearance of BPAF was extensive with 7HC in PHH suspensions with 82% depletion and T1/2 ~86 minutes (Table 6, Figure 4). HepG2 cells revealed an apparent biphasic depletion with initial fast and delayed slow disappearance of BPAF with T1/2 ~31 and 1,600 minutes, respectively. Percent depletion of BPAF in HepG2 cells was 36% for 240 min and 66% after 24 hours. This extensive depletion of parent BPAF demonstrates Tox21 cell lines are in fact competent for xenobiotic metabolism by conversion parent chemicals to alternative but form, likely to be sulfation based on our observations of Phase II metabolism data. This non-zero parent chemical depletion has the potential to impact the interpretation of Tox21 assay data. HEK293 showed a similar biphasic depletion of BPAF with T1/2 ~43 and 1,900 minutes, respectively. MCF7 further mirrored the biphasic depletion of BPAF revealing the fastest initial metabolic clearance rate of across these three Tox21 cell lines with T1/2 ~23 minutes, and the slowest second phase with T1/2 ~9,700 minutes. These data were consistent with the extensive BPAF sulfate formation shown in Figure 2.
Figure 4. Clearance of BPAF by PHH and three Tox21 cell lines.
HepG2, HEK293 and MCF7 cells were incubated at 50,000 cells/well for 24 hours individually in comparison to PHHs (50,000 cells/well) incubated with 1.0 μM BPAF at 0, 15, 30, 60, 90, 150 and 240 minutes. LC-MS analysis was performed to monitor the clearance of BPAF parent compound at 1.0 μM to derive apparent half-life (t1/2). BPAF clearance in PHHs (A); BPAF clearance in HepG2 cells (B); BPAF clearance in HEK293 cells (C); BPAF clearance in MCF7 cells (D).
Metabolic clearance was by far the least extensive for BPS among these 3 compounds (i.e., 7HC, BPAF, and BPS). As shown in Table 6 and Figure 5, depletion of BPS was 59% after 240 minutes in PHH suspensions with correspondingly slower T1/2 ~205 minutes. In HepG2, significantly lower depletion of 17% was observed with BPS after the initial 240 minutes, and 39% turnover of BPS after 24 hours with overall half-life T1/2 ~326 minutes. Compared with HepG2, BPS turnover in HEK293 was only 9.5% after 240 minutes and 36% after 24 hours and an overall half-life that was not calculated due to insufficient trend within the dataset. By comparison, MCF7 produced the most extensive BPS depletion among Tox21 cell lines with ~30% depletion, which was largely achieved in the initial 240 minutes and ranked as the fastest BPS depletion half-life of T1/2 ~120 minutes. These observations were largely consistent with metabolite formation data.
4. Discussion
The Tox21 high throughput screening program has been criticized for a perceived lack of ‘metabolic competence’ across the suite of bioactivity assays screening thousands of environmental and pharmaceutical chemicals. However, to date this speculative critique has not been experimentally confirmed or characterized. Given the importance of biotransformation and internal exposure profiles to toxicological outcomes, the perceived yet poorly characterized lack of xenobiotic metabolism in Tox21 cell culture models, and the broad impact of Tox21 assay data made available to the public for thousands of chemicals, it is important to characterize and contextualize the capacity of Tox21 cell culture models for ‘metabolic competence’. In this study, we established a framework to benchmark ‘metabolic competence’ using the hepatic ‘gold standard’ of pooled-donor PHH suspensions, and characterized 9 commonly used Tox21 cell culture models for major drug metabolizing enzyme activities (e.g., CYP3A, CYP1A, CYP2B, UGT, SULT). The results highlight both anticipated and surprising results, which have implications to the field of in vitro toxicology and in vitro to in vivo translation of bioactivity assay data.
As we considered how best to approach the challenges in characterizing ‘metabolic competence’ within cell culture models used for Tox21 assays, we focused on CYP, UGT, and SULT metabolism assays. The rationale focused on their preeminent role in xenobiotic metabolism and clearance in humans, and our need to address the predominant modes of metabolic activation of chemicals to toxic metabolites that historically involve hepatic metabolism (Walsh J.S., 2006). Xenobiotic metabolism is known to both detoxify and activate toxicological responses, and can arise from both hepatic and extrahepatic metabolism (Jhajra et al., 2012). The enzymes commonly involved in metabolic activation often include UGTs (e.g., acyl glucuronide electrophiles), peroxidases, monoamine oxidases, and most often cytochromes P450 (CYPs).
CYPs form a variety of reactive metabolites including epoxides, quinones, heterocyclics, halocarbons, hydrazines, and free radical ions, and are attributed to bioactivation roles with well-known toxicants such as benzo(a)pyrene, aflatoxin B1, benzene, dimethylnitrosamine, carbon tetrachloride, and acetaminophen. In humans, CYPs drive the majority of xenobiotic metabolism clearance via CYP3A, CYP1A, CYP2C, enzymes, and to a less prevalent extent via CYP2B, CYP2E, and CYP2D enzymes. These enzymes typically initiate Phase I metabolism to transform lipophilic chemicals towards more hydrophilic forms poised for Phase II metabolism and excretion. This rationale guided our approach to evaluate, major CYP, UGT, and SULT metabolite formations, and metabolic clearance in context with PHHs suspensions. PHH suspensions integrate active/passive uptake transport, intrinsic production of requisite enzyme cofactors at physiological proportions (e.g., NADPH, UDPGA, PAPS), and functional expression of a physiologically-relevant spectrum of xenobiotic metabolizing enzymes produced in human liver within the initial 1–2 hours of incubation. The differentiation state of PHHs in culture can be inferred based on their CYP450 activities that dynamically change based on culture conditions. We observed that Tox21 cell lines were largely devoid of reference CYP450 metabolite formation relative to PHH suspensions using established clinical drug substrates for CYP3A4/5, CYP1A2, and CYP2B6 as sentinels to broader ‘metabolic competence’ (Jackson et al., 2016; Lu et al., 2015). It is important to note that while these specific activities (i.e., metabolite formation of acetaminophen, hydroxybupropion, and 1-hydroxymidazolam) are well-established for human liver enzymes, their specificity for metabolizing enzymes with non-human origin Tox21 cell lines CHO-K1 (hamster), GH3TRE (rat), and C3H3T1/2 (mouse) are likely more generalized reflections of CYP3A, CYP1A, and CYP2B metabolism as some conservation of metabolism is known across a CYP subfamily. Another consideration to contextualize these findings is that many of these Tox21 cell lines are not of hepatic lineage, thus lower activities for major xenobiotic metabolizing enzymes were anticipated given the importance of hepatic metabolism in drug clearance. Extrahepatic human tissues often vary in the degree and proportions of CYP expression, which can impact the specificity of metabolite formation due to substrate overlap among enzymes. A final point of context is that CYPs involved in endogenous metabolism such as CYP19A1 (i.e., aromatase catalyzing conversion of androgens to estrogens) with high specificity for endogenous substrates and narrow ranges of substrate recognition are most likely operative in Tox21 culture models, but generally do not serve as rate-determining xenobiotic metabolism/clearance. These considerations guided our rationale to evaluate a broad spectrum of xenobiotic metabolism substrate concentrations over shorter and longer (i.e., 24 hours) incubations with Tox21 cultures. Overall, these data definitively establish a profound lack of CYP-dependent ‘xenobiotic metabolism’, as defined within the rationale of this study, and establish a benchmarking framework to evaluate CYP-related ‘metabolic competence’ with in vitro toxicology models.
The most surprising finding within this study was the substantial capacity of Tox21 cell culture models to produce Phase II sulfate metabolites of 7HC, BPAF, and BPS that in some instances rivaled PHH suspensions. These data were further supported by metabolic clearance data indicating significant loss of parent chemical over 240 minutes and 24 hour exposures. These data demonstrated a clear ‘competence’ to appreciably biotransform phenolic chemicals. These data also showed a strong bias towards sulfation in Tox21 cell lines compared to PHH suspensions with more balanced glucuronidation and sulfation. The extents of metabolic clearance (i.e., disappearance of parent chemical over time) mirrored the extents of Phase II metabolite formation to sulfation/glucuronidation in most data sets. These data suggest that Phase II metabolism is driving the loss of parent BPS, BPAF, and 7HC in these cultures, but further study with inhibitors towards a mass balance evaluation would be required to confirm these coincident observations. It is interesting to note that with Tox21 cultures, though proficient for sulfation of BPAF and BPS akin to PHH suspensions, appeared to reach a plateau of parent chemical depletion with 1 μM exposures across multiple cell lines. This could reflect additional rate-determining factors such as free fraction protein binding (e.g., 10% FBS) or active transport processes that compartmentalize parent chemicals and limit the extent of turnover. In addition, UGT and SULT enzymes often display substrate affinity Km values in the higher micromolar to low millimolar range, which may also play a role in the observed limit of further BPS and BPAF depletion between 240 minutes and 24 hours. For context, Ishii and Tamura reported the apparent Km of bisphenol A for sulfotransferase activity was 74 μM, and the depletion of bisphenol A was only 4% after 5 days incubations with 10 μM exposure (Ishii and Tamura, 2003). Given the range of exposures and shorter (< 24 hours in most assays) exposure times for Tox21 assay systems, we could hypothesize from these observations that some phenolic and aromatic hydroxyl compounds, along with other sulfotransferase substrates, may deplete to some extent during Tox21 assays. Future analysis is required to assess the potential impact of these findings across the thousands of Tox21 chemicals evaluated to date. Depletion, along with the potential for skewed metabolite exposure profiles favoring sulfation, could result in shifted potencies that under- or over-estimate the potential for human biological activity and parent chemical responses in humans. These data highlight the challenge in ascribing Tox21 assay results to nominal exposure concentrations of parent chemicals that presume a lack of ‘metabolic competence’. Furthermore, a more nuanced definition of ‘metabolic competence’ reflective of tissue-specific biotransformation is needed. As we consider the fit-for-purpose use of different types of in vitro liver models, both HepG2 and PHHs likely have a useful role to fulfill. For high throughput screening, HepG2 are a practical and inexpensive model system with some degree of CYP1A metabolism, and surprisingly robust sulfation capacity. However, their profoundly limited CYP450 metabolism and hepatic receptor signaling pose fundamental challenges for translational hepatotoxicity predictions. Suspension cultures of PHHs remain the ‘gold standard’ for accurate estimations of human metabolism outcomes, but their extremely limited viability time frame (i.e., a few hours due to the absence of epithelial-like cell-cell and cell matrix interactions) essentially reflects the responses of dying cells that are incapable of toxicological modeling without more advanced culture configurations (e.g., sandwich cultures, 3D spheroids, microphysiological systems). Future studies to evaluate the in vitro disposition of Tox21 compounds should further improve and refine our understanding of the impacts of in vitro exposure profiles and ‘skewed’ metabolism on the translation of assay results to humans.
In conclusion, we have characterized and benchmarked a profound deficiency of major human CYP450 drug metabolism activities with 9 cell line backgrounds used in Tox21 assays relative to 50-donor pooled suspensions of primary human hepatocytes. Thus, compounds requiring hepatic CYP450 bioactivation would likely be poorly modeled with existing Tox21 assay systems. This approach establishes a framework for evaluation of human ‘metabolic competence’ with in vitro systems using pooled-donor suspensions of PHHs. Focused evaluation of phenolic and aromatic hydroxyl-containing compounds 7HC, BPAF, and BPS towards Phase II metabolism revealed a surprisingly robust capacity for sulfation over glucuronidation metabolism, which were consistent with metabolic clearance assay data. These results indicate a more nuanced definition of ‘metabolic competence’ is required as we expand the use of Tox21 assay data, and these findings likely have implications for the interpretation and quantitative translation of a subset of chemicals within existing Tox21 assay data appreciably metabolized by sulfotransferase enzymes.
Supplementary Material
Acknowledgments
We thank Drs. Esra Mutlu and Nisha Sipes for their helpful reviews of this manuscript. We want to thank Dr. Raymond Tice and John Bucher for their guidance in exploring these xenobiotic metabolism challenges for the Tox21 Program, and also thank Jui-Hua Hsieh for her assistance with Tox21 ER agonist data.
Quantitation of CYP activity assays was performed by RTI (research Triangle Park, NC) for the National Toxicology Program, National Institute of Environmental Health Sciences, National Institutes of Health, U.S. Department of Health and Human Services, under contract HHSN273201400022C.
NIH IRP project ES103318-04 (2019); Biomolecular Screening and Alternative Approaches for the National Toxicology Program; https://intramural.nih.gov/search/index.taf.
The authors received no specific funding for this work.
Abbreviations:
- ABT
aminobenzotriazole
- ACTT
American Type Culture Collection
- AOPI
acridine orange propidium iodide
- BPA
bisphenol A
- BPAF
bisphenol AF
- BPAF G
bisphenol AF glucuronide
- BPAF S
bisphenol AF sulfate
- BPS
bisphenol S
- BPS G
bisphenol S glucuronide
- BPS S
bisphenol S sulfate
- BPS 2S
bisphenol S 2 sulfate conjugates
- CYPs
cytochromes P450
- DMSO
dimethyl sulfoxide
- FBS
fetal bovine serum
- 7HC
7-hydroxycoumarin
- 7HCG
7-hydroxycoumarin glucuronide
- 7HCS
7-hydroxycoumarin sulfate
- LC-MS
liquid chromatography coupled to mass spectrometry
- MCF7
MCF-7aroERE cells
- MI
midazolam
- NAMs
new approach methods
- NTP
National Toxicology Program
- OMP
omeprazole
- PHHs
primary human hepatocytes
- RIF
rifampicin
- SD
standard deviation
- SULT
sulfotransferase
- Tox21
the Toxicology in the 21st Century Program
- UGT
UDP-glucuronosyltransferase
Footnotes
Declaration of Competing Interest
Authors declare that they have no conflicts of interest that might be relevant to the content of the manuscript.
References
- Collins FS, Gray GM, and Bucher JR, 2008. Toxicology. Transforming Environmental Health Protection. Science 319, 906–907. 10.1126/science.1154619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Extrahepatic Drug-Metabolizing Enzymes and Their Significance Part III. General Principles of ADME. Drug Transporters and Other Mechanisms of Transport Shalu Jhajra Ninad Ramesh Varkhede Deepak Suresh Ahire Bukke Vidyasagar Naik Bhagwat Prasad Jyoti Paliwal Saranjit Singh, October 2012, Wiley, 10.1002/9780470921920.edm028. [DOI] [Google Scholar]
- Fischer FC, Abele C, Henneberger L, Klüver N, König M, Mühlenbrink M, Schlichting R, Escher BI, 2020. Cellular Metabolism in High-Throughput In Vitro Reporter Gene Assays and Implications for the Quantitative In Vitro- In Vivo Extrapolation. Chem Res Toxicol. 33, 1770–1779. 10.1021/acs.chemrestox.0c00037 [DOI] [PubMed] [Google Scholar]
- Gerin B, Dell’Aiera S, Richert L, Smith S, Chanteux H, 2013. Assessment of cytochrome P450 (1A2, 2B6, 2C9 and 3A4) induction in cryopreserved human hepatocytes cultured in 48well plates using the cocktail strategy. Xenobiotica 43, 320–335. 10.3109/00498254.2012.719088. [DOI] [PubMed] [Google Scholar]
- Guengerich FP, 1997. Role of cytochrome P450 enzymes in drug-drug interactions. Adv. Pharmacol 43, 7–35. 10.1016/s0009-2797(97)00068-9. [DOI] [PubMed] [Google Scholar]
- Hewitt NJ, Lecluyse EL, Ferguson SS, 2007. Induction of hepatic cytochrome P450 enzymes: methods, mechanisms, recommendations, and in vitro-in vivo correlations. Xenobiotica 37, 1196–1224. 10.1080/00498250701534893 [DOI] [PubMed] [Google Scholar]
- Hsieh JH, Huang R, Lin JA, Sedykh A, Zhao J, Tice RR, Paules RS, Xia M, Auerbach SS, 2017. Correction: Real-time cell toxicity profiling of Tox21 10K compounds reveals cytotoxicity dependent toxicity pathway linkage. PLoS One 12, e0181291. 10.1371/journal.pone.0181291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R, Sakamuru S, Martin MT, Reif DM, Judson RS, Houck KA, Casey W, Hsieh JH, Shockley KR, Ceger P, Fostel J, Witt KL, Tong W, Rotroff DM, Zhao T, Shinn P, Simeonov A, Dix DJ, Austin CP, Kavlock RJ, Tice RR, Xia M, 2014. Profiling of the Tox21 10K compound library for agonists and antagonists of the estrogen receptor alpha signaling pathway. Sci Rep. 4, 5664–5672. 10.1038/srep05664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii Y, Tamura HO, 2003. Sulfoconjugation of bisphenol A in a human neuroblastoma cell line, NB-1. J Health Sci. 49, 319–323 10.1248/jhs.49.319 [DOI] [Google Scholar]
- Jackson JP, Li L, Chamberlain ED, Wang H, Ferguson SS, 2016. Contextualizing Hepatocyte Functionality of Cryopreserved HepaRG Cell Cultures. Drug Metab Dispos. 44, 1463–1479. 10.1124/dmd.116.069831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafert-Kasting S, Alexandrova K, Barthold M, Laube B, Friedrich G, Arseniev L, Hengstler JG, 2006. Enzyme induction in cryopreserved human hepatocyte cultures. Toxicology 220, 117–125. 10.1016/j.tox.2005.12.013. [DOI] [PubMed] [Google Scholar]
- Kavlock RJ, Austin CP, and Tice RR, 2009.Toxicity Testing in the 21st Century: Implications for Human Health Risk Assessment. Risk Anal 29, 485–497. 10.1111/j.1539-6924.2008.01168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson B, Moeller T, Banks P, Cali JJ, 2011. Automated triplexed hepatocyte-based viability and CYP1A and −3A induction assays. J Biomol Screen. 16, 895–902. 10.1177/1087057111411482. [DOI] [PubMed] [Google Scholar]
- Lee KS, Kim SK, 2013. Direct and metabolism-dependent cytochrome P450 inhibition assays for evaluating drug-drug interactions. J Appl Toxicol. 33, 100–108. 10.1002/jat.1720. [DOI] [PubMed] [Google Scholar]
- Li M, Yang Y, Yang Y, Yin J, Zhang J, Feng Y, Shao B, 2013. Biotransformation of bisphenol AF to its major glucuronide metabolite reduces estrogenic activity. PLoS One 8, 1–11. 10.1371/journal.pone.0083170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao C, Liu F, Guo Y, Moon HB, Nakata H, Wu Q, Kannan K, 2012. Occurrence of eight bisphenol analogues in indoor dust from the United States and several Asian countries: implications for human exposure. Environ Sci Technol. 46, 9138–9145. 10.1021/es302004w. [DOI] [PubMed] [Google Scholar]
- Lu JT, Einhorn S, Venkatarangan L, Miller M, Mann DA, Watkins PB, LeCluyse E, 2015. Morphological and Functional Characterization and Assessment of iPSC-Derived Hepatocytes for In Vitro Toxicity Testing, Toxicol Sci. 147, 39–54. 10.1093/toxsci/kfv117. [DOI] [PubMed] [Google Scholar]
- Mao J, Tay S, Khojasteh CS, Chen Y, Hop CE, Kenny JR, 2016. Evaluation of Time Dependent Inhibition Assays for Marketed Oncology Drugs: Comparison of Human Hepatocytes and Liver Microsomes in the Presence and Absence of Human Plasma. Pharm Res. 33, 12041219. 10.1007/s11095-016-1865-9. [DOI] [PubMed] [Google Scholar]
- National Toxicology Program (NTP) (2008). Available: http://ntp.niehs.nih.gov/ntp/htdocs/Chem_Background/ExSumPdf/BisphenolAF_093008_508.pdf#search=BisphenolAF. Accessed 10 July 2013
- Ramaiahgari SC, Waidyanatha S, Dixon D, DeVito MJ, Paules RS, Ferguson SS, 2017. From the Cover: Three-Dimensional (3D) HepaRG spheroid model with physiologically relevant xenobiotic metabolism competence and hepatocyte functionality for liver toxicity screening. Toxicol Sci. 159, 124–136. 10.1093/toxsci/kfx122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Ruan T, Wang T, Liu R, Jiang G, 2012. Distribution and preliminary exposure assessment of bisphenol AF (BPAF) in various environmental matrices around a manufacturing plant in China. Environ Sci Technol. 46, 13136–13143. 10.1021/es303960k. [DOI] [PubMed] [Google Scholar]
- Tice RR, Austin CP, Kavlock RJ, Bucher JR, 2013. Improving the human hazard characterization of chemicals: a Tox21 update. Environ Health Perspect. 121,756–765. 10.1289/ehp.1205784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker DK, Hayes Bouknight S, Brar SS, Kissling GE, Fenton SE, 2018. Evaluation of Prenatal Exposure to Bisphenol Analogues on Development and Long-Term Health of the Mammary Gland in Female Mice. Environ Health Perspect.126087003–17. 10.1289/EHP3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waidyanatha S,. Mathews JM, Patel P,R, Black SR, Snyder RW, Fennell TR, 2015. Disposition of bisphenol AF, a bisphenol A analogue, in hepatocytes in vitro and in male and female Harlan Sprague-Dawley rats and B6C3F1/N mice following oral and intravenous administration. Xenobiotica 45, 811–819. 10.3109/00498254.2015.1021732 [DOI] [PubMed] [Google Scholar]
- Walsh JS, 2006. Metabolic Activation-Role in Toxicity and Idiosyncratic Reactions. In: Borchardt RT, Kerns EH, Hageman MJ, Thakker DR, Stevens JL (eds) Optimizing the “Drug-Like” Properties of Leads in Drug Discovery. Biotechnology: Pharmaceutical Aspects, vol IV. Springer, New York, NY. [Google Scholar]
- Wang L, Zhang Z, Xu X, Zhang D, Wang F, Zhang L, 2015. Simultaneous determination of four trace level endocrine disrupting compounds in environmental samples by solid-phase microextraction coupled with HPLC. Talanta.142, 97–103. 10.1016/j.talanta.2015.04.043. [DOI] [PubMed] [Google Scholar]
- Wetmore BA, Wambaugh JF, Ferguson SS, Sochaski MA, Rotroff DM, Freeman K, Clewell HJ 3rd, Dix DJ, Andersen ME, Houck KA, Allen B, Judson RS, Singh R, Kavlock RJ, Richard AM, Thomas RS, 2012. Integration of dosimetry, exposure, and high-throughput screening data in chemical toxicity assessment. Toxicol Sci.125, 157–174. 10.1093/toxsci/kfr254. [DOI] [PubMed] [Google Scholar]
- Yamasaki K, Takeyoshi M, Yakabe Y, Sawaki M, Takatsuki M, 2003. Comparison of the reporter gene assay for ER-alpha antagonists with the immature rat uterotrophic assay of 10 chemicals. Toxicol Lett. 142, 119–131. 10.1016/s0378-4274(03)00019-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.