Abstract
The MCM proteins are essential for the initiation of DNA replication. We have isolated an MCM3-associated protein (MCM3AP) in a two-hybrid screen using MCM3. Here we demonstrate that MCM3AP is an acetyltransferase which acetylates MCM3 and that chromatin-bound MCM3 is acetylated in vivo. The MCM3 acetylase, MCM3AP, is also chromatin-bound. This study also indicates that MCM3AP contains putative acetyl CoA binding motifs conserved within the GCN5-related N-acetyltransferase superfamily. Mutation of those motifs significantly inhibits the MCM3 acetylase activity. Over-expression of MCM3AP inhibits DNA replication, whereas mutation of the acetylase motifs abolishes this effect, suggesting that acetylation plays a role in DNA replication. Taken together, we suggest that MCM3 acetylation is a novel pathway which might regulate DNA replication.
INTRODUCTION
An essential feature of eukaryotic DNA replication is that DNA synthesis occurs once and only once per cell cycle. Although the detailed mechanisms that govern this regulation have not been clarified, several proteins are reported to be involved. After mitosis, origin recognition complex and Cdc6 recruit MCM proteins to form a pre-replication complex in G1 phase (for review see Tye, 1999; Ritzi and Knippers, 2000), and DNA replication is initiated upon activation of the complex by cyclin–Cdk and CDC7 kinases (Bousset and Diffley, 1998; Donaldson et al., 1998; Jiang et al., 1999). MCM proteins are essential for initiation of DNA replication and a sub-complex of three human MCM proteins, MCM4, 6 and 7, has weak DNA helicase activity at least in vitro (Ishimi, 1997; You et al., 1999). As an archaeal protein related to the MCM family also has DNA helicase activity (Kelman et al., 1999), MCM proteins may be required for strand unwinding during the initiation step of DNA replication. A recent study indicates that MCM proteins are essential for the elongation of DNA replication in addition to its initiation (Labib et al., 2000).
Histone acetyltransferases are implicated in the activation of RNA polymerase II-dependent transcription (Pineiro et al., 1991). Histone acetylation in a particular region of chromatin promotes destabilization of histone–DNA interactions in the nucleosome, resulting in increased accessibility of the chromatin to the transcription machinery. Targeted histone acetylation of a particular region is achieved by recruitment of acetyltransferases to the responsive promoters (for review see Cheung et al., 2000; Sterner and Berger, 2000). Recent studies indicate that some histone acetyltransferases acetylate non-histone proteins and modify the function of their substrate proteins (Gu and Roeder, 1997; Chen et al., 1999; Sartorelli et al., 1999; Martinez-Balbas et al., 2000). P/CAF is an acetyltransferase which acetylates non-histone protein, MyoD (Sartorelli et al., 1999) and E2F1 (Martinez-Balbas et al., 2000) in addition to the core histones. Acetylated MyoD shows increased affinity for its DNA target. Acetylation of E2F1 by P/CAF has three functional consequences on E2F1: increased DNA-binding ability, increased activation potential, and increased protein half-life. Thus, protein acetylation, like phosphorylation, is one of the post-translational modifications that regulates protein function (Kouzarides, 2000).
In the present study we focused on the possibility that acetylation might play a role for initiation of DNA replication in addition to RNA transcription. Here we demonstrate that endogenous MCM3 is acetylated and that an MCM3-associated protein (originally called Map 80, but called MCM3AP in this paper), which was isolated by two-hybrid screening using MCM3 as bait (Takei and Tsujimoto, 1998), is an MCM3 acetyltransferase.
RESULTS AND DISCUSSION
MCM3 is acetylated in vivo
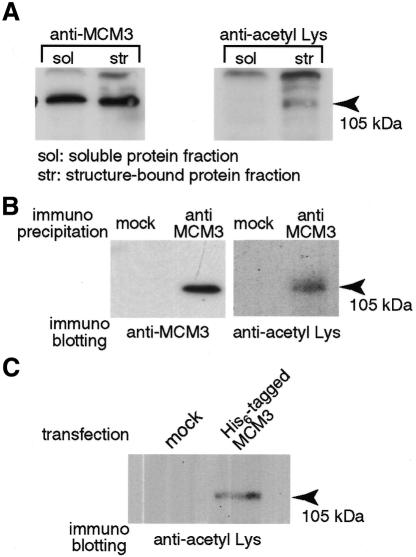
To observe endogenous MCM3 acetylation, protein fractions were prepared from soluble proteins (including dissolved membranes) and structure-bound proteins. MCM3 is known to bind to chromatin in G1 phase of the cell cycle and its binding is resistant to low concentrations of detergent but not to high concentrations of salt buffer. Thus, free MCM3 is fractionated into the soluble protein fraction whereas chromatin-bound MCM3 is fractionated into the structure-bound protein fraction (Coverley et al., 1998). Using an antibody which recognizes acetylated Lys, an acetylated protein which exhibited the same mobility as MCM3 on SDS–PAGE was observed in the structure-bound protein fraction but not in the soluble protein fraction (Figure 1A, lanes 3 and 4). These two fractions have equivalent amounts of MCM3 as demonstrated using an MCM3-specific antibody (Figure 1A, lanes 1 and 2). These results indicate that the anti-acetyl Lys antibody used in this assay cannot bind to MCM3 nonspecifically and suggest that MCM3 is acetylated endogenously. To confirm the in vivo acetylation of MCM3 further, we immunoprecipitated MCM3 and then probed with the anti-acetyl Lys antibody. Figure 1B showed that acetylated MCM3 can be detected in the anti-MCM3 immunoprecipitates. We can also detect acetylated MCM3 in the anti-acetyl Lys immunoprecipitates (data not shown). The results shown in Figure 1C also indicate in vivo acetylation of MCM3. His6-tagged MCM3 was expressed and purified from 293T cells to examine whether it is acetylated or not. The His6-tagged MCM3 was also detected with the anti-acetyl Lys antibody (Figure 1C). Taken together, these results indicate that MCM3 is acetylated endogenously.
Fig. 1. In vivo acetylation of MCM3. (A) The soluble protein fraction and the structure-bound protein fraction prepared from 5 × 105 asynchronous cells were analyzed by 7.5% SDS–PAGE. The proteins were blotted to PVDF membrane. The membranes were incubated with indicated antibody, respectively. The bound antibody was detected by horseradish peroxidase-labeled secondary antibody and visualized by ECL PLUS kit. (B) HeLa cell extracts prepared from 1 × 107 cells were incubated with protein A–Sepharose and with anti-MCM3 antibody or normal rabbit IgG (mock). After 1 h incubation, column-bound proteins were analyzed by SDS–PAGE (7.5%) and blotted to PVDF membrane. (C) MCM3/pcDNA3.1 plasmid or mock vector was transfected to 293T cells, respectively. After culture for 20 h the cells were harvested and resuspended in hypotonic buffer containing 1% NP40, 0.5 M NaCl, and 7 mM Na butyrate. Cell extracts were incubated with a TALON column and the column was washed extensively with same buffer. Bound proteins were extracted from the column with 100 mM imidazole and 7 mM Na butyrate in hypotonic buffer and analyzed by SDS–PAGE. The analyzed proteins were blotted to PVDF membrane.
HeLa cells were arrested in late G1 phase and M phase with mimosine and nocodazole, respectively. Chromatin-bound MCM3 disappeared in nocodazole treated cells (M phase) and appeared in mimosine treated cells (late G1 phase) since chromatin-binding of MCM proteins is tightly regulated by the cell cycle (Figure 2; anti-MCM3) (Ritzi and Knippers, 2000). Acetylated MCM3 was still observed in the structure-bound protein fraction of mimosine treated cells but not in the soluble protein fraction of mimosine treated cells or in either fraction of nocodazole treated cells, consistent with the observations in Figure 1A. Moreover, the MCM3AP is also detected only in the structure-bound protein fraction of mimosine treated cells (Figure 2, anti-MCM3AP).
Fig. 2. Effects of cell cycle stage on MCM3 acetylation. The soluble protein fraction and the structure-bound protein fraction prepared from 5 × 105 cells (for anti-acetyl Lys antibody) or from 1 × 105 cells (for other antibodies) treated with nocodazole or mimosine were analyzed by 7.5% SDS–PAGE. The proteins were blotted to PVDF membrane. The membranes were incubated with the indicated antibody, respectively.
MCM3AP acetylates MCM3
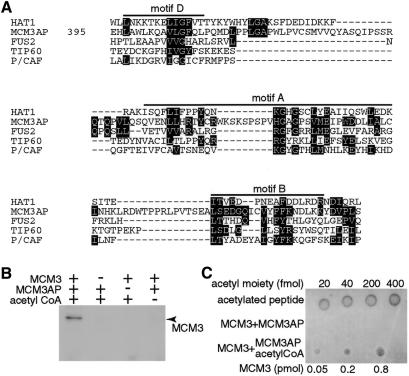
The fact that MCM3AP co-localizes with acetylated MCM3 prompted us to consider the possibility that MCM3AP is an acetyltransferase. Examination of the MCM3AP sequence reveals that this protein has sequence similarity to the GCN5-related N-acetyltransferase (GNAT) superfamily (Neuwald and Landsman, 1997; Dutnall et al., 1998). Figure 3A shows that MCM3AP shares conserved sequence motifs with members of the GNAT superfamily. The GNAT superfamily includes almost all acetyl CoA binding proteins, N-acetyltransferases, metabolic enzymes, and detoxification and drug resistance enzymes. Structural analyses of yeast HAT1 (Dutnall et al., 1998) and human P/CAF (Clements et al., 1999) indicate that the conserved sequence motifs in Figure 3 form part of the acetyl CoA binding site. MCM3AP has a unique insertion of sequence at amino acids 460–470 in the center of motif A. This unique sequence of MCM3AP, amino acids 460–470, contains three putative phosphorylation sites, 464S, 466S and 468S. Two of these are SP sequences, which may imply that MCM3AP activity may be regulated in a cell cycle specific fashion.
Fig. 3. MCM3AP-mediated acetylation of MCM3. (A) The HAT domain of yeast HAT1 (HAT1), human putative tumor suppressor (FUS2), human Tat interactive protein (60 kDa) (TIP60), and human P/CAF (P/CAF) were compared with MCM3AP. Conserved amino acids between HAT and MCM3AP were indicated by inverted characters. The alignment of MCM3AP was adjusted manually on the basis of classification of amino acid residues: aromatic residues (F, W and Y); negative charged residues (D, E); positive charged residues (H, K, R); aliphatic residues (I, L, and V). (B) MCM3 was incubated with MCM3AP and acetyl CoA or only one of them at 30°C. After incubation the samples were analyzed by SDS–PAGE and blotted to PVDF membrane. (C) After incubation with MCM3AP in the presence or absence of acetyl CoA, the indicated amount of MCM3 was spotted onto nitrocellulose membrane. Acetylated histone H4 peptide was spotted as a standard to evaluate the amount of acetyl moiety.
We next tried to establish if MCM3AP has intrinsic acetyltransferase activity. As shown in Figure 3B, recombinant MCM3 was detected by the anti-acetyl Lys antibody only when it was incubated with both MCM3AP and acetyl CoA. The extent of MCM3 acetylation under our conditions was 50–240 fmol/pmol MCM3, estimated with acetylated histone H4 N-terminal peptide as standard (Figure 3C). MCM3AP also acetylates histones to a much lesser extent than MCM3 (data not shown).
We generated a mutation in the amino acid sequence of MCM3AP, 471HGAG to 471AAAA, which corresponds to the center of motif A to confirm that the conserved sequence observed in MCM3AP is important for the acetyltransferase activity of MCM3AP. The same protein amounts of wild type and mutant MCM3AP were examined for their MCM3 acetylase activity with 14C-labeled acetyl CoA under the same conditions. Acetyltransferase activity towards MCM3 was severely decreased by this mutagenic change (Figure 4A). These results confirm that MCM3 is acetylated by MCM3AP and indicate that the conserved sequence included in MCM3AP is important for its acetyltransferase activity. Figure 4B shows that the mutant MCM3AP used in Figure 4A can still bind to MCM3. In the presence of MCM3–Gal4 DNA binding domain, the fusion of mutant MCM3AP with the Gal4 activation domain activated the expression of a reporter gene, HIS3, as well as the wild type MCM3AP fusion.
Fig. 4. Effects of mutation at amino acids included in putative acetyl CoA binding site. (A) His6-tagged wild-type (wt) or mutant (mut, 471HGAG to AAAA) MCM3AP was incubated with or without MCM3 in the presence of 14C-labeled acetyl CoA. The same amount of wt and mutant MCM3AP were examined for acetyltransferase activity under the same conditions. (B) Y190 yeast strains transfected with the indicated plasmids were cultured for 4 days at 30°C on -LWH SD plates containing 25 mM 3-AT. BD; Gal4 binding domain fusion, AD; Gal4 activation domain fusion. (C) GFP-tagged wt or mutant MCM3AP were expressed in 293T cells. The proportion of GFP positive cells that were also BrdU positive is indicated. TOTO3 positive images were used to identify non transfected cells. The extent of BrdU positive cells in TOTO3 positive but GFP negative images is also indicated as ‘not transfected’.
The possibility that the acetylation activity affects DNA replication was assessed since MCM3 is an essential factor for DNA replication. Expression of wild type MCM3AP decreased the number of DNA-replicating cells, whereas mutant MCM3AP examined in Figure 4A and B showed no inhibition compared to vector-transfected cells (Figure 4C). When mutant MCM3AP or vector alone were transfected, 35% of cells were labeled with 5-bromo-2′-deoxyuridine (BrdU), whereas, when wild type MCM3AP was transfected, ∼20% of cells were labeled. These results indicate that the acetylase-activity of MCM3AP can inhibit DNA replication.
In summary, we demonstrate that endogenous MCM3 is acetylated and that the acetylated component of MCM3 is chromatin-bound. We also indicate here that an MCM3AP is MCM3 acetyltransferase which is able to inhibit DNA replication. Thus, the present study provides a link between acetylation and DNA replication by identifying an enzyme which may be dedicated to acetylation of replication proteins. We propose to change the name ‘MCM3-associated protein’ to ‘MCM3 acetylating protein (MCM3AP)’. Biochemical comparison of acetylated MCM3 and non-acetylated MCM3 should determine precise roles of the MCM3 acetylation.
METHODS
Cells, plasmids and recombinant proteins
HeLa cells and 293T cells were cultured in 10% FCS in DMEM. The MCM3:pcDNA3.1 (Invitrogen), MCM3AP:pEGFP (Clontech) and mutant MCM3AP:pEGFP C2 were transfected into 293T cells using LipofectAMINE PLUS reagent (Gibco-BRL). To generate mutant MCM3AP, we used a QuickChangeTM Site-Directed Mutagenesis Kit purchased from Stratagene, and all assays were carried out according to the manufacturer’s instruction. The full-length MCM3AP inserted in pAcHLT was used as the template. The DNA sequence of mutant MCM3AP was confirmed.
For expression of human MCM3 and wild type and mutant MCM3AP in Sf9 cells, we used pAcHLT vector (Pharmingen). These plasmids were co-transfected with BaculoGold DNA (Pharmingen) into Sf9 insect cell line with Superfectin transfection reagent (Gibco-BRL) to generate recombinant baculoviruses. The recombinant viruses were co-cultured with Sf9 at 27°C. After 48 h these cells were harvested and crushed by sonication in sonication buffer (10 mM HEPES-Na pH 7.4, 1 mM MgCl2, 2 mM 2-mercaptoethanol, 5 mM imidazole, and protease inhibitors). The cell extracts were obtained as a centrifugation supernatant at 10 000 g for 20 min. The extracts were applied to a TALON Superflow metal affinity column (Clontech) equilibrated with the sonication buffer. The columns were washed extensively with wash buffer (20 mM HEPES-Na pH 7.4, 1 mM MgCl2, 2 mM 2-mercaptoethanol, 10 mM imidazole, 10% glycerol and protease inhibitors) including 500 mM NaCl or 0.1% Triton X-100, sequentially. The fractions eluted with wash buffer including 70 mM imidazole were diluted 10 times with the sonication buffer and applied on a new TALON Superflow metal affinity column. The columns were washed and eluted as above.
To make mutant MCM3AP inserted in pGAD424 (Clontech), mutant MCM3AP cDNA was cut out from pAcHLT then inserted in pGAD424 at BamHI and SalI sites. The yeast strain Y190 was purchased from Clontech. Its genotype is his3-200, lys2-801 and LYS2::GAL1UAS-HISTATA-HIS3. The method for transfection of plasmids to Y190 was described previously (Takei and Tsujimoto, 1998).
In Figure 4B, cells were labeled with 50 µM BrdU for 1.5 h, 20 h after transfection, then fixed with 2% paraformaldehyde and denatured in 50 mM NaOH for 5 min. Incorporated BrdU was probed with anti-BrdU antibody (Amersham Pharmacia) and visualized by Texas Red labeled secondary antibody. Chromatin was stained by TOTO3 dye. Cells were observed with confocal microscopy and TOTO3 positive images were recognized as cells. The GFP positive and BrdU positive cells were counted to estimate the proportion of DNA-replicating cells. At least 100 GFP positive cells in each sample were observed and the experiment was repeated three times.
Drug treatment and cell fractionation
Exponentially growing HeLa cells were cultured with 0.5 mM mimosine or 50 ng/ml nocodazole in the growing medium. After incubation for 25 h the cells were washed with ice cold PBS and used for preparation of cell extracts.
Exponentially growing HeLa cells or drug-treated cells were harvested and washed with ice-cold hypotonic buffer (10 mM HEPES-KOH pH 7.3, 5 mM KCl, 1.5 mM MgCl2, 1 mM DTT and protease inhibitors). The cells were resuspended in the hypotonic buffer containing 1% NP40 and incubated on ice for 15 min. After incubation the cells were centrifuged at 15 000 r.p.m. for 5 min. The supernatant was stored at –80°C as the soluble protein fraction. The precipitate was washed with hypotonic buffer containing 1% NP40 and resuspended in hypotonic buffer containing 1% NP40 and 0.5 M NaCl. After incubation for 15 min on ice the cells were centrifuged at 15 000 r.p.m. for 5 min. The supernatant was stored at –80°C as the structure-bound protein fraction.
For immunoprecipitation of MCM3, HeLa cells were broken by sonication in hypotonic buffer containing 1% NP40 and 0.5 M NaCl. The cells were centrifuged at 15 000 r.p.m. for 5 min. The supernatant was diluted by hypotonic buffer 5 times to decrease the concentration of detergent and salt and then incubated with antibodies and protein A.
Acetyltransferase assay
For acetylation of MCM3, 0.5 µg of recombinant MCM3 was incubated with 0.1 µg of MCM3AP and 20 µM acetyl CoA in the acetylation buffer consisting of 50 mM Tris–HCl pH 8.0, 0.1 mM EDTA, 10 mM Na-butyrate, 1 mM DTT, 10% glycerol. After incubation at 30°C for 1 h the samples were separated by SDS–PAGE and transferred to a PVDF membrane at a constant voltage of 15 V for 1 h. The membrane was blocked with 3% BSA and 0.25% Triton X-100 in PBS and probed with anti-acetyl Lys antibody (Cell Signaling Ltd.) in 5% BSA and 0.25% Triton X-100 in PBS. The bound antibodies were detected with peroxidase-conjugated secondary antibody (Amersham Pharmacia) using 1/5000 dilution in 3% BSA and 0.25% Triton X-100 in PBS and the protein bands were visualized by the ECL PLUS kit (Amersham Pharmacia).
For acetylation with 14C acetyl CoA, 5 µg of recombinant MCM3 was incubated with MCM3AP (0.5 µg) in the presence of 14C acetyl CoA (Amersham Pharmacia) in the acetylation buffer. After 1 h incubation at 30°C, samples were analyzed by SDS–PAGE and blotted to PVDF membrane. The membrane was exposed to X-ray film for 5–7 days at –80°C.
In Figure 3C, after acetylation the reaction mixture was directly spotted onto nitrocellulose membrane. The membrane was dried and incubated with 3% BSA in PBS, then probed with anti-acetyl Lys antibody (Upstate biotechnology) in 3% BSA in PBS. The bound antibodies were detected with peroxidase-conjugated secondary antibody (Amersham Pharmacia) using 1/5000 dilution in 3% BSA in PBS and visualized by the ECL PLUS kit.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Marion Ritzi and Rolf Knippers for gift of anti-MCM3 antibody, and David Santamaria and Cristina Pelizon for helpful comments. M.S. was supported by a Cancer Research Campaign studentship. A.T. and G.T. were supported in part by a Grant for the Organized Research Combination System from the Science and Technology Agency of Japan. This work was supported by the Cancer Research Campaign and the Louis-Jeantet Foundation.
REFERENCES
- Bousset K. and Diffley, J.F. (1998) The Cdc7 protein kinase is required for origin firing during S phase. Genes Dev., 12, 480–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Lin, R.J., Xie, W., Wilpitz, D. and Evans, R.M. (1999) Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell, 98, 675–686. [DOI] [PubMed] [Google Scholar]
- Cheung W.L., Briggs, S.D. and Allis, C.D. (2000) Acetylation and chromosomal functions. Curr. Opin. Cell. Biol., 12, 326–333. [DOI] [PubMed] [Google Scholar]
- Clements A., Rojas, J.R., Trievel, R.C., Wang, L., Berger, S.L. and Marmorstein, R. (1999) Crystal structure of the histone acetyltransferase domain of the human PCAF transcriptional regulator bound to coenzyme A. EMBO J., 18, 3521–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coverley D., Wilkinson, H.R., Madine, M.A., Mills, A.D. and Laskey, R.A. (1998) Protein kinase inhibition in G2 causes mammalian Mcm proteins to reassociate with chromatin and restores ability to replicate. Exp. Cell Res., 238, 63–69. [DOI] [PubMed] [Google Scholar]
- Donaldson A.D., Fangman, W.L. and Brewer, B.J. (1998) Cdc7 is required throughout the yeast S phase to activate replication origins. Genes Dev., 12, 491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutnall R.N., Tafrov, S.T., Sternglanz, R. and Ramakrishnan, V. (1998) Structure of the histone acetyltransferase Hat1: a paradigm for the GCN5-related N-acetyltransferase superfamily. Cell, 94, 427–438. [DOI] [PubMed] [Google Scholar]
- Gu W. and Roeder, R.G. (1997) Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell, 90, 595–606. [DOI] [PubMed] [Google Scholar]
- Ishimi Y. (1997) A DNA helicase activity is associated with an MCM4, -6, and -7 protein complex. J. Biol. Chem., 272, 24508–24513. [DOI] [PubMed] [Google Scholar]
- Jiang W., McDonald, D., Hope, T.J. and Hunter, T. (1999) Mammalian Cdc7-Dbf4 protein kinase complex is essential for initiation of DNA replication. EMBO J., 18, 5703–5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelman Z., Lee, J.K. and Hurwitz, J. (1999) The single minichromosome maintenance protein of Methanobacterium thermoautotrophicumΔH contains DNA helicase activity. Proc. Natl Acad. Sci. USA, 96, 14783–14788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. (2000) Acetylation: a regulatory modification to rival phosphorylation? EMBO J., 19, 1176–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K., Tercero, J.A. and Diffley, J.F. (2000) Uninterrupted MCM2-7 function required for DNA replication fork progression. Science, 288, 1643–1647. [DOI] [PubMed] [Google Scholar]
- Martinez-Balbas M.A., Bauer, U.M., Nielsen, S.J., Brehm, A. and Kouzarides, T. (2000) Regulation of E2F1 activity by acetylation. EMBO J., 19, 662–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald A.F. and Landsman, D. (1997) GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem. Sci., 22, 154–155. [DOI] [PubMed] [Google Scholar]
- Pineiro M., Gonzalez, P.J., Hernandez, F. and Palacian, E. (1991) Interaction of RNA polymerase II with acetylated nucleosomal core particles. Biochem. Biophys. Res. Commun., 177, 370–376. [DOI] [PubMed] [Google Scholar]
- Ritzi M. and Knippers, R. (2000) Initiation of genome replication: assembly and disassembly of replication-competent chromatin. Gene, 245, 13–20. [DOI] [PubMed] [Google Scholar]
- Sartorelli V., Puri, P.L., Hamamori, Y., Ogryzko, V., Chung, G., Nakatani, Y., Wang, J.Y. and Kedes, L. (1999) Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Mol. Cell, 4, 725–734. [DOI] [PubMed] [Google Scholar]
- Sterner D.E. and Berger, S.L. (2000) Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev., 64, 435–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei Y. and Tsujimoto, G. (1998) Identification of a novel MCM3-associated protein that facilitates MCM3 nuclear localization. J. Biol. Chem., 273, 22177–22180. [DOI] [PubMed] [Google Scholar]
- Tye B.K. (1999) MCM proteins in DNA replication. Annu. Rev. Biochem., 68, 649–686. [DOI] [PubMed] [Google Scholar]
- You Z., Komamura, Y. and Ishimi, Y. (1999) Biochemical analysis of the intrinsic Mcm4-Mcm6-Mcm7 DNA helicase activity. Mol. Cell. Biol., 19, 8003–8015. [DOI] [PMC free article] [PubMed] [Google Scholar]