Abstract
The Jun activating binding protein (JAB1) specifically stabilizes complexes of c-Jun or JunD with AP-1 sites, increasing the specificity of target gene activation by AP-1 proteins. JAB1 is also known as COP9 signalosome subunit 5 (CSN5), which is a component of the COP9 signalosome regulatory complex (CSN). Over the past year, JAB1/CSN5 has been implicated in numerous signaling pathways including those that regulate light signaling in plants, larval development in Drosophila, and integrin signaling, cell cycle control, and steroid hormone signaling in a number of systems. However, the general role of the CSN complex, and the specific role of JAB1/CSN5, still remain obscure. This review attempts to integrate the available data to help explain the role of JAB1/CSN5 and the COP9 signalosome in regulating multiple pathways (in this review, both JAB1 and CSN5 terminologies are used interchangeably, depending on the source material).
Introduction
The Jun activating binding protein (JAB1) was first identified by Claret and co-workers because of its interaction with the activation domain of c-Jun (Claret et al., 1996). JAB1 stabilizes complexes of the transcription factors c-Jun or JunD at their specific AP-1 transcription factor binding sites, increasing the specificity of target gene activation. A homologous protein was identified in Arabidopsis as a component of the COP9 signalosome (CSN) (Kwok et al., 1998), and subsequently termed COP9 signalsome subunit 5 (CSN5) (Deng et al., 2000). JAB1/CSN5 is an ∼40 kDa soluble protein. Like other CSN subunits, JAB1/CSN5 is highly conserved between the Plant and Animal Kingdoms, with the homologous proteins from Arabidopsis and humans sharing >60% amino acid identity. Unlike other CSN subunits, a probable CSN5 homolog is present in Saccharomyces cerevisiae (M. Glickman, personal communication), implying that at least in yeast, CSN5 can function independently of the CSN. CSN5 contains an MPN (MPR1–PAD1–Nterm) domain in its N-terminus (Hofmann and Bucher, 1998). This domain is common to several of the subunits of the CSN, to the regulatory lid of the proteasome and to the translation initiation factor eIF3, as well as being present in several unknown proteins (Glickman et al., 1998; Hofmann and Bucher, 1998; Wei et al., 1998). The function of this domain is still unknown, although it may have a role in protein–protein interactions.
In addition to its established interactions with c-Jun and subunits of the CSN, JAB1/CSN5 also has been shown to interact with a variety of other proteins. Common themes of possible significance are the frequent occurrence of interactions between JAB1/CSN5 and its partner proteins in the context of multi-protein complexes, and the apparent connection of these interactions to ubiquitin-dependent protein degradation pathways.
The COP9 signalosome
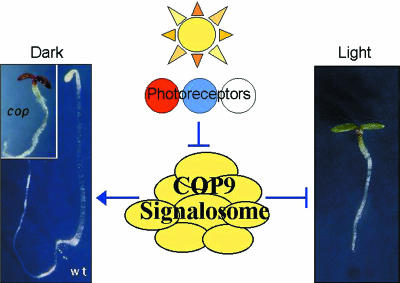
Before delving further into the data available for CSN-independent interactors, it is important to understand the COP9 signalosome and the presumed role of CSN5 within this complex. This complex was first isolated from plants as an essential regulator of light-mediated development (Chamovitz et al., 1996; reviewed in Karniol and Chamovitz, 2000). The CSN is ∼450 kDa in mass and comprises eight core subunits termed CSN1–CSN8, in order of descending subunit size. As mentioned above, the individual subunits are highly conserved across diverse species. The complex is essential for both plant and animal development. In both Arabidopsis and Drosophila, mutations in different subunits of the CSN lead to multiple pleiotropic defects that culminate in developmental arrest and death. In Arabidopsis these phenotypes include constitutive photomorphogenesis (causing plants to grow in the dark as if exposed to light), cellular and subcellular differentiation defects, stunted stature and the deregulation of numerous genes (Figure 1).
Fig. 1. Model of COP9 signalosome (CSN) function in photomorphogenesis. In the absence of light signals, the CSN represses photomorphogenesis (Light) leading to dark-grown (Dark) growth patterns (wt, left). Part of the repression is mediated through the degradation of the HY5 transcription factor (not shown). Light signals transduced from the photoreceptors lead to a release of this repression. Mutations in the CSN lead to constitutive photomorphogenesis (cop) in the dark. A representative dark-grown cop mutant, and dark- and light-grown wild-type (wt), Arabidopsis seedlings are shown.
Within the CSN complex, CSN5 interacts with at least four other subunits, CSN1, CSN2, CSN4 and CSN7 (reviewed in Kapelari et al., 2000). In Arabidopsis and Drosophila, CSN5 is present both in the CSN complex and in CSN-independent forms (Kwok et al., 1998; Freilich et al., 1999). It has yet to be determined whether these two forms are present in mammals. Both forms appear to have functional significance as, at least in Arabidopsis, mutations in other loci can affect their distribution differently (Kwok et al., 1998). For example, mutations in several other CSN subunits lead to a complete loss of the 450 kDa CSN-dependent form of CSN5, but maintain the smaller CSN-independent forms. Conversely, mutations in two other Arabidopsis loci, COP1 and DET1, lead to a loss of the smaller, CSN-independent forms of CSN5 while maintaining the intact CSN-dependent form (Kwok et al., 1998). Interestingly, though perhaps counter-intuitively, plants with null mutations in either COP1 or DET1 are phenotypically indistinguishable from mutants for CSN subunits.
The connection of COP1 to the CSN complex has led to a potential connection of the CSN to the proteasome. COP1 is a Zn-Ring finger, WD-40 domain protein that has been suggested to function as an E3 ubiquitin ligase for specific proteins (Osterlund et al., 2000). In dark-grown plants COP1 localizes to the nucleus, where it binds to, and perhaps mediates the proteasome-dependent degradation of, the HY5 transcription factor. In the light, on the other hand, COP1 is found in the cytoplasm, and HY5 protein accumulates in the nucleus. In mutants with a defective CSN complex, COP1 remains cytoplasmic in the dark, HY5 accumulates and light-regulated genes are transcribed (Osterlund et al., 2000). Together with the reported interaction between subunits of the CSN and the proteasome (Kwok et al., 1999) and the copurification of the two complexes or their subunits (Karniol et al., 1998, Seeger et al., 1998), these findings indicate that the CSN, and thus CSN5, may be involved in the regulation of protein degradation.
JAB1 interactors
The recently identified JAB1/CSN5 interactors include a number of diverse proteins that play roles in many cell processes (see Table I). It could be claimed that JAB1/CSN5 is simply promiscuous. At least one study has shown that JAB1 interacts with the DNA binding domainGal4 alone to activate both the HIS3 and LacZ reporter genes (O. Gabrielsen, personal communication). This does not rule out, though, that JAB1 specifically interacts in yeast with other proteins to get higher levels of reporter gene activation. As detailed in Table I, the JAB1 interactions have undergone additional biochemical controls including in vitro pull-downs and immunoprecipitations, using either recombinant JAB1/CSN5 or the endogenous protein. The true test for these interactions, however, is their biological relevance, and many studies have been carried out to illustrate the pertinence of these protein–protein interactions. Clearly the list of interactions is long, and the literature is therefore confusing. Here we group the findings into a classification scheme that may help in conceptualizing a model for CSN5/JAB1 action (Table I).
Table I. Proteins that interact with JAB1/CSN5.
| JAB1/CSN5 interactor | Function | Method | Reference |
|---|---|---|---|
| COP9 signalsome subunits | |||
| CSN1 | COP9 signalosome subunit 1 | 2 hyb, pur | Kwok et al. (1998); Kapelari et al. (2000) |
| CSN2 | COP9 signalosome subunit 2 | 2 hyb, fb, pur | Freilich et al. (1999); Kapelari et al. (2000) |
| CSN4 | COP9 signalosome subunit 4 | 2 hyb, pur | Serino et al. (1999) |
| CSN7 | COP9 signalosome subunit 7 | fb, pur | Kapelari et al. (2000) |
| Cell cycle regulators | |||
| p27Kip1 |
CDK inhibitor |
2 hyb, pd, rip |
Tomoda et al. (1999) |
| MIF1 |
cytokine, macrophage migration inhibitory factor |
2 hyb, pd, rip, eip |
Kleeman et al. (2000) |
| Regulators of AP-1 transcription | |||
| cJun | oncoprotein and subunit of the AP-1 transcription factor | 2 hyb | Claret et al. (1996) |
| LFA-1 | integrin, adhesion receptor | 2 hyb, pd, rip, eip | Bianchi et al. (2000) |
| LHR | G-coupled lutropin/choriogonadotropin receptor | 2 hyb, pd, rip | Li et al. (2000) |
| PR | progesterone receptor | 2 hyb, pd | Chauchereau et al. (2000) |
| SRC-1 | steroid receptor co-activator | 2 hyb, pd, rip | Chauchereau et al. (2000) |
| Bcl-3 | oncoprotein, IκB family | 2 hyb, pd, rip | Dechend et al. (1999) |
2 hyb, yeast two-hybrid interaction trap; pur, copurification; fb, filter binding assay; pd, in vitro pull-down assay; rip, co-immunoprecipitation with recombinant JAB1/CSN5; eip, co-immunoprecipitation with endogenous JAB1/CSN5.
The JAB1–p27Kip1 connection: regulation of the cell cycle via protein degradation
As a cyclin dependent kinase (CDK) inhibitor, p27Kip1 is involved in inducing G1 cell cycle arrest. Indeed, ectopic introduction of p27Kip1 efficiently induces G1 arrest of proliferating mouse fibroblasts (Tomoda et al., 1999). Co-expression of JAB1 rescues cells from p27Kip1-induced G1 arrest, and ectopic over-expression of JAB1 drives a marked number of serum-starved (i.e. G1 arrested) cells to progress to S phase in a JAB1-dose-dependent manner. Thus, ectopic over-expression of JAB1 both downregulates p27Kip1 in vivo to neutralize its inhibitory action, and reduces the serum dependence of the cells (Figure 2).
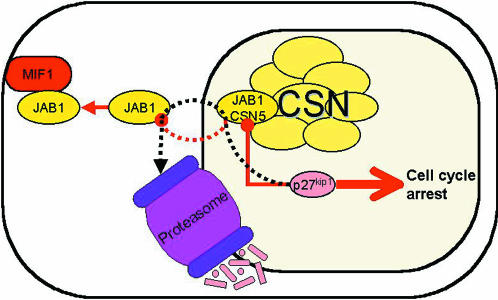
Fig. 2. Model of JAB1/CSN5 function in p27Kip1 mediated cell cycle control. Thin lines ending with filled circles represent inhibitory interactions. Thin lines ending with closed arrow heads represent stimulatory interactions. Dotted lines show protein movement. Binding of p27Kip1 to JAB1 leads to cell cycle progression, and the concomitant export of p27Kip1 from the nucleus and its degradation. Binding of JAB1 to MIF allows the nuclear accumulation of p27Kip1 and cell cycle arrest. The protein–protein interactions within the CSN are not specified (reviewed in Kapelari et al., 2000). CSN, COP9 signalosome.
Studies in which an HA-tagged JAB1 expression vector was introduced into proliferating fibroblasts showed that JAB1 is capable of markedly reducing endogenous p27Kip1, as well as of exogenously expressed HA-p27Kip1 (Tomoda et al., 1999). Inhibitors of the 26S proteasome prevented this downregulation. Together, these results indicate that JAB1 exerts its effects on p27Kip1 via the 26S proteasome, rather than through inhibition of p27Kip1 expression. The physical interaction of JAB1 with p27Kip1 is necessary for this regulation, as a p27Kip1 mutant lacking Thr187 cannot complex with JAB1 and is more resistant than normal p27Kip1 to JAB1-mediated downregulation. Furthermore, deletion of the nuclear localization signal sequence of p27Kip1 rendered the molecule resistant to the action of JAB1. As Thr187 is a substrate for CDKs (Vlach et al., 1997), it is likely that transportation of p27Kip1 into the nucleus and phosphorylation of Thr187 are prerequisites for the marking of p27Kip1 for degradation, as well as for its binding by JAB1. JAB1 appears to interact with p27Kip1 in the nucleus, and to accelerate p27Kip1 degradation by bringing it to the degradation machinery, or by facilitating phosphorylation of Thr187 (Tomoda et al., 1999).
The data presented above demonstrate that a physical JAB1-p27Kip1 interaction links JAB1 to the cell-cycle-dependent proteolytic machinery. The physiological relevance of this interaction is highlighted by the study of interleukin signaling (Boussiotis et al., 2000). In vivo immune tolerance, and its in vitro counterpart, anergy, are defined as the inability of T cells to produce the interleukin IL-2 and to undergo clonal expansion after stimulation with competent antigen-presenting cells. Upregulation of p27Kip1 causes inhibition of IL-2 transcription and blocks clonal expansion of anergic/tolerant T cells, both in vitro and in vivo. Increased p27Kip1 may inhibit IL-2 transcription simply through its association with JAB1, as this results in the translocation of JAB1 to the cytoplasm. This would keep JAB1 from performing its role in the stabilization of c-Jun protein binding to AP-1 sites, thereby resulting in the failure of AP-1 transactivation. Indeed, defective AP-1 transactivation is a specific molecular marker of anergy. Also, p27Kip1 is associated with JAB1 in cytoplasmic extracts from anergic, but not from productively stimulated, i.e. immune reactive, cells (Boussiotis et al., 2000). IL-2 receptor-mediated signals prevent the induction of the anergic state by preventing the accumulation, and promoting the degradation, of p27Kip1. Therefore, p27Kip1 acts as an ‘anergy factor’ whose function may involve sequestration of JAB1.
Additional evidence for a functional role of the JAB1-p27Kip1 interaction was provided by a study of the tyrosine kinase growth receptor HER-2/neu (Yang et al., 2000). Amplification or over-expression of this receptor is frequently found in human cancers, and may stimulate cell proliferation through tyrosine kinase signaling, possibly by downregulating p27Kip1. Indeed, over-expression or activation of HER-2/neu enhances the ubiquitin-mediated degradation of p27Kip1 while also increasing the export of JAB1 from the nucleus to the cytoplasm. Thus, given the interaction between JAB1 and p27Kip1, HER2/neu activation may promote the export of JAB1 to the cytoplasm which, in turn, might facilitate p27Kip1 degradation.
The recent discovery of a JAB1-cytokine interaction is illuminating in terms of understanding JAB1 control of cell cycle through p27Kip1. Macrophage migration inhibitory factor (MIF) is a cytokine that has anti-inflammatory growth-inhibitory activities. Exogenously applied MIF binds endogenous intracellular JAB1, and the MIF–JAB1 complex is found in the cytosol, enriched near the peripheral plasma membrane (Kleemann et al., 2000). Functionally, MIF has an inhibitory effect on JAB1. For example, JAB1-induced activation of an AP-1-dependent reporter gene was inhibited in cells cotransfected with both MIF and JAB1, and MIF interfered with JAB-1 activation of c-Jun N-terminal kinase (JNK) activity. Furthermore, MIF also antagonizes JAB1-dependent cell-cycle regulation. Whereas JAB1 suppresses p27Kip1 levels, MIF causes a JAB1-dependent increase of p27Kip1 levels. MIF does not directly bind p27Kip1, nor does it stimulate p27Kip1 mRNA or protein synthesis. Rather, it stabilizes the p27Kip1 protein. Collectively, these data suggest that the MIF-mediated effects are equivalent to p27Kip1-mediated growth arrest, and that that they are mediated through inhibition of JAB1-dependent degradation of p27Kip1 (Kleemann et al., 2000) (Figure 2).
Additional JAB1 interactions that regulate AP-1 mediated transcription
The above examples involve nuclear-cytoplasmic translocation of JAB1. The subcellular redistribution of JAB1 is a recurring theme, as JAB1 also appears to function at the plasma membrane.
In normal COS7 cells, transiently expressed JAB1 maintains a diffuse distribution. In experiments in which these cells also transiently express the adhesion receptor LFA-1, a fraction of JAB1 was enriched in the same peripheral membrane regions as this ectopically expressed integrin, consistent with an integrin–JAB1 interaction that had been found using the yeast two-hybrid assay (Bianchi et al., 2000). As the CSN has never been shown to localize to peripheral membranes, it can be assumed that this form of JAB1, like that involved in MIF signaling, is CSN-independent (Figure 3).
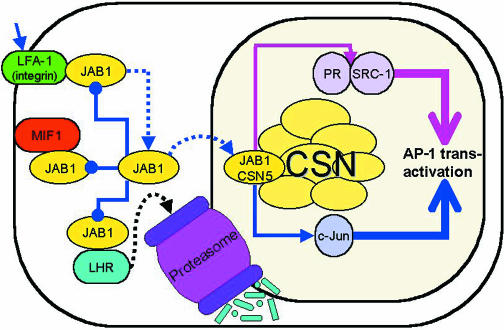
Fig. 3. Model of JAB1/CSN5 function in regulating AP-1 mediated transcription. Line colors relate to specific functions: blue, c-Jun mediated transactivation; pink, hormone receptor mediated transactivation. Thin lines ending with filled circles represent inhibitory interactions. Thin lines ending with closed arrow heads represent stimulatory interactions. Dotted lines show protein movement. JAB1/CSN5 mediates AP-1 transactivation through c-Jun. Interaction of JAB1 with LFA-1, MIF or LHR inhibits AP-1 transactivation. Extracellular substrate binding to LFA-1 releases JAB1 which redistributes to the nucleus leading to AP-1 transactivation. Binding of JAB1/CSN5 to PR and SRC-1 also induces AP-1 transactivation. The protein–protein interactions within the CSN are not specified (reviewed in Kapelari et al., 2000). CSN, COP9 signalosome.
When the doubly-transfected COS7 cells were treated with an artificial or physiological ligand for LFA-1, the membrane-associated JAB1 was lost and gradually shifted from the cytoplasm back to the nucleus, illustrating that the interaction is specific. Furthermore, the resulting increase in the JAB1 nuclear pool was paralleled by an increase in the binding of nuclear AP-1 complexes to an AP-1 site, and by the stimulation of transcriptional activity of an AP-1-driven luciferase reporter gene (Bianchi et al., 2000). Consistent with these findings, over-expression of two different JAB1 antisense constructs resulted in both a clear reduction of the endogenous JAB1 levels, and a reduction of the activation of the AP-1 reporter gene induced by LFA-1 stimulation. Taken together, these results indicate that endogenous JAB1 is physiologically involved in the regulation of AP-1 transcriptional activity. Together with the fact that integrins cooperate with growth factors to induce AP-1 transactivating function (Tremble et al., 1995), these data support the hypothesis that one pathway by which stimuli delivered through LFA-1 modulate AP-1 transcription involves the redistribution of JAB1 from an extranuclear to a nuclear compartment. In this scenario, the integrin serves to sequester JAB1 from its transcriptional targets in the nucleus (Bianchi et al., 2000).
JAB1 sequestering by ER-associated proteins is also used to downregulate AP-1 mediated transcription. In their study of the interaction between JAB1 and the rat G-coupled lutropin/choriogonadotropin receptor (rLHR), Li et al. (2000) found that over-expression of LHR reduces the ability of JAB1 to potentate the c-Jun-mediated transactivation of an AP-1-luciferase reporter gene. As in the case of LFA-1, this is presumably due to interference with the JAB1–c-Jun interaction. However, JAB1 was further shown to affect the turnover of the intracellular, immature, 68 kDa precursor of LHR in the endoplasmic reticulum. Binding of JAB1 to this form of LHR promotes degradation of this precursor, providing another link between JAB1 and ubiquitin-mediated protein degradation through the proteasome. JAB1 does not appear to interact with the mature, 85 kDa, cell surface LHR (Li et al., 2000). Thus, the JAB1 interaction with LHR may have two consequences: the sequestering of JAB1 from c-Jun and the subsequent downregulation of AP-1 regulated genes; and the degradation of the LHR precursor through the proteasome (Figure 3).
JAB1 is also capable of modulating the transcriptional transactivation activity of nuclear receptors (Chauchereau et al., 2000). This was clearly demonstrated for the progesterone receptor (PR). While the PR is capable of exerting hormone-dependent transactivation by itself, this ability was markedly enhanced by the addition of JAB1. JAB1 was also shown to be capable of potentiating the hormone-dependent transactivation properties of other nuclear receptors including the glucocorticoid, mineralocorticoid, androgenic and estrogenic receptors, as well as the transcription factor NF-κB, although its physical interaction with all of these proteins remains to be demonstrated (Chauchereau et al., 2000).
Since the steroid receptor co-activator 1 (SRC-1) is a common co-activator for nuclear receptors, it appeared conceivable that JAB1’s effect on their transactivation activities may be mediated by SRC-1. Indeed, JAB1 and SRC-1 physically interact in vitro and in yeast (Chauchereau et al., 2000) (Figure 3). The possibility that the interaction of JAB1 with the PR, in particular, might affect its transactivation function by stabilizing its interaction with SRC-1 was examined using a mammalian three-hybrid system. Addition of JAB1 to cells co-transfected with a reporter gene driven by a Gal4-upstream activation sequence, a DNA binding domainGal4-PR expression vector, and a vector encoding SRC-1-activation domain increased the hormone-dependent PR–SRC-1 -driven reporter gene transcription, suggesting that JAB1 stabilizes the PR–SRC-1 complexes. This result was confirmed by in vitro studies using 3H-labeled PR, and 35S-labeled SCR-1, and immunoprecipitation with anti-PR. Addition of increasing amounts of unlabeled JAB1 led to a clear increase in SRC-1 coimmunoprecipitation. JAB1 also affects the transactivation activity of other transcription factors. For example, co-transfection of cells with NF-κB and JAB1 strongly enhanced NF-κB activity on a target gene (Chauchereau et al., 2000). This effect of JAB1 may also be mediated through SRC-1 since NF-κB is known to interact with SRC-1 (Na et al., 1998). The Bcl-3 oncoprotein, a member of the IκB multigene family, modulates the activities of NF-κB/Rel transcription factors. JAB1 interacts with Bcl-3 and increases the DNA binding activity of NF-κB–Bcl-3 heterodimer to the NF-κB DNA binding site. However this did not result in the formation of a quaternary complex of JAB1 with NF-κB and the NF-κB DNA binding site, nor could the binding of JAB1 to Bcl-3 facilitate NF-κB dependent gene activation (Dechend et al., 1999).
JAB1 or CSN5 – in or out of the complex?
How do the many JAB1 functions relate to the CSN? Surprisingly, this question has been largely ignored in the various JAB1 studies. The biochemical function of the CSN remains obscure, though accumulating evidence places the CSN at the juncture of kinase signaling and ubiquitin-dependent protein degradation. Although an early report claimed that the CSN phosphorylates a variety of proteins including c-Jun (Seeger et al., 1998), this activity now appears to be mediated by a CSN-associated kinase (Naumann et al., 1999).
Many of the CSN subunits have been shown to affect cell-cycle associated signaling pathways (reviewed in Karniol and Chamovitz, 2000). For example, human CSN1 was originally identified as a high-copy suppressor of JNK activity. In Schizosaccharomyces pombe, this protein is involved in the DNA-integrity checkpoint in S-phase progression. CSN6 was identified as interacting with HIV-PR, which serves as a cell cycle regulator for HIV. CSN2 was originally identified as thyroid receptor interacting protein 15 (TRIP15) and has also been shown to regulate interferon signaling pathways (Cohen et al., 2000).
These cell cycle- and kinase pathway-associated roles are complementary to the JAB1 functions described above. However this still leaves open the question of the role of the JAB1 monomer as opposed to the CSN5 subunit of the CSN complex. Perhaps one of the many roles of the CSN is to act as a scaffolding complex for JAB1/CSN5 and its nuclear substrates. In this scenario, the specific CSN subunits might control JAB1/CSN5 activity by selectively recruiting other proteins for regulation by JAB1/CSN5. Alternatively, these proteins may serve to receive signals that release JAB1/CSN5 from the CSN.
In assessing the data discussed here, one must keep in mind the caveat that the various JAB1-induced phenotypes described were obtained by ectopic over-expression of JAB1. None of these experiments were concerned with which form (CSN-associated or CSN-independent) of the exogenously applied JAB1/CSN5 existed in the cell. Nor did they consider the effect of the treatment on CSN activity. One could speculate that most of this excess protein is found independent of the CSN, in which case it would obviously interfere with the equilibrium between the two forms. If so, this would imply that the ‘active’ form of JAB1/CSN5 in these experiments is the CSN-independent form, and that the CSN may sequester this form. As the CSN is also similar to the regulatory lid of the proteasome, and JAB1/CSN5 appears to present some proteins to the proteasome, perhaps a competition between the proteasome and the CSN for JAB1/CSN5 association is essential for JAB1/CSN5 activity. Genetic approaches aimed at interfering specifically with JAB1/CSN5 binding to the CSN will be invaluable for elucidating the roles of JAB1/CSN5.

Daniel A. Chamovitz & Daniel Segal

Acknowledgments
Acknowledgements
We thank Professor Odd S. Gabrielsen of the University of Oslo, and Dr Michael Glickman of The Technion for communicating results prior to publication, and Professor Albrecht G. von Arnim of the University of Tennessee for insightful comments. Our research on the COP9 signalosome is supported by a grant from the Israel Science Foundation (519/00-1).
References
- Bianchi E., Denti, S., Granata, A., Bossi, G., Geginat, J., Villa, A., Rogge, L. and Pardi, R. (2000) Integrin LFA-1 interacts with the transcriptional co-activator JAB1 to modulate AP-1 activity. Nature, 404, 617–621. [DOI] [PubMed] [Google Scholar]
- Boussiotis V.A., Freeman, G.J., Taylor, P.A., Berezovskaya, A., Grass, I., Blazar, B.R. and Nadler, L.M. (2000) p27Kip1 functions as an anergy factor inhibiting interleukin 2 transcription and clonal expansion of alloreactive human and mouse helper T lymphocytes. Nature Med., 6, 290–297. [DOI] [PubMed] [Google Scholar]
- Chamovitz D.A., Wei, N., Osterlund, M.T., von Arnim, A.G., Staub, J.M., Matsui, M. and Deng, X.-W. (1996) The COP9 complex, a novel multisubunit nuclear regulator involved in light control of a plant developmental switch. Cell, 86, 115–121. [DOI] [PubMed] [Google Scholar]
- Chauchereau A., Georgiakaki, M., Perrin-Wolff, M., Milgrom, E. and Loosfelt, H. (2000) JAB1 interacts with both the progesterone receptor and SRC-1. J. Biol. Chem., 275, 8540–8548. [DOI] [PubMed] [Google Scholar]
- Claret F.X., Hibi, M., Dhut, S., Toda, T. and Karin, M. (1996) A new group of conserved coactivators that increase the specificity of AP-1 transcription factors. Nature, 383, 453–457. [DOI] [PubMed] [Google Scholar]
- Cohen H., Azriel, A., Cohen, T., Meraro, D., Hashmueli, S., Bech-Otschir, D., Kraft, R., Dubiel, W. and Levi, B.Z. (2000) Interaction between ICSBP and CSN2 (TRIP15) – a possible link between IRF signaling and the COP9/signalosome. J. Biol. Chem., 275, 39081–39089. [DOI] [PubMed] [Google Scholar]
- Dechend R., Hirano, F., Lehmann, K., Heissmeyer, V., Ansieau, S., Wulczyn, F.G., Scheidereit, C. and Leutz, A. (1999) The Bcl-3 oncoprotein acts as a bridging factor between NF-κB/Rel and nuclear co-regulators. Oncogene, 18, 3316–3323. [DOI] [PubMed] [Google Scholar]
- Deng X.W. et al. (2000) Unified nomenclature for the COP9 signalosome and its subunits: an essential regulator of development. Trends Genet., 16, 202–203. [DOI] [PubMed] [Google Scholar]
- Freilich S., Oron, E., Kapp, Y., Nevo-Caspi, Y., Orgad, S., Segal, D. and Chamovitz, D.A. (1999) The COP9 signalosome is essential for development of Drosophila melanogaster. Curr. Biol., 9, 1187–1190. [DOI] [PubMed] [Google Scholar]
- Glickman M.H., Rubin, D.M., Coux, O., Wefes, I., Pfeifer, G., Cjeka, Z., Baumeister, W., Fried, V.A. and Finley, D. (1998) A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell, 94, 615–623. [DOI] [PubMed] [Google Scholar]
- Hofmann K. and Bucher, P. (1998) The PCI domain: A common theme in three multi-protein complexes. Trends Biochem. Sci., 23, 204–205. [DOI] [PubMed] [Google Scholar]
- Kapelari B., Bech-Otschir, D., Hegerl, R., Schade, R., Dumdey, R. and Dubiel, W. (2000) Electron microscopy and subunit-subunit interaction studies reveal a first architecture of COP9 signalosome. J. Mol. Biol., 300, 1169–1178. [DOI] [PubMed] [Google Scholar]
- Karniol B. and Chamovitz, D.A. (2000) The COP9 signalosome: from light signaling to general developmental regulation and back. Curr. Opin. Plant. Biol., 3, 387–393. [DOI] [PubMed] [Google Scholar]
- Karniol B., Yahalom, A., Kwok, S.F., Tsuge, T., Matsui, M., Deng, X.-W. and Chamovitz, D.A. (1998) The Arabidopsis homolog of an eIF3 complex subunit associates with the COP9 complex. FEBS Lett., 439, 173–179. [DOI] [PubMed] [Google Scholar]
- Kleemann R. et al. (2000) Intracellular action of the cytokine MIF to modulate AP-1 activity and the cell cycle through JAB1. Nature, 408, 211–216. [DOI] [PubMed] [Google Scholar]
- Kwok S.F., Solano, R., Tsuge, T., Chamovitz, D.A., Ecker, J.R., Matsui, M. and Deng, X.W. (1998) Arabidopsis homologs of a c-Jun coactivator are present both in monomeric form and in the COP9 complex and their abundance is differentially affected by the pleiotropic cop/det/fus mutations. Plant Cell., 10, 1779–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok S.F., Staub, J.S. and Deng, X.-W. (1999) Characterization of two subunits of the Arabidopsis 19S proteasome regulatory complex and its possible interaction with the COP9 complex. J. Mol. Biol., 285, 85–95. [DOI] [PubMed] [Google Scholar]
- Li S., Liu, X. and Ascoli, M. (2000) p38JAB1 binds to the intracellular precursor of the lutropin/choriogonadotropin receptor and promotes its degradation. J. Biol. Chem., 275, 13386–13393. [DOI] [PubMed] [Google Scholar]
- Na S.Y., Lee, S.K., Han, S.J., Choi, H.S., Im, S.Y., Lee, J.W. (1998) Steroid receptor coactivator-1 interacts with the p50 subunit and coactivates nuclear factor κB-mediated transactivations. J. Biol. Chem., 273, 10831–10834. [DOI] [PubMed] [Google Scholar]
- Naumann M., Bech-Otschir, D., Huang, X., Ferrell, K. and Dubiel, W. (1999) COP9 signalosome-directed c-Jun activation/stabilization is independent of JNK. J. Biol. Chem., 274, 35297–35300. [DOI] [PubMed] [Google Scholar]
- Osterlund M.T., Hardtke, C.S., Wei, N. and Deng, X.W. (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature, 405, 462–466. [DOI] [PubMed] [Google Scholar]
- Seeger M., Kraft, R., Ferrell, K., Bech-Otschir, D., Dumdey, R., Schade, R., Gordon, C., Naumann, M. and Dubiel, W. (1998) A novel protein complex involved in signal transduction possessing similarities to 26S proteasome subunits. FASEB J., 12, 469–478. [PubMed] [Google Scholar]
- Serino G., Tsuge, T., Kwok, S., Matsui, M., Wei, N., Deng, X.W. (1999) Arabidopsis cop8 and fus4 mutations define the same gene that encodes subunit 4 of the COP9 signalosome. Plant Cell, 10, 1967–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremble P., Damsky, C.D. and Werb, Z. (1995) Components of the nuclear signaling cascade that regulate collagenase gene expression in response to integrin-derived signals. J. Cell Biol., 129, 1707–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoda K., Kubota, Y. and Kato, J. (1999) Degradation of the cyclin-dependent-kinase inhibitor p27Kip1 is instigated by JAB1. Nature, 398, 160–165. [DOI] [PubMed] [Google Scholar]
- Vlach J., Hennecke, S. and Amati, B. (1997) Phosphorylation-dependent degradation of the cyclin-dependent kinase inhibitor p27Kip1. EMBO J., 16, 5334–5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei N., Tsuge, T., Serino, G., Dohmae, N., Takio, K., Matsui, M. and Deng, X.W. (1998) Conservation of the COP9 complex between plants and mammals and its relationship to the 26S proteasome regulatory complex. Curr. Biol., 8, 919–922. [DOI] [PubMed] [Google Scholar]
- Yang H.Y., Zhou, B.P., Hung, M.C. and Lee, M.H. (2000) Oncogenic signals of HER-2/neu in regulating the stability of the cyclin-dependent kinase inhibitor p27. J. Biol. Chem., 275, 24735–24739. [DOI] [PubMed] [Google Scholar]