Abstract
Ku is a conserved heterodimeric DNA-binding protein that plays critical roles in DNA repair and telomere homeostasis. In Saccharomyces cerevisiae, deletion of YKU70 or YKU80 results in an inability to grow at 37°C. This is suppressed by overexpression of several components of telomerase (EST1, EST2 and TLC1). We show that overexpression of EST2 or TLC1 in yku80 mutants does not restore efficient DNA repair, or restore normal telomere function, as measured by telomere length, single-stranded G-rich strand or transcriptional silencing. Instead, yku80 mutants activate a Rad53p-dependent DNA-damage checkpoint at 37°C and this is suppressed by overexpression of EST2 or TLC1. Indeed, deletion of genes required for Rad53p activation also suppresses the yku80 temperature sensitivity. These results suggest that activation of the DNA-damage checkpoint in yku mutants at 37°C does not result from reduced telomere length per se, but reflects an alteration of the telomere structure that is recognized as damaged DNA.
INTRODUCTION
Ku is a heterodimeric DNA-binding protein that functions in mammals and yeast in the repair of DNA double-strand breaks by non-homologous end-joining (NHEJ; reviewed in Smith and Jackson, 1999; Tuteja and Tuteja, 2000). Ku is believed to bind to DNA ends to prevent unnecessary DNA degradation, as well as to facilitate DNA ligation by juxtaposing DNA ends and recruiting DNA ligases (Bliss and Lane, 1997; Pang et al., 1997; McElhinny et al., 2000; Teo and Jackson, 2000). Ku has also been shown to play an essential and direct role in telomere homeostasis. Telomeres are specialized structures at the ends of eukaryotic chromosomes that, in Saccharomyces cerevisiae, are heterogeneous C1–3A:TG1–3 repeats of ∼300 base pairs. Deletion of YKU70 or YKU80 results in telomere shortening and loss of transcriptional silencing of genes placed near the telomere (Boulton and Jackson, 1996b, 1998; Porter et al., 1996; Tsukamoto et al., 1997; Nugent et al., 1998). In addition, whereas the G-rich strand of telomeres is single-stranded only during S-phase in wild-type yeast, yku70 or yku80 mutants accumulate long single-stranded telomeric regions throughout the cell-cycle (Gravel et al., 1998; Polotnianka et al., 1998).
Saccharomyces cerevisiae mutants in which YKU70 or YKU80 is deleted are unable grow at 37°C (Feldmann and Winnacker, 1993; Boulton and Jackson, 1996a,b; Feldmann et al., 1996; Barnes and Rio, 1997). Barnes and Rio (1997) suggested that this might be because of an inability to repair spontaneous double-strand breaks generated in vivo. More recently, however, Nugent et al. (1998) found that overexpression of EST2 (the catalytic subunit of telomerase), TLC1 (the telomerase RNA component) or EST1 (a targetting subunit of telomerase) can suppress this temperature sensitivity, suggesting that this phenotype is linked to the role of Ku in telomere homeostasis rather than to its role in NHEJ. We show here that overexpression of EST2 or TLC1 does not restore the normal telomeric regulation to a yku80 mutant strain. Instead, we find that a DNA-damage checkpoint is activated in yku80 mutants grown at elevated temperatures, and that this checkpoint is suppressed by overexpression of EST2 or TLC1. Finally, we show that deletion of several checkpoint genes that do not rescue telomere length in yku80 mutants, but suppress the checkpoint activation, also partly rescues the temperature sensitive phenotype.
RESULTS AND DISCUSSION
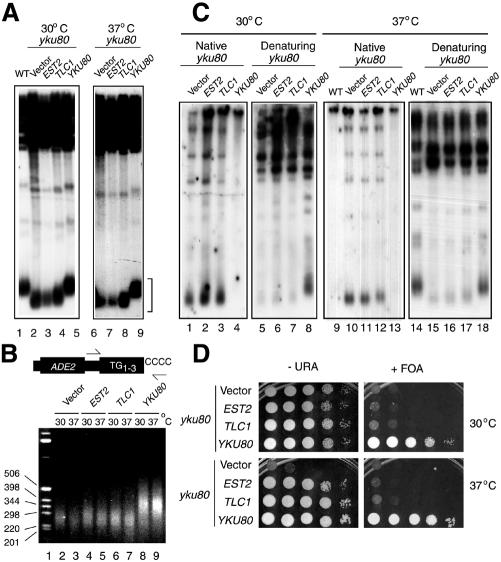
We performed a multicopy suppressor screen for genes that, when overexpressed, suppress the yku80 mutant temperature-sensitive phenotype (see Methods). We found that overexpression of either EST2 or TLC1 restored the ability of yku80 mutants to grow at 37°C to levels close to those of wild-type yeast. Other weaker suppressors of this temperature-sensitive phenotype were also identified but are not hitherto known components of telomerase. Overexpression of these genes also restored the ability of yku70 mutants to grow at 37°C (data not shown). The identification of telomerase subunits as strong suppressors of the yku80 mutant temperature-sensitive phenotype suggested that this phenotype might be due to a telomere defect that could be restored by overexpression of yeast telomerase catalytic activity. Analysis of telomere length by Southern blotting, however, showed that yku80 mutants overexpressing EST2 or TLC1 still had short telomeres and that there were no gross changes in telomere length when strains were grown at 30 or 37°C (Figure 1A). We also analysed the length of a telomere in which the ADE2 gene had been inserted using a novel method described recently (Forstemann et al., 2000). In this method, genomic DNA was treated with terminal deoxynucleotidyl transferase (TdT) in the presence of dCTP, and the ADE2-marked telomere was then amplified by PCR using a primer specific for a sequence distal to the subtelomeric ADE2 gene and a primer complementary to the dC tail. Figure 1B shows that using this method, no gross changes in telomere length were detected in yku80 mutants overexpressing EST2 or TLC1. In addition, these PCR products were cloned into pGEMT and the length of TG repeats determined by DNA sequencing. The average length of the G-rich telomeric strand in the yku80 mutant grown at 37°C was 137 ± 33 (average length ± SD, n = 5). In yku80 mutant strains overexpressing EST2 or TLC1, the length was 126 ± 50 and 148 ± 49, respectively. In contrast, telomeres in wild-type yeast were 246 ± 32. Collectively, these data reveal that there are no marked changes in the length of the TG-rich strand of the telomere.
Fig. 1. (A) Southern blot of XhoI-digested genomic DNA probed with Y′-specific sequences of yeast telomeres. Genomic DNA was prepared from strains grown at 30°C (left) or 37°C (right) for 18 h, from wild-type (lane 1) or yku80 mutant strains containing either the YEP13 (2µ) plasmid (lanes 2, 6), YEP13–EST2 (lanes 3, 7), –TLC1 (lanes 4, 8) or –YKU80 (lanes 5, 9). Identical results (with the exception of yku80 with YEP13) were obtained with strains grown for >100 generations. Telomeric restriction fragments are indicated by a square bracket. (B) Telomere PCR. Genomic DNA was isolated, denatured and tailed with dCTP using TdT. The G-rich strand is specifically amplified with a primer complementary to the dC tail and a primer specific for a sequence distal of the subtelomeric ADE2 gene. The bottom panel shows PCR products analysed on an ethidium bromide-stained agarose gel. (C) In-gel hybridization of XhoI-digested genomic DNA probed with a radiolabelled oligonucleotide corresponding to the CA-repeat sequence of yeast telomeres, in native or denaturing conditions. Quantitation of the single-strand telomeric regions in yku80 mutants with or without EST2 or TLC1 overexpression showed no reproducible differences. (D) TPE was tested using a strain containing a telomere-proximal URA3 gene. Yeast strains were spotted in 10-fold serial dilutions on plates with (right) or without (left) FOA, and grown at 30°C (top) or 37°C (bottom) for 3 days.
Next, we tested the single-stranded regions in the G-rich strand of the telomere in native hybridizations of genomic DNA prepared from unsynchronized cultures (Gravel et al., 1998). In yku80 mutants, but not in strains complemented with full-length YKU80, these single-stranded regions were easily detected (Figure 1C, lanes 1 and 4, respectively). Lanes 2 and 3 show that these single-stranded regions persisted in yku80 strains overexpressing EST2 or TLC1 (Figure 1C, lanes 5–8 show the results of probing under denaturing conditions). These single-stranded regions were unchanged when cultures were grown at 37°C (Figure 1C, lanes 9–18). Notably, the single-strand regions were unaffected by overexpression of EST2 or TLC1.
We also analysed the status of transcriptional silencing of a URA3 gene placed near the telomere of chromosome VII by plating cells on medium containing 5-fluoro-orotic acid (FOA). yku80 mutants but not wild-type cells were unable to silence the URA3 gene and hence died on FOA plates (Figure 1D). It has been reported previously that overexpression of EST1, EST2 or TLC1 can partly complement the yku80 mutant silencing defect (Evans et al., 1998). Using a different strain background, we found that overexpression of EST2 or TLC1 did not efficiently restore telomeric silencing at 30, 34 or 37°C (Figure 1D and data not shown). Moreover, we have found that overexpression of EST2 or TLC1 suppressed the temperature sensitivity of a sir4/yku80 mutant strain (data not shown), indicating that suppression can be achieved independently of effects on telomeric silencing. Together, these results show that the disrupted telomeric structure in yku80 mutants is not restored to that of wild-type yeast strains by overexpression of EST2 or TLC1.
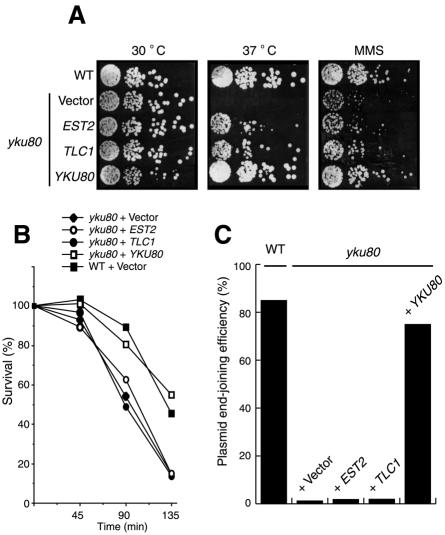
We next considered the possibility that spontaneous DNA strand-breaks generated but unrepaired in the yku80 mutant might be particularly deleterious when strains are grown at elevated temperatures, and that this repair deficiency is alleviated by overexpression of telomerase components. We therefore tested the ability of EST2 or TLC1 to suppress the sensitivity of a yku80 mutant to the DNA-damaging agents methylmethanesulfonate (MMS) or bleomycin. Notably, yku80 mutants overexpressing either EST2 or TLC1 were as sensitive as yku80 mutants to bleomycin (data not shown) or MMS (Figure 2A and B). EST2 or TLC1 overexpression also did not restore NHEJ activity in yku80 mutants, as measured by a plasmid repair assay (Figure 2C). These data indicate that overexpression of EST2 or TLC1 does not restore the ability of yku80 mutant cells to repair DNA strand-breaks.
Fig. 2. (A) Strains were spotted in 5-fold serial dilutions onto plates without (left and middle) or with 0.005% MMS (right) and grown at 30°C (left and right) or 37°C (middle) for 3 days. (B) Exponentially growing cells were exposed to 0.05% MMS for the times indicated and then plated onto YPD. Percentage cell survival was plotted against time in MMS. (C) Plasmid repair assay was performed by transforming either supercoiled or XbaI-linearized pRS413 into various yeast strains (described in Figure 1A). The number of HIS+ colonies obtained with linear versus supercoiled DNA was taken as a measure of NHEJ efficiency.
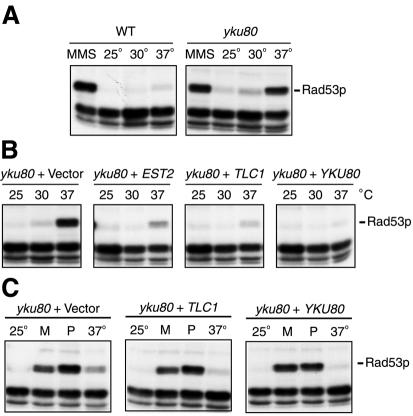
Another possibility that we considered was that yku80 strains accumulate DNA damage at the restrictive temperature and that this triggers a DNA damage-signalling cascade that leads to cell cycle arrest. Thus, we tested whether the DNA-damage response was activated upon growth at elevated temperatures by use of an Rad53p in situ autophosphorylation assay (Pellicioli et al., 1999; see Methods). As shown previously, several radiolabelled bands that migrate faster than Rad53p in an SDS–polyacrylamide gel and are unchanged by treatment with MMS are detected, and serve as loading controls. The predominant radioactive species detected in extracts from strains treated with MMS (Figure 3A) corresponds to that of autophosphorylated Rad53p, as shown by its absence in rad53 mutants or as a slower migrating band in strains bearing epitope-tagged Rad53p (Pellicioli et al., 1999 and data not shown). Figure 3A (left panel) shows that in wild-type yeast, Rad53p autophosphorylation was stimulated by MMS, but no stimulation was observed when yeast were grown at 25, 30 or 37°C. In contrast, in yku80 mutants, Rad53p autophosphorylation was not only stimulated by MMS, but also by growth at 37°C, and to a lesser extent at the semi-permissive temperature of 30°C (Figure 3A, right panel; the levels of Rad53p were unaffected as judged by western blotting; data not shown). We next tested the effect of overexpression of EST2 or TLC1 on Rad53p activation in yku80 mutants grown at 25, 30 or 37°C. Notably, when these genes were overexpressed in yku80 strains, we observed partial suppression of the temperature-dependent activation of Rad53p autophosphorylation (Figure 3B). This seems to be specific to growth at elevated temperatures since Rad53p activation in response to the DNA-damaging agents MMS (M) or phleomycin (P) was not altered by the overexpression of EST2 (data not shown) or TLC1 (Figure 3C).
Fig. 3. (A) Autoradiograph of the Rad53p in situ autophosphorylation assays. Wild-type or yku80 mutant strains were grown in the presence of 0.02% MMS at 30°C for 2 h, or at 25, 30 or 37°C for 5 h. (B) yku80 mutant strains containing YEP13 (2µ), YEP13–EST2, –TLC1 or –YKU80 were grown at 25, 30 or 37°C for 5 h. (C) Strains were grown at 25 or 37°C, or with 0.005% MMS (M) or 20 µg/ml phleomycin (P), and Rad53p autophosphorylation was assayed as above.
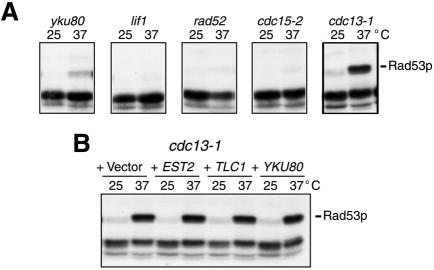
Stimulation of Rad53p autophosphorylation has hitherto only been reported in response to DNA-damaging agents or blocked DNA replication (Pellicioli et al., 1999). The above results therefore raised the possibility that, as a consequence of the NHEJ defect of the yku80 mutant strain, spontaneous unrepaired DNA strand-breaks generated by growth at elevated temperatures might directly lead to Rad53p activation. If this is the case, we reasoned that Rad53p might also be activated at 37°C in other DNA-repair mutants such as lif1 (defective in NHEJ; Herrmann et al., 1998) or rad52 (defective in homologous recombination). As shown in Figure 4A, there was no detectable activation of Rad53p in these strains at 37°C, indicating that the Rad53p response in yku80 mutants is unlikely to result from spontaneous DNA strand breaks being generated at the non-permissive temperature.
Fig. 4. (A) Rad53p in situ autophosphorylation of strains grown at 25 or 37°C. (B) Rad53p in situ autophosphorylation of cdc13-1 mutant strains containing YEP13 (2µ), YEP13–EST2, –TLC1 or –YKU80, and grown at either 25 or 37°C.
Unlike yku80 mutants, however, the lif1 and rad52 strains are not temperature-sensitive, so we therefore tested for Rad53p activation in two other temperature-sensitive mutants: cdc15-2 (Schweitzer and Philippsen, 1991) and cdc13-1 (Garvik et al., 1995). Cdc15p functions in the mitotic exit pathway and thus cdc15-2 mutants arrest at the end of anaphase. Unlike yku80 mutants, the cdc15-2 mutant strain did not activate Rad53p at the non-permissive temperature of 37°C (Figure 4A). Cdc13p protects the telomere from degradation and mediates the access of telomerase to the telomere. The cdc13-1 temperature-sensitive mutant showed marked elevation of Rad53p autophosphorylation at 37°C (Figure 4A). cdc13-1 mutants have been shown previously to form extensive single-stranded telomeric regions when grown at non-permissive temperatures (Garvik et al., 1995; Lydall and Weinert, 1997). Notably, however, whereas overexpression of EST2 or TLC1 suppressed the temperature-dependent activation of Rad53p in yku80 mutants, overexpression of these genes or of YKU80 did not suppress the temperature-dependent activation of Rad53p in cdc13-1 mutants (Figure 4B).
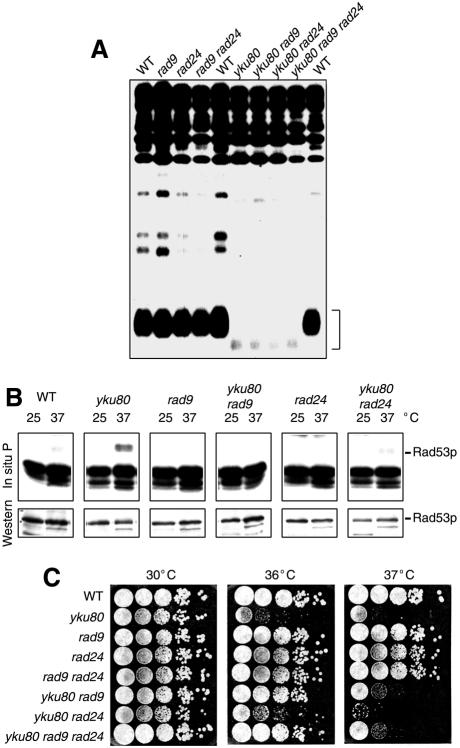
One explanation for the above data is that in yku80 mutant strains exposure to 37°C leads to the induction of a DNA-damage signalling cascade. If this is so, we reasoned that mutation of genes known to be required for activation of Rad53p might allow survival of yku80 mutants at 37°C. Extensive genetic and biochemical analyses in S. cerevisiae have suggested that Rad9p and Rad24p form part of two distinct DNA-damage sensing pathways that additively activate Mec1p, which in turn activates Rad53p by phosphorylation (reviewed in Lowndes and Murguia, 2000). We therefore tested the effect of deleting RAD9, RAD24, or both genes, in wild-type and yku80 mutant strains. There were no gross changes in telomere length in rad9/yku80, rad24/yku80 or rad9/rad24/yku80 mutants compared with the yku80 mutants (Figure 5A). Moreover, like wild-type strains, there was no apparent activation of Rad53p in rad9 or rad24 strains at 37°C (Figure 5B). However, the temperature-dependent activation of Rad53p in the rad9/yku80 strain and, to a lesser extent, the rad24/yku80 strain was reduced compared to the yku80 strain (Figure 5B), indicating that activation of Rad53p was mediated by both Rad9p and Rad24p.
Fig. 5. (A) Southern blot analysis of telomeres from various yeast strains. (B) Top, Rad53p in situ autophosphorylation of strains grown at 25 or 37°C. Bottom, Western blot analysis of Rad53p was performed on 12% polyacrylamide gels. (C) Strains were spotted in 10-fold serial dilutions onto YPAD plates and grown at 30, 36 or 37°C.
The above data suggested that exposure to 37°C in the yku80 mutant activates a signalling pathway that is dependent primarily on Rad9p and to a lesser extent on Rad24p, which in turn leads to chronic activation of Rad53p and sustained growth arrest. We therefore reasoned that, if this is the case, reduction of the Rad53p activation by deletion of RAD9 or RAD24 might suppress the temperature sensitive phenotype of the yku80 mutant. As shown in Figure 5C, this is indeed the case—deletion of RAD9, and to a much lesser extent RAD24, resulted in partial suppression of the yku80 temperature-sensitivity. It is noteworthy, however, that only weak suppression is observed at 37°C suggesting that temperature-sensitive lethality is not only due to telomeric activation of the DNA-damage checkpoint and cell-cycle arrest. The role of Ku in DNA damage checkpoints is indeed a complex one as illustrated in the complex interactions that take place during adaptation in response to a double-strand break (Lee et al., 1998).
CONCLUSION
These data suggest that when yku80 mutants are grown at 37°C, they activate a DNA-damage signalling pathway akin to that triggered in response to DNA strand breaks. This pathway can be partly suppressed by overexpression of telomerase components EST2 or TLC1, or by deletion of RAD9 or RAD24, thereby allowing growth at elevated temperatures. It is tempting to speculate that the telomeric single-stranded regions in the yku mutants initiate the signalling cascade. However, the single-stranded G-rich extensions are equally detectable when yku80 mutant cells are grown at 30 or 37°C (Gravel et al., 1998 and Figure 1C). In addition, although overexpression of EST2 or TLC1 alleviates the yku80 temperature sensitivity, it has no apparent effect on the extent of the single-stranded regions or the length of the G-strand. This suggests that the changes that occur at the telomere at non-permissive temperatures must be more subtle; perhaps involving alterations in the in vivo accessibility of the single-stranded regions or interactions between the telomere and other proteins, or nuclear structures.
Our data therefore lend support to models in which telomere length itself is often a poor indicator of telomere-induced crisis or checkpoint arrest, and suggest that other features are likely to be critical. More specifically, at least in instances where the telomeres are critically short, such as in the yku80 mutant, it may be that components of yeast telomerase play additional telomere-specific genome-protective roles by preventing inappropriate activation of the DNA-damage checkpoint pathway. Such a model is consistent with one put forward recently by Blackburn (2000) in which the telomere is proposed to switch stochastically between a capped and uncapped state in a manner that is determined, not by telomere length per se, but by preservation of the physical integrity of the telomere. Further studies investigating the effect of telomerase components on telomere chromatin structure and accessibility may provide vital insights into how telomere ‘capping’ might prevent the recognition of chromosome ends as DNA double-strand breaks.
METHODS
Yeast strains. W303 (MATα ade2-1 leu2-3,112 his3-11, 15 trp1 ura3-1 can1-100) and isogenic derivatives were used for all experiments. Deletion strains were made by one-step gene disruption of haploid strains using p80::KANX (constructed by J. Downs), pLIF1::URA3 (Teo and Jackson, 2000), pRAD52::TRP1 (gift from D. Weaver). Haploid strains for Figure 5 were generated from dissection of the rad9::HIS3 rad24::URA3 yku80::KANX diploid to generate single, double and triple mutants. TPE was tested in W303 strains in which the URA3 gene was inserted near the telomere of chromosome VII (gift from D. Gottschling). Telomere PCR was tested in W303 yku80::KANX strains in which the ADE2 gene was inserted at the telomere of the right arm of chromosome V (gift of D. Shore).
Temperature sensitivity multicopy suppressor screen. We found that the yku80 isw2 mutant strain was markedly more temperature sensitive than either the yku80 or isw2 (gift from T. Tsukiyama) mutant strains. We therefore performed multicopy suppressor screens by transforming the yku80 isw2 double mutant with a YEP13 2µ library of plasmids in which random fragments of genomic DNA had been cloned (gift from K. Nasmyth). Transformants were plated onto minimal medium and incubated for 8 h at 30°C before growth at 37°C for 3 days. Plasmids from temperature-resistant colonies were shuttled into Escherichia coli and retested for their ability to suppress the temperature-sensitive phenotype by transforming into yku80 mutants. All positive clones were found to suppress the temperature sensitivity of the yku80 single mutant and subsequent experiments were conducted in this strain.
Telomere PCR. Telomere PCR experiments were conducted as described in Forstemann et al. (2000). Briefly, genomic DNA was prepared from yku80 mutant cultures in which the ADE2 gene is placed at the telomere of the right arm of chromosome V. The phenotype of yku80 mutant strains overexpressing EST2 or TLC1 is similar to that observed in W303 strains without the ADE2 insertion (data not shown). Genomic DNA was then denatured and tailed with terminal deoxynucleotidy transferase (TdT) in the presence of dCTP. PCR was then performed using a primer specific for a sequence distal of the subtelomeric ADE2 gene and a primer complementary to the dC tail. The PCR products were analysed in a 2% agarose 1× TAE gel and following gel electrophoresis telomeric bands were purified and cloned into pGEMT. Plasmids were sequenced using the T7 primer by automated DNA sequencing.
MMS sensitivity experiments and in vivo plasmid repair assay. Exponentially growing cells were treated with 0.05% MMS for the times indicated and dilutions of cells were plated onto YPD agar plates. Survival relative to untreated cells was plotted relative to exposure time in MMS. Plasmid repair assays were performed as described previously (Boulton and Jackson, 1996a).
In situ Rad53p autophosphorylation assay. Yeast whole cell extracts were prepared from TCA-treated cells and analysed as described (Pellicioli et al., 1999). Briefly, yeast extracts were separated by 10% SDS–PAGE and blotted onto PVDF membrane. After denaturation and renaturation of proteins, an in situ autophosphorylation assay was performed by incubating the blot with [γ-32P]ATP. The blot was then washed extensively and subjected to autoradiography.
Acknowledgments
ACKNOWLEDGEMENTS
We thank members of the S.P.J. laboratory for stimulating discussions and critical comments, Chris Maddren for DNA sequencing and E. Louis, D. Shore, T. Tsukiyama, N. Lowndes, D. Weaver, A. Amon, D. Gottschling and J. Haber for strains and reagents. S.T. is funded by the Royal Society Dorothy Hodgkin Fellowship and this work is supported by the Cancer Research Campaign.
REFERENCES
- Barnes G. and Rio, D. (1997) DNA double-strand-break sensitivity, DNA replication, and cell cycle arrest phenotypes of Ku-deficient S. cerevisiae. Proc. Natl Acad. Sci. USA, 94, 867–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn E.H. (2000) Telomere states and cell fates. Nature 408, 53–56. [DOI] [PubMed] [Google Scholar]
- Bliss T.M. and Lane, D.P. (1997) Ku selectively transfers between DNA molecules with homologous ends. J. Biol. Chem., 272, 5765–5773. [DOI] [PubMed] [Google Scholar]
- Boulton S.J. and Jackson, S.P. (1996a) S. cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J., 15, 5093–5103. [PMC free article] [PubMed] [Google Scholar]
- Boulton S.J. and Jackson, S.P. (1996b) Identification of a S. cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res., 24, 4639–4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton S.J. and Jackson, S.P. (1998) Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J., 17, 1819–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans S.K., Sistrunk, M.L., Nugent, C.I. and Lundblad, V. (1998) Telomerase, Ku, and telomeric silencing in S. cerevisiae. Chromosoma, 107, 352–358. [DOI] [PubMed] [Google Scholar]
- Feldmann H. and Winnacker, E.L. (1993) A putative homologue of the human autoantigen Ku from S. cerevisiae. J. Biol. Chem., 268, 12895–12900. [PubMed] [Google Scholar]
- Feldmann H., Driller, L., Meier, B., Mages, G., Kellerman, J. and Winnacker, E.L. (1996) HDF2, the second subunit of the Ku homologue from S. cerevisiae. J. Biol. Chem., 271, 27765–27769. [DOI] [PubMed] [Google Scholar]
- Forstemann K., Hoss, M. and Lingner, J. (2000) Telomerase-dependent repeat divergence at the 3′ ends of yeast telomeres. Nucleic Acids Res., 28, 2690–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvik B., Carson, M. and Hartwell, L. (1995) Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol., 15, 6128–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S., Larrivee, M., Labrecque, P. and Wellinger, R.J. (1998) Yeast Ku as a regulator of chromosomal DNA end structure. Science, 280, 741–744. [DOI] [PubMed] [Google Scholar]
- Herrmann G., Lindahl, T. and Schar, P. (1998) S. cerevisiae LIF1: a function involved in DNA double-strand break repair related to mammalian XRCC4. EMBO J., 17, 4188–4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.E., Moore, J.K., Holmes, A., Umezu, K., Kolodner, R.D. and Haber, J. (1998) Saccharomyces Ku70, Mre11/Rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell, 94, 399–409 [DOI] [PubMed] [Google Scholar]
- Lowndes N.F. and Murguia, J.R. (2000) Sensing and responding to DNA damage. Curr. Opin. Genet. Dev., 10, 17–25. [DOI] [PubMed] [Google Scholar]
- Lydall D. and Weinert, T. (1997) Use of cdc13-1-induced DNA damage to study effects of checkpoint genes on DNA damage processing. Methods Enzymol., 283, 410–424. [DOI] [PubMed] [Google Scholar]
- McElhinny N.S.A., Snowden, C.M., McCarville, J. and Ramsden, D.A. (2000) Ku recruits the XRCC4-ligase IV complex to DNA ends. Mol. Cell. Biol., 20, 2996–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent C.I., Bosco, G., Ross, L.O., Evans, S.K., Salinger, A.P., Moore, J.K., Haber, J.E. and Lundblad, V. (1998) Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr. Biol., 8, 657–660. [DOI] [PubMed] [Google Scholar]
- Pang D., Yoo, S., Dynan, W.S., Jung, M. and Dritschilo, A. (1997) Ku proteins join DNA fragments as shown by atomic force microscopy. Cancer Res., 57, 1412–1415. [PubMed] [Google Scholar]
- Pellicioli A., Lucca, C., Liberi, G., Marini, F., Lopes, M., Plevani, P., Romano, A., Di Fiore, P.P. and Foiani, M. (1999) Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J., 18, 6561–6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polotnianka R.M., Li, J. and Lustig, A.J. (1998) The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr. Biol., 8, 831–834. [DOI] [PubMed] [Google Scholar]
- Porter S.E., Greenwell, P.W., Ritchie, K.B. and Petes, T.D. (1996) The DNA-binding protein Hdf1p (a putative Ku homologue) is required for maintaining normal telomere length in S. cerevisiae. Nucleic Acids Res., 24, 582–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer B. and Philippsen, P. (1991) CDC15, an essential cell cycle gene in S. cerevisiae, encodes a protein kinase domain. Yeast, 7, 265–273. [DOI] [PubMed] [Google Scholar]
- Smith G.C. and Jackson, S.P. (1999) The DNA-dependent protein kinase. Genes Dev., 13, 916–934. [DOI] [PubMed] [Google Scholar]
- Teo S.H. and Jackson, S.P. (2000) Lif1p targets the DNA ligase Lig4p to sites of DNA double-strand breaks. Curr. Biol., 10, 165–168. [DOI] [PubMed] [Google Scholar]
- Tsukamoto Y., Kato, J. and Ikeda, H. (1997) Silencing factors participate in DNA repair and recombination in S. cerevisiae. Nature, 388, 900–903. [DOI] [PubMed] [Google Scholar]
- Tuteja R. and Tuteja, N. (2000) Ku autoantigen: a multifunctional DNA-binding protein. Crit. Rev. Biochem. Mol. Biol., 35, 1–33. [DOI] [PubMed] [Google Scholar]