Abstract
In the present study, we utilize the well-characterized Escherichia coli lac repressor/operator system to demonstrate that protein binding can lead to demethylation at the binding sites in the chromosome. Similar to the findings using the episome, we found that the presence of LacI in the cells can lead to demethylation of methylated lacO in the chromosome and the LacI inhibitor, isopropyl-β-d-thiogalactopyranoside (IPTG), can prevent demethylation of the methylated lacO. The lacO sites become progressively more demethylated over time with the presence of LacI, supporting the role of protein occupancy in demethylation targeting. These results validate our earlier conclusions using a stable episomal system, and establish for the first time that protein binding can specify sites of demethylation in the chromosome.
INTRODUCTION
The pattern of DNA methylation in the mammalian genome is the result of the dynamic processes of methylation and demethylation. Significant changes in DNA methylation pattern occur during early embryonic development (Monk et al., 1987). It is generally believed that the methylation status is stable in adult somatic tissues. However, some randomness in addition to the consistent pattern at various loci has been observed in somatic tissues and cell lines as well as in embryonic cells, when the methylation pattern in each cell is examined with the high-resolution bisulfite genomic sequencing method (Macleod et al., 1994; Warnecke et al., 1998; Devereux et al., 1999; Warnecke and Clark, 1999; Vu et al., 2000). It is possible that DNA methylation and demethylation occur frequently at some sites and that the pattern of DNA methylation changes based on the activity of these two processes at these sites in each cell.
DNA demethylation can occur through a replication-dependent passive process, an active process by demethylase, or a combination of the passive and active processes. We have demonstrated previously, using a stable episomal system, that binding of nuclear antigen-1 (EBNA1) from Epstein–Barr Virus (EBV) to its replication origin oriP can lead to demethylation at the binding sites (Hsieh, 1999). Similarly, binding of the Escherichia coli lac repressor (LacI) to the lac operator (lacO) can also lead to demethylation of previously methylated lacO sites (Lin et al., 2000). This protein-specified DNA demethylation may involve a replication-dependent first-strand demethylation followed by a demethylase, which actively demethylates the second DNA strand (Hsieh, 1999). A 5-methylcytosine DNA glycosylase (5-MCDG) purified from chicken embryo demethylates hemimethylated DNA substrates also has the G/T mismatch DNA glycosylase activity (Jost, 1993; Zhu et al., 2000). This suggests the possibility that DNA repair machinery may be involved in the second step of the protein-mediated DNA demethylation. We further demonstrated that isopropyl-β-d-thiogalactopyranoside (IPTG) can modulate lacO demethylation by inhibiting LacI binding to lacO in the episomal system (Lin et al., 2000). This strongly suggests that occupancy of the binding site is critical for the protein-specified demethylation process. The interaction between a DNA-binding protein and its binding site could play an essential role in specifying sites of demethylation as proposed previously (Singer et al., 1979). The concentration and affinity of the DNA-binding proteins, their interaction with other proteins, and the available binding sites in the cell may be critically important for the protein-specified demethylation pathway. Protein–DNA and protein–protein interactions in each cell may play a key role in the demethylation process, and variations between these interactions may be responsible for some of the minor differences in the methylation pattern in each cell.
Experiments using an integrated, methylated adenine phosphoribosyltransferase (APRT) gene showed that an Sp1 site provides a signal for demethylation in mouse ES cells (Brandeis et al., 1994). However, it was not clear what the signal and mechanism of demethylation might be. Although we have shown that the protein-specified demethylation occurs on the stable episome (Hsieh, 1999; Lin et al., 2000), it has not been demonstrated that this process can occur in the chromosome. To test whether the protein-specified demethylation can occur in the chromosome, the same binding proteins and DNA sequences used on the stable episome can be tested in the chromosome. It is unclear how oriP would function when integrated into the chromosome because of its function as a replication origin. Therefore, we have chosen to integrate the lacO sites into the human chromosome to determine whether the protein-specified demethylation can occur in the genome.
RESULTS
Methylation is stably maintained in the lacO sites after integration into the human genome
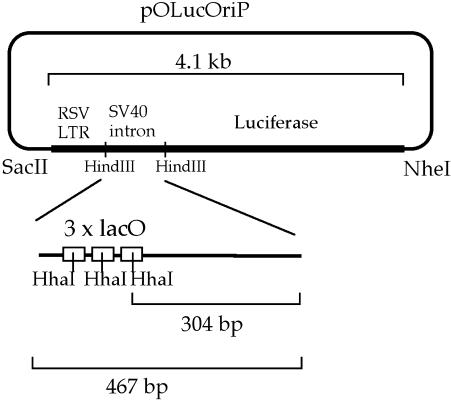
In order to test whether the presence of LacI can lead to demethylation of the lacO sequence as observed on the stable episome (Lin et al., 2000), cell lines with lacO sites integrated into the chromosome first have to be generated. Moreover, these lacO sequences have to be inserted in the genome in a methylated state and have to remain methylated after integration without going through rearrangement. An in vitro methylated SacII–NheI fragment from the plasmid, pOLucOriP, used in the stable episomal study (Lin et al., 2000) was integrated into the 293/EBNA1 cells by cotransfecting a puromycin expression vector, pLXSP. Four independent puromycin-resistant cell clones were analyzed for methylation status by probing the Southern blot of HindIII–HhaI double-digested genomic DNA from these cell clones with the 467 bp HindIII fragment. A single 467 bp band should be observed if the inserts remained methylated at all three HhaI sites within the lacO sequences. Demethylation of any copy of the insert at one or more of the three HhaI sites within the lacO sequence would lead to the detection of additional smaller size bands than the 467 bp band (Figure 1) on the Southern blot. A single 467 bp fragment was detected in HindIII–HhaI double-digested DNA from all four cell clones more than 20 days post transfection (Figure 2). This indicates that all three HhaI sites within the lacO sequences of all integrants remained methylated in these clones through multiple generations. Also, the SV40 intron containing the three lacO sites was not grossly rearranged in these cell lines.
Fig. 1. Illustration of pOLucOriP. The SacII–NheI fragment of 4.1 kb includes the RSV LTR, the SV40 intron harboring three copy of the lacO sequences, and the luciferase gene. The 304 bp band was desribed erroneously as 378 bp in a previous study (Lin et al., 2000).
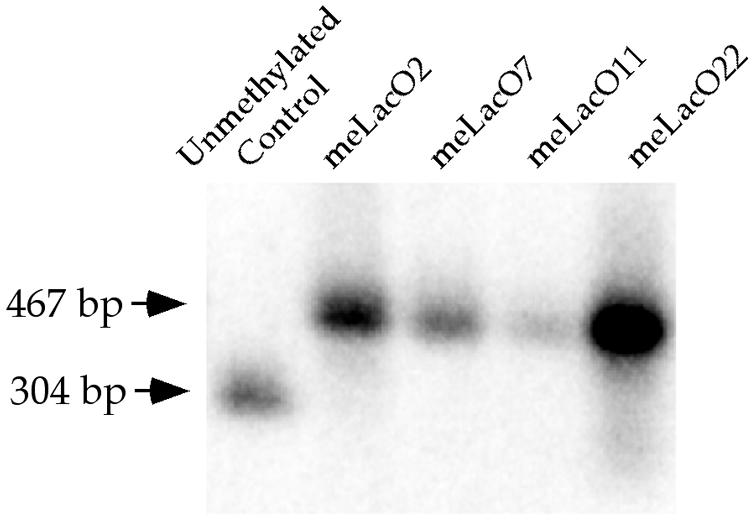
Fig. 2. Methylation status of the integrated lacO sites. Southern blot of HindIII–HhaI double-digested genomic DNA harvested from 22 to 32 days after transfection. The unmethylated control DNA is digested to completion; therefore, a 304 bp band is detected. The DNA from cell clones meLacO2, meLacO7, meLacO11, and meLacO22 remained methylated at the HhaI sites in the lacO sequence; therefore, only the 467 bp fragment is detected.
Stable expression of LacI does not lead to demethylation of the lacO sites when IPTG is present
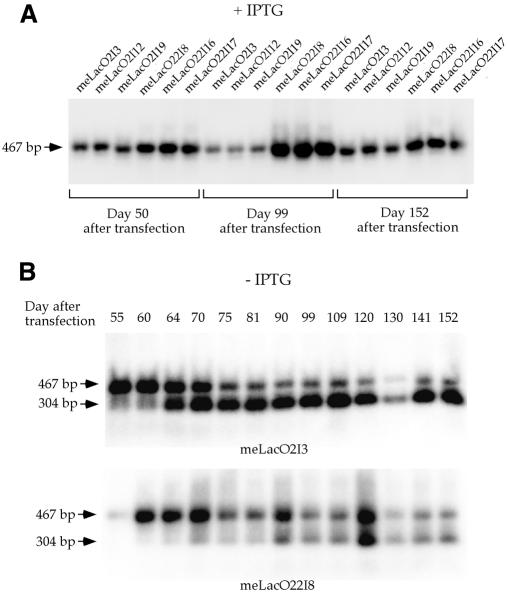
When LacI expression vector was transiently transfected into cells that harbor the lacO sequences in the genome, only a very low percentage of the lacO become demethylated (data not shown). This may be due to only a fraction of the cells being transfected with pCMVlacI and the limited level and relatively short duration of LacI expression from the transiently transfected plasmid. A higher percentage of the lacO sites on the stable episome become demethylated over time in the presence of LacI in previous experiments (Lin et al., 2000). It is possible that long-term expression of LacI can lead to demethylation in a higher percentage of the lacO sites integrated into the chromosome over time. However, the stable expression of LacI at a high level may lead to demethylation of the lacO sites even when IPTG, a LacI inhibitor, is present in the cells. To test this, the linearized pCMVlacI was integrated into both meLacO2 and meLacO22 cells with a DNA fragment containing the hygromycin gene. These two cell lines were chosen for this experiment because they contain a higher copy number of lacO integrants than meLacO7 and meLacO11 (data not shown); therefore, a small percentage of lacO demethylation can be detected without overloading the gel with total genomic DNA. It is known that methylated lacO sites on the episome can become demethylated when LacI is present in the cells, and IPTG can prevent this demethylation event. To prevent demethylation of the methylated lacO sites in the genome by the integration of LacI into the chromosome, IPTG was added to a final concentration of 5 mM at 5 h prior to transfection and at every media change thereafter. Twenty-four hygromycin resistant cell clones were isolated from each cell line. LacI expression in these cell clones was verified by immunofluoresence staining using the polyclonal rabbit anti-LacI antibody (Stratagene). Six cell clones (three from each of the meLacO2 and meLacO22 cell lines), meLacO2I3, meLacO2I12, meLacO2I19, meLacO22I8, meLacO22I16 and meLacO22I17, expressing LacI at a high level were used for further experiments. The IPTG treatment of these cell clones was continued through the entire experiment. Methylation status of the lacO sites was assessed by probing the Southern blot of HindIII–HhaI double-digested DNA, harvested 50, 99 and 152 days after LacI was transfected into the cell lines using the 467 bp HindIII fragment. A single 467 bp band was observed in all three cell clones from each cell line (Figure 3A). This clearly showed that demethylation of the lacO sites did not occur over a 152-day interval with constitutive expression of LacI when IPTG is present.
Fig. 3. Methylation status of the lacO sites in the chromosome with and without IPTG presence when LacI was stably expressed. (A) Three independent LacI expressing cell clones derived from each of the meLacO2 and meLacO22 were treated with IPTG from 5 h before LacI transfection to 50, 99 and 152 days after transfection. All DNA was double-digested with HindIII and HhaI. A single 467 bp band was detected in the HindIII–HhaI double-digested DNA from these cell clones, indicating that no demethylation occurred with IPTG presence even though LacI was stably expressed in the cells. (B) Progression of demethylation in the lacO sites with constitutive LacI expression when IPTG was absent. Southern blot of HindIII–HhaI digested DNA from cell clone meLacO2I3 and meLacO22I8 shows that the intensity of the 304 bp band representing the demethylated lacO sites increased over time while the intensity of the 467 bp band representing the methylated lacO sites decreased over time.
LacI binding can lead to demethylation of the lacO sites, and it is a progressive process when LacI is expressed constitutively
Demethylation of the lacO sites is expected to occur when IPTG is withdrawn from the cultures, while the lacO sites should stay methylated in the cells that continue to receive IPTG. Cells from each of the six cell clones, meLacO2I3, meLacO2I12, meLacO2I19, meLacO22I8, meLacO22I16 and meLacO22I17, were seeded in multiple tissue culture plates. IPTG was withdrawn from some of the cultures while it was maintained at a final concentration of 5 mM in other cultures as controls. Genomic DNA was harvested from these cells at 5 days after IPTG withdrawal, and a small fraction of the cells was replated. The same harvesting and replating procedure was repeated every 5 to 10 days after the first harvest. A total of 13 harvests were done over a 102-day interval (this was 152 days after LacI integration). The DNA harvested was double-digested with HindIII and HhaI for methylation analysis. No demethylation at the three HhaI sites within the lacO sequence was observed throughout the experimental interval of 102 days with IPTG present in the media as shown above. In contrast, 467 bp and 304 bp fragments were detected upon Southern blotting of HindIII–HhaI double-digested DNA from all 13 harvests of meLacO2I3 and meLacO22I8 after IPTG was withdrawn (Figure 3B). Furthermore, the relative intensity of the 304 bp band to the 467 bp band increased from the first harvest to the thirteenth harvest (Figure 3B). DNA harvested from meLacO2I12, meLacO2I19, meLacO22I16 and meLacO22I17 showed similar results (data not shown).
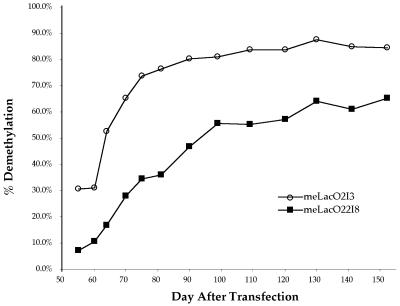
Quantitation of radioactivity in the 467 bp band and the 304 bp band detected in each DNA harvest revealed that demethylation at the three HhaI sites within the lacO sequences increased over time. Demethylation at these sites increased from 30.6% at 5 days after IPTG withdrawal (55 days after LacI integration) to 84.7% at 102 days after IPTG withdrawal (152 days after LacI integration) in cell line meLacO2I3 (Figure 4). It increased from 7.4% at 5 days after IPTG withdrawal to 65.6% at 102 days after IPTG withdrawal for cell line meLacO22I8 (Figure 4). Cell lines meLacO2I12, meLacO2I19, meLacO22I16 and meLacO22I17 showed different absolute percentages of demethylation but similar relative increases from day 5 after IPTG withdrawal to day 102 after IPTG withdrawal (data not shown).
Fig. 4. Percentage of demethylation in the lacO sites increases over time. The percentage of demethylation is derived from quantitation of the radioactivity in the 304 bp band and the 467 bp band in each lane of the Southern blot in Figure 3B after correcting for the fraction of the probe that can hybridize to each band.
These findings indicate that LacI binding to the lacO sites in the chromosome can lead to demethylation within these sequences. Over time, more lacO sites in the chromosome become demethylated if LacI was present for binding to these sequences. The meLacO2 and meLacO22 cell lines have lacO sequences integrated at different sites based on the differences in HindIII digestion patterns detected on Southern blot (data not shown). All three clones with LacI expression derived from each of the meLacO2 and meLacO22 cell lines showed similar results. This suggests that LacI mediated demethylation of lacO can occur at different chromosomal sites.
DISCUSSION
In this study, we demonstrated that: (i) methylated lacO sites remained methylated at different integration sites after integration into the human chromosomes; (ii) transient expression of LacI can lead to a low level of lacO demethylation in the chromosome; (iii) the methylated lacO sites did not become demethylated over a long interval with LacI expression when IPTG was present; (iv) methylated lacO sites became progressively more demethylated with LacI expression when IPTG was absent; and (v) methylated lacO sequences inserted at different chromosomal sites showed similar demethylation.
This study clearly showed that protein binding to DNA can lead to demethylation at the binding sites in the human chromosome similarly to what was observed on the episome previously (Lin et al., 2000). It has always been an important question whether the phenomenon observed on the stable episome is representative of what occurs in the chromosome. Although the oriP based stable episome replicates once per cell cycle and acquires nucleosomal structure (Stanfield-Oakley and Griffith, 1996), the lack of a positional effect can be a flaw due to the lack of higher order chromatin structure, despite being an advantage of the episomal system. It was shown previously that LacI binding leads to demethylation of the lacO sequences on a stable episome in human cells (Lin et al., 2000). Here we found that methylated lacO sequences in the human chromosome became demethylated when LacI was expressed in the cells. However, the lacO sequences in the cells expressing the same amount of LacI remained methylated when 5 mM IPTG was present in the tissue culture media. This illustrated that LacI binding to lacO sequence in the genome can lead to demethylation at the binding sites. This provides clear evidence that protein binding can target binding sites for demethylation in the genome. It is likely that the Sp1 binding to sites upstream of the APRT gene can target the binding sites as well as the region for demethylation by recruiting other factors to the region, thereby interfering with the remethylation of the in vitro methylated transgene as observed in the study by Brandeis et al. (1994).
In previous studies using the episomal system, there was strong evidence that demethylation of lacO sites is primarily determined by the occupancy of the lacO sites by LacI (Lin et al., 2000). In this study, only a fraction of the lacO sites become demethylated at any time point, and the percentage of demethylated lacO sites increased over the 98 days of the experimental interval. Furthermore, cell lines with the same lacO integration site, but different LacI integration sites, showed different amounts of demethylation with the same trend of increase over the experimental interval. These findings suggest that the amount of LacI in the cells plays a role in the percentage of lacO demethylation and the occupancy of lacO by LacI protein is important for lacO demethylation.
This study showed that a DNA-binding protein can specify demethylation at its binding site in the genome. DNA surrounding the lacO sites did not become demethylated regardless of the presence or absence of LacI (data not shown). This indicates the importance of the interaction between DNA-binding proteins and their binding sites in the demethylation process. The concentration and affinity of the DNA-binding proteins, their interaction with other proteins, and the available binding sites in the cell may be critically important for the dynamic process of demethylation. It is possible that demethylation can be one of the consequences of any alteration of the delicate balance between DNA and DNA-binding proteins. The variation in occupancy of the binding protein at its binding site in each cell may lead to the minor differences in methylation pattern from one cell to another. The dynamics of protein–DNA and protein–protein interaction may indirectly influence the DNA methylation pattern.
METHODS
Plasmids
The pOLucOriP has been described previously (Lin et al., 2000). A 4.1 kb SacII–NheI fragment containing the RSV LTR promoter, the SV40 intron with three copies of the lacO sites, and the luciferase gene was used for integration (Figure 1). The pCMVlacI, which has also been described previously, was used to supply LacI in human cells (Brown et al., 1987). The hygromycin resistance gene was isolated by a SalI–NruI double-digestion from plasmid pHyg (Sugden et al., 1985). Plasmid pLXSP was used for the puromycin resistance.
In vitro DNA methylation
The SacII–NheI DNA fragment from pOLucOriP was methylated at all CpG sites using the SssI-methylase (New England Biolabs) in vitro under the conditions recommended by the manufacturer. After methylation, DNA was extracted with phenol–chloroform and dialyzed with nitrocellulose filters (Millipore). The methylation status was confirmed by HhaI restriction enzyme digestion.
Cell line and transfection
The 293/EBNA1 cells that were used previously (Hsieh, 1994) were used in all experiments. The calcium phosphate transfection method (Wigler et al., 1979; Hsieh, 1994) was used throughout the study. The methylated SacII–NheI fragment containing the lacO sites (Figure 1) was integrated by cotransfecting 1 µg of the DNA fragment and 150 ng of the linearized puromycin expression plasmid, pLXSP, into 106 293/EBNA1 cells. Puromycin resistant clones were selected by 2.5 µg/ml of puromycin beginning at 48 h after transfection. Twenty-four puromycin resistant clones were isolated, and the successful integration and the methylation status of the inserted DNA fragment were confirmed by Southern blotting. These clones are designated meLacO followed by a number.
The LacI-expressing cell lines were generated by cotransfection of 2 µg of linearized pCMVlacI (Brown et al., 1987) and 100 ng of a 2 kb SalI–NruI DNA fragment containing the hygromycin resistance gene from pHyg (Sugden et al., 1985) into the meLacO2 and the meLacO22 cells in the presence of 5 mM IPTG. The hygromycin resistant cell clones from each cell line were selected using 200 µg/ml hygromycin from 2 days after transfection. Twenty-four clones from each cell line were isolated, and the expression of LacI was tested by immunofluorescent staining as described previously (Grawunder et al., 1996).
DNA recovery and analysis
When cells reached confluence in the experiments, 2.5% of the cells were replated on a 100-mm-diameter tissue culture plate, and genomic DNA was harvested from the remaining cells by the standard SDS lysis/proteinase K/phenol/chloroform method. Approximately 10 µg of the genomic DNA harvested from each cell line in the experiment was digested with HindIII alone or double-digested with HindIII and HhaI to assess the methylation status. Digested DNA was fractionated on 1% agarose gel, Southern transferred, and probed with the 467 bp HindIII fragment unless otherwise indicated. Southern blots were exposed to phosphoimaging screen and the radioactivity was quantitated with a phosphoimager (Bio-Rad FX). The amount of radioactivity in the 304 bp band was corrected for the size of the fragment because only a portion of the probe can hybridize to this fragment. The percentage of demethylation occurred at these HhaI sites within the lacO can be estimated by dividing the radioactivity in the 304 bp band by the total radioactivity in the 467 bp and the 304 bp bands.
Acknowledgments
ACKNOWLEDGEMENTS
The authors would like to thank F. Chedin, J. Castle and M.R. Lieber for critical reading of the manuscript. This work was supported by NIH grant GM54781.
REFERENCES
- Brandeis M., Frank, D., Keshet, I., Siegfried, Z., Mendelsohn, M., Nemes, A., Temper, V., Razin, A. and Cedar, H. (1994) Sp1 elements protect a CpG island from de novo methylation. Nature, 371, 435–438. [DOI] [PubMed] [Google Scholar]
- Brown M., Figge, J., Hansen, U., Wright, C., Jeang, K.-T., Khoury, G., Livingston, D.M. and Roberts, T.M. (1987) lac repressor can regulate expression from a hybrid SV40 early promoter containing a lac operator in animal cells. Cell, 49, 603–612. [DOI] [PubMed] [Google Scholar]
- Devereux T.R., Horikawa, I., Anna, C.H., Annab, L.A., Afshari, C.A. and Barrett, J.C. (1999) DNA methylation analysis of the promoter region of the human telomerase reverse transcriptase (hTERT) gene. Cancer Res., 59, 6087–6090. [PubMed] [Google Scholar]
- Grawunder U., Finnie, N., Jackson, S.P., Riwar, B. and Jessberger, R. (1996) Expression of DNA-dependent protein kinase holoenzyme upon induction of lymphocyte differentiation and V(D)J recombination. Eur. J. Biochem., 241, 931–940. [DOI] [PubMed] [Google Scholar]
- Hsieh C.-L. (1994) Dependence of transcriptional repression on CpG methylation density. Mol. Cell. Biol., 14, 5487–5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C.-L. (1999) Evidence that protein binding specifies sites of DNA demethylation. Mol. Cell. Biol., 19, 46–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost J.-P. (1993) Nuclear extracts of chicken embryos promote an active demethylation of DNA by excision repair of 5-methyldeoxycytidine. Proc. Natl Acad. Sci. USA, 90, 4684–4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin I.G., Tomzynski, T.J., Ou, Q. and Hsieh, C.-L. (2000) Modulation of DNA binding protein affinity directly affects target site demethylation. Mol. Cell. Biol., 20, 2343–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macleod D., Charlton, J., Mullins, J. and Bird, A.P. (1994) Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes Dev., 8, 2282–2292. [DOI] [PubMed] [Google Scholar]
- Monk M., Boubelik, M. and Lehnert, S. (1987) Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development, 99, 371–382. [DOI] [PubMed] [Google Scholar]
- Singer J., Roberts-Ems, J., Luthardt, E.W. and Riggs, A.D. (1979) Methylation of DNA in mouse early embryos, teratocarcinoma cells and adult tissues of mouse and rabbit. Nucleic Acids Res., 20, 2369–2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanfield-Oakley S.A. and Griffith, J.D. (1996) Nucleosomal arrangement of HIV-1 DNA: maps generated from an integrated genome and an EBV-based episomal model. J. Mol. Biol., 256, 503–516. [DOI] [PubMed] [Google Scholar]
- Sugden B., Marsh, K. and Yates, J. (1985) A vector that replicates as a plasmid and can be efficiently selected in B-lymphoblasts transformed by Epstein-Barr virus. Mol. Cell. Biol., 5, 410–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu T.H., Li, T., Nguyen, D., Nguyen, B.T., Yao, X.M., Hu, J.F. and Hoffman, A.R. (2000) Symmetric and asymmetric DNA methylation in the human IGF2-H19 imprinted region. Genomics, 64, 132–143. [DOI] [PubMed] [Google Scholar]
- Warnecke P.M. and Clark, S.J. (1999) DNA methylation profile of the mouse skeletal alpha-actin promoter during development and differentiation. Mol. Cell. Biol., 19, 164–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnecke P.M., Mann, J.R., Frommer, M. and Clark, S.J. (1998) Bisulfite sequencing in preimplantation embryos: DNA methylation profile of the upstream region of the mouse imprinted H19 gene. Genomics, 51, 182–190. [DOI] [PubMed] [Google Scholar]
- Wigler M., Sweet, R., Sim, G.K., Wold, B., Pellicer, A., Lacy, E., Maniatis, T., Silverstein, S. and Axel, R. (1979) Transformation of mammalian cells with genes from prokaryotes and eukaryotes. Cell, 16, 777–785.222468 [Google Scholar]
- Zhu B., Zheng, Y., Hess, D., Angliker, H., Schwarz, S., Siegmann, M., Thiry, S. and Jost, J.P. (2000) 5-methylcytosine-DNA glycosylase activity is present in a cloned G/T mismatch DNA glycosylase associated with the chicken embryo DNA demethylation complex. Proc. Natl Acad. Sci. USA, 97, 5135–5139. [DOI] [PMC free article] [PubMed] [Google Scholar]