Abstract
In chromosomal rearrangements of acute myeloid leukaemia patients the mixed lineage leukaemia (MLL) gene, a human homolog of the Drosophila gene trithorax, is frequently fused to AF10. Here we describe the identification and a functional characterization of the Drosophila homolog dAF10. We show that dAF10 functions in heterochromatin-dependent genomic silencing of position effect variegation, a phenomenon associated with chromosomal rearrangements that cause mosaic expression of euchromatic genes when relocated next to heterochromatin. We also demonstrate that dAF10 can associate with the heterochromatin protein 1 (HP1) in vitro and in vivo. The results indicate that dAF10 is an HP1-interacting component of the heterochromatin-dependent gene silencing pathway, which either contributes to the stability of the heterochromatin complex or to its function.
INTRODUCTION
Leukaemogenesis correlates with alterations in chromatin structure brought about by chromosomal translocations (Saha et al., 1998). One gene, which is rearranged in 20% of acute myeloid leukaemias (AMLs), is termed mixed lineage leukaemia (MLL) (Bower et al., 1994). It codes for a human homolog of trithorax (trx), a Drosophila gene encoding a member of the trithorax group (trx-G) proteins which are necessary to maintain the spatially correct expression of homeotic genes during both Drosophila and vertebrate embryogenesis (Kennison, 1995; Paro, 1995; Yu et al., 1995). Trx-G proteins as well as their functional counterparts, the Polycomb group of proteins (Pc-G) act not only in the context of homeotic genes but also during oogenesis, neural development and cell proliferation (Gebuhr et al., 2000). Trx-G and Pc-G proteins comprise a long-term memory system of cells to pass on a particular developmental fate through many rounds of cell division by an epigenetic mechanism that probably involves chromatin remodelling (Gebuhr et al., 2000). Although the MLL protein has not been shown to remodel chromatin, its C-terminal region was found to interact with components of the SNF/SWI complex, a chromatin remodelling system which maintains the active state of transcription (Gebuhr et al., 2000). In acute leukaemia patients, the C-terminal region of MLL is lost due to reciprocal translocations at chromosomal locus 11q23, which result in fusion proteins composed of the N-terminal portion of MLL extending into structurally and functionally unrelated fusion partners (Look, 1997; Adler et al., 1999).
Translocations that cause AML show remarkable specificity for hematopoetic cells blocked in defined stages of differentiation. This property suggests that the different oncogenes produced by the translocations specifically interfere with transcriptional networks which normally function in concert with growth factors to regulate hematopoesis. This conclusion is reinforced by the results of genetically manipulated mice which showed significant effects on normal hematopoesis in response to translocation-targeted transcription factors (reviewed in Look, 1997). To date fifteen Mll fusion genes that result from chromosomal translocations have been cloned and characterized (reviewed in Look, 1997; Adler et al., 1999). Two of these are CBP and AF10 (Chaplin et al., 1995), they have been shown to carry oncogenic potential also in the context of fusion partners different from MLL (Borrow et al., 1996; Dreyling et al., 1996). CBP is a transcriptional coactivator that acts as a multifunctional adapter protein to regulate transcription through acetylation of chromatin and recruitment of basal transcription factors (Kitabayashi et al., 1998), whereas the molecular function of the second, AF10, is unknown.
Here we present a functional analysis of the Drosophila homolog of AF10, referred to as dAF10. We show that dAF10 contains a pentameric protein–protein interaction motif and can associate in vitro with HP1, the Drosophila heterochromatin protein 1 (Paro and Hogness, 1991) which is encoded by the gene Su(var)2-5. HP1 takes part in a gene silencing phenomenon, termed position effect variegation (PEV), which refers to mosaic expression of euchromatic genes if they become relocated next to heterochromatin due to chromosomal rearrangements (Spofford, 1967). PEV is suppressed by HP1 mutations (Clark and Elgin, 1992). We show that dAF10 functions like HP1 as a suppressor of PEV and interacts, by genetic means, with Su(var)2-5. The results suggest that dAF10 participates in chromatin mediated gene silencing.
RESULTS AND DISCUSSION
Structure and transcript pattern of the Drosophila AF10 homolog
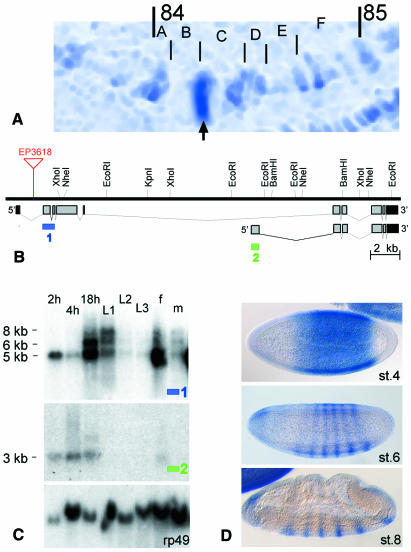
Cloning of the Drosophila homolog dAF10, was initiated by a database search (Berkeley Drosophila Genome project, BDGP; http//www.fruitfly.org/). This search found the corresponding annotated transcription unit (CG1070) (Adams et al., 2000), which maps into region 84C1-2 on the right arm of chromosome 3 (Figure 1A). Sequence analysis of both the genomic DNA and various EST clones (SD04152, SD09324, LP02873 and LP08584) confirmed the chromosomal location as outlined by the BDGP (http//www.fruitfly.org/) and revealed that dAF10 encodes different splicing variants of which the two major forms (see below) could be assigned unambiguously. The structure of the dAF10 gene and the two major dAF10 transcripts are summarized in Figure 1B.
Fig. 1. Genomic organization and the spatiotemporal expression pattern of dAF10. (A) Polytene chromosome in situ hybridization showing that dAF10 is located on the right arm of chromosome 3 in region 84C1–84C2, indicated by an arrow. (B) Schematic representation of the genomic organization of dAF10, the mapping of the two major dAF10 transcripts and location of the P-element EP3618 in the first intron. Position of fragments used as probes for northern blot analysis (C) are indicated (1,2; blue or green boxes). (C) Developmental northern blot probed with antisense RNA of fragments 1 and 2 [see blue or green boxes in (B)] and ribosomal protein rp49 antisense RNA (top to bottom). Total RNA of 2, 4 and 18 h old embryos, first (L1), second (L2) and third (L3) instar larvae, and adult females (f) and males (m) were loaded. Transcript sizes are indicated. (D) In situ hybridization pattern of whole-mount preparations from embryos (stages 4, 6, 8; Campos-Ortega and Hartenstein, 1997) as revealed by antisense RNA corresponding to both the 3 and 5 kb cDNAs. Lateral views (anterior is left, dorsal up) showing that expression is restricted along the anterior–posterior axis of embryos (stage 4), and subsequently resolves into a series of stripes during cellular blastoderm and early gastrulation (stages 6 and 8).
In order to establish the temporal pattern of the dAF10 expression, we performed developmental northern blot analysis (Figure 1C). dAF10 codes for four different transcripts with distinct temporal expression profiles during the Drosophila life cycle. One of two major transcripts, ∼5 kb in length, is constitutively expressed with high levels during embryogenesis and in adult females. The second major transcript of ∼3 kb is restricted to embryogenesis and females. In addition, two splicing variants of the transcript (6 and 8 kb) were found from later stages of embryogenesis onwards (Figure 1C).
Whole-mount in situ hybridization to oocytes and embryos revealed that the 5 and 3 kb transcripts are expressed maternally. Both splicing variants were found in nurse cells and later equally distributed in oocytes (data not shown). They are maintained in fertilized eggs and decease gradually until the preblastoderm stage. Zygotic dAF10 expression is initiated during syncytial blastoderm (stage 4; stageing according to Campos-Ortega and Hartenstein, 1997) and develops a stripe pattern similar to pair-rule segmentation genes (Figure 1D). A detailed description of the dAF10 expression patterns will be described elsewhere (B. Linder, N. Gerlach and H. Jäckle, in preparation) since embryonic dAF10 function is beyond the scope of this study.
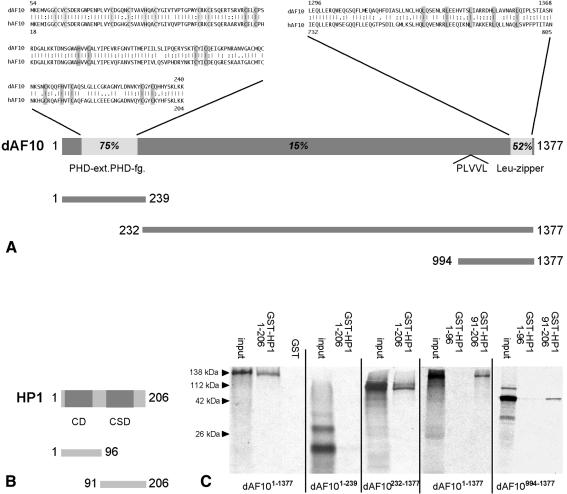
The 3 and 5 kb transcripts were recovered in two corresponding full-size cDNA clones. The 5 kb transcript encodes a 4131 bp open reading frame (ORF) corresponding to a 1377 amino acids polypeptide (DDBJ/EMBL/GenBank accession No. AF281145) which contains a PHD-finger next to an extended PHD-finger domain and a leucine zipper motif as observed with human AF10 (Linder et al., 2000) (Figure 2A). The 3 kb transcript starts with an alternative exon (Figure 1B) and codes for an 826 amino acid protein (DDBJ/EMBL/GenBank accession No. AF281146), which lacks the N-terminal PHD-finger/extended PHD-finger domains. Thus, only the larger of the two major transcripts codes for the human AF10 homolog. However, both variants contain the leucine zipper motif and a PLVVL pentamer motif (Smothers and Henikoff, 2000) found in a subset of proteins that interact with HP1 in vitro (Murzina et al., 1999; Ryan et al., 1999; Smothers and Henikoff, 2000) (Figure 2A). This observation suggests that dAF10 may bind to HP1 and function in an HP1-dependent manner.
Fig. 2. (A) Sequence alignment of the Drosophila and human AF10 proteins showing the conserved N-terminal PHD- (light grey) and extended PHD-finger (dark grey) motifs and the C-terminal leucine zipper region. Sequence identities are 75 and 52%, respectively. Consensus Cys-His-residues of the PHD-fingers and Leu of the leucine zipper region are highlighted. The chromo shadow domain-binding peptide (PLVVL) is indicated. In vitro translated sub-fragments of dAF10 for pull down assays are shown below. (B) Schematic presentation of the protein structure of HP1 with chromo domain (CD) and chromo shadow domain (CSD) marked in grey; regions used for the protein–protein interaction experiments are indicated below. (C) GST pull-down assays showing an interaction between dAF10 and HP1–GST fusion protein. In vitro translated dAF10 and fragments thereof are indicated below the panel, GST fusion proteins on top. Note that the chromo shadow domain of HP1 binds to the PLVVL-containing C-terminal region of dAF10. Apparent molecular weights are indicated on the left.
Interaction of HP1 and dAF10 in vitro and in vivo
We examined a possible dAF10::HP1 interaction via pull-down assays involving a GST–HP1 fusion protein and in vitro translated, labelled dAF10. HP1 binds via its chromo shadow domain to full-size dAF10 and to the subfragment that contains the PLVVL motif (Figure 2B and C). These findings suggest that dAF10 may function in an HP1-dependent gene silencing pathway.
HP1 is encoded by the Su(var)2-5, a suppressor of PEV. This silencing phenomenon is seen with the inversion mutation In(1)wmh4, which results in a relocation of the white (w) gene next to heterochromatin. In this location, w expression is silenced in some ommatidia of the Drosophila eye but not in others (Elgin, 1996). Males which contain the Su(var)2-5 mutation in addition to the In(1)wmh4 mutant suppress PEV, causing w expression in a significant larger area of the eye. If the interaction between HP1 and dAF10 is indeed functional, the derepression effect Su(var)2-5 should be enhanced in flies which carry a mutant dAF10 allele in addition to Su(var)2-5 (see below).
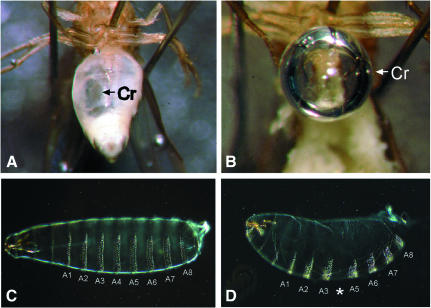
In order to apply the PEV assay, we generated mutations of the dAF10 gene by taking advantage of a P-element insertion within the first intron of the gene (EP3618, see Figure 1B). Homozygous EP3618 individuals are semi-lethal, meaning that up to 70% of the individuals develop into viable and fertile adults. Such individuals exert two characteristic defects: they are unable to fly and about half of them develop a severe dilation of the diventricular crop, a cuticle-lined muscular organ of the upper digestive tract (Strasburger, 1935) (Figure 3A and B). The offspring of those flies, which have a strongly reduced lifespan of about one week only, are embryonic-lethal. They develop a segmentation phenotype (see Figure 3C and D), suggesting that maternal expression of dAF10 in females (see above) is essential for embryogenesis.
Fig. 3. Phenotype of dAF10 mutants. (A and B) Homozygous EP3618 flies show a severe dilation of the diventricular crop (Cr). Note that due to the lack of muscles, this organ is only cuticle lined and thus unable to contract; it appears as a gas filled bubble (B). (C and D) Larval cuticle preparations of wildtype (C) and homozygous dAF10 mutant embryos laid by heterozygous females (D). Note the loss of abdominal segment 4 (asterisk) and pattern defects in the terminal regions (head and non-segmented tail region) in mutant embryos.
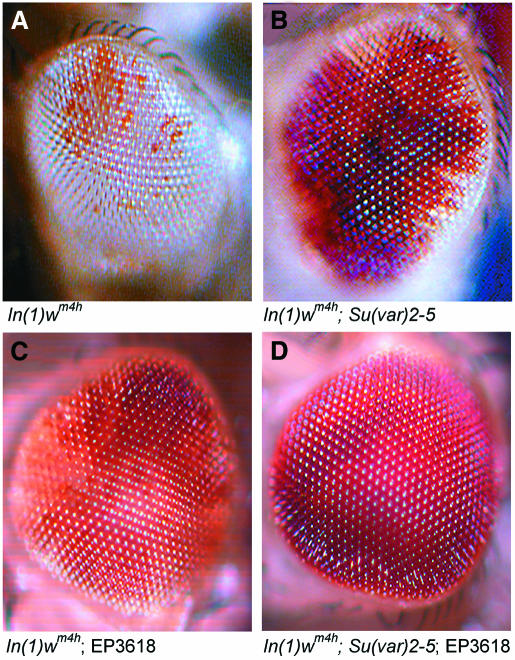
We examined w expression in the eyes of In(1)wmh4 males (Figure 4A) which carried the EP3618 insertion mutant, a dAF10 lack-of-function allele generated by imprecise excision of the EP3618 element (B. Linder, N. Gerlach and H. Jäckle, in preparation) or the wildtype EP3618 chromosome from which the EP element had been excised precisely. In individuals which carry the dAF10 lack-of-function or the the EP3618 insertion allele, the w silencing area in the eye was reduced as observed in In(1)wmh4 individuals bearing the Su(var)2-5 allele (Figure 4B,C), whereas no effect on w silencing had been observed with males bearing the chromosome from which the EP element had been excised precisely (data not shown). These observations suggest that like HP1, dAF10 is a functional repressor component of wildtype heterochromatin, which renders the inverted w gene inactive in most but not all ommatidia. Furthermore, in In(1)wmh4 males which are double-heterozygous for a mutant dAF10 and Su(var)2-5 alleles, silencing of w expression was completely suppressed. The resulting wildtype-like w expression (Figure 4D) indicates that dAF10 and HP1 are components of the same gene silencing pathway.
Fig. 4. Genetic interaction of dAF10 and HP1 as revealed by PEV suppression. (A) Eye of a male that carries the inversion mutation In(1)wmh4. Note white expression in the minority of ommatidia, which escaped heterochromatic silencing. (B) Eye of a male that carries the inversion In(1)wmh4 and the Su(var)2-5 mutation. Note suppression of silencing as reflected in an increased number of white expressing ommatidia. (C) Eye of a male that carries the inversion In(1)wmh4 and the dAF10 mutation. Note that white expression expands over more ommatitia in a manner similar to what is observed in the Su(var)2-5 mutation (compare B and C), indicating that dAF10 is a suppressor of PEV. (D) Suppression of PEV in an eye of a male that carries the inversion In(1)wmh4, the Su(var)2-5 and the dAF10 mutations. Note wildtype-like white expression, indicating that both HP1 and dAF10 act in heterochromatic gene silencing.
Our results establish that dAF10 and HP1 can associate in vitro and that both components act in heterochromatin-induced gene silencing. It has been proposed that this type of silencing results from a co-operative assembly of heterochromatin as multimeric complexes proceeding from a pre-existing block of heterochromatin (Locke et al., 1988). HP1 action involves the recognition of an epigenetic methylation mark at lysine 9 of the N-terminus of histone (Lachner et al., 2001) and is likely to cause chromatin remodelling involving homo- and/or heterophilic protein–protein interactions (Cavalli and Paro, 1998). Furthermore, human HP1 was found to be associated with the lamin B receptor, a component of the inner nuclear membrane (Ye and Worman, 1996). These observations are consistent with the argument that HP1 is a constitutive component of heterochromatin, which initiates silencing at distinct sites and may also function in the subnuclear localization of heterochromatin.
SPECULATION
dAF10 exerts a spatiotemporally restricted expression profile. This finding, the in vitro association between HP1 and dAF10 and their genetic interaction in suppression of PEV suggest that dAF10 is a component of a distinct heterochromatin-dependent silencing process. dAF10 may either contribute to the stability of the heterochromatin complex or serve as an attachment site for other proteins to join the silencing complex. Interestingly, the human AF10 is frequently fused with the trithorax homolog MLL of AML patients (Bower et al., 1994). It is therefore tempting to speculate that combining MLL with AF10 in a chimeric MLL–AF10 fusion protein causes a switch in cellular memory. The protein may attach to HP1 and cause gene silencing instead of maintaining target gene expression.
METHODS
Molecular analysis of dAF10. ESTs used for sequencing the cDNA of dAF10 were SD04152, LP08584, SD09324, LP02873 (BDGP; http//www.fruitfly.org/). Total RNA was isolated using RNAzol (Gibco-BRL, Germany) according to the manufacturer’s instructions. 30 µg of each RNA fraction were separated on a 1% formaldehyde gel. For synthesis of the antisense RNA probes and blotting we used the Strip-EZ RNA and Northern Max Kit from Ambion (Germany). Northern blots were hybridized with dAF10 probes corresponding to the following regions: bp 1–1295 of the 5 kb transcript, bp1-564 (alternative exon) of the 3 kb transcript (Figure 1B) and rp49 as a loading control. Digoxigenin-labelled antisense LP08584 RNA was used for in situ hybridization of chromosome squashes (Hartmann and Jäckle, 1995) and whole-mount preparations of ovaries and embryos (Hauptmann and Gerster, 1996).
Generation and analysis of dAF10 mutants. P-element line EP3618 was obtained from the Hungarian stock centre (BDGP; http//www.fruitfly.org/). In order to correlate the EP3618-mutant phenotype with the dAF10 gene, we remobilized the P-element by crossing EP3618 flies with flies containing a transposase-expressing transgene. Mobilization caused two precise excisions of the P-element, resulting in a reversion of the EP3618-mutant phenotype to wildtype. Thus, the P-element insertion is the cause of the mutant phenotype. In addition, we obtained three lethal lines, each carrying a deletion of the dAF10 gene. Embryos homozygous for the dAF10 deletions were embryonic lethal, showing the same segmentation phenotype as seen with embryos from females homozygous for the EP3618 allele. A detailed molecular description of the alleles and their effects on embryonic development will be presented elsewhere (B. Linder, N. Gerlach and H. Jäckle, in preparation). For the PEV modifier assay the following stocks were used: In1wm4h; Su(var)2-5(5) and In1wm4h; Su(var)2-5(4) (Umeå stock centre). Larval cuticle were prepared according to Nüsslein-Volhard and Wieschaus (1980).
Protein–protein interaction assay. GST protein expression, their purification and the pull-down assays were performed as described (Linder et al., 2000). PCR fragments covering the full ORFs of dAF10 (aa 1–1377), the PHD- and extended PHD-finger (aa 1–239) and fragments of the C-terminus (aa 232–1377, aa 994–1377) were cloned into pcDNA-HisB (Invitrogen) for in vitro translation. PCR fragments covering the ORF of HP1 (aa 1–206), the chromo- (aa1-96) and chromo shadow domain (aa91–206) were cloned into and expressed from pGEX-4T3 (Amersham).
Acknowledgments
ACKNOWLEDGEMENTS
We thank R. Pflanz, G. Dowe, M. Gonzalez-Gaitan and Wendy Gerber for help during various parts of the work. Joel C. Eissenberg kindly provided the HP1 cDNA. The work was supported by the Max-Planck Society.
REFERENCES
- Adams M.D. et al. (2000) The genome sequence of Drosophila melanogaster. Science, 287, 2185–2195. [DOI] [PubMed] [Google Scholar]
- Adler H.T., Chinery, R., Wu, D.Y., Kussick, S.J., Payne, J.M., Fornace, A.J.Jr and Tkachuk, D.C. (1999) Leukemic HRX fusion proteins inhibit GADD34-induced apoptosis and associate with the GADD34 and hSNF5/INI1 proteins. Mol. Cell. Biol., 10, 7050–7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrow J. et al. (1996) The translocation t(8;16)(p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nature Genet., 14, 33–41. [DOI] [PubMed] [Google Scholar]
- Bower P., Parry, M., Carter, D.M., Lillington, D.M., Amess, J., Lister, T.A., Evans, G. and Young, B. (1994) Prevalence and clinical correlations of MLL gene rearrangements in AML-M4/5. Blood, 84, 3776–3780. [PubMed] [Google Scholar]
- Campos-Ortega J.A. and Hartenstein, V. (1997). The Embryonic Development of Drosophila melanogaster. 2nd edn. Springer-Verlag, Berlin).
- Cavalli G. and Paro, R. (1998) Chromo-domain proteins: linking chromatin structure to epigenetic regulation. Curr. Opin. Cell Biol., 10, 354–360. [DOI] [PubMed] [Google Scholar]
- Chaplin T., Bernard, O., Beverloo, H., B.Saha, V., Hagemeijer, A., Berger, R. and Young, B.D. (1995) The t(10;11) translocation in acute myeloid leukaemia (M5) consistently fuses the leucine zipper motif of AF10 onto the HRX gene. Blood, 86, 2073–2076. [PubMed] [Google Scholar]
- Clark R.F. and Elgin, S.C. (1992) Heterochromatin protein 1, a known suppressor of position-effect variegation, is highly conserved in Drosophila. Nucleic Acids Res., 20, 6067–6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyling M.H., Martinez-Climent, J.A., Zheng, M., Mao, J., Rowley, J.D. and Bohlander, S.K. (1996) The t(10;11)(p13;q14) in the U937 cell line results in the fusion of the AF10 gene and CALM, encoding a new member of the AP-3 clathrin assembly protein family. Heterochromatin and gene regulation in Drosophila. Proc. Natl Acad. Sci. USA, 93, 4804–4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin S.C.R. (1996) Heterochromatin and gene regulation in Drosophila. Curr. Opin. Genet. Dev., 6, 193–202. [DOI] [PubMed] [Google Scholar]
- Gebuhr T.C., Blutman, S.J. and Magnuson, T. (2000) Pc-G/trx-G and the SWI/SNF connection: Developmental gene regulation through chromatin remodeling. Genesis, 26, 189–197. [DOI] [PubMed] [Google Scholar]
- Hartmann C. and Jäckle, H. (1995) Spatiotemporal relationships between a novel Drosophila stripe expressing gene and known segmentation genes by simultaneous visualisation of transcript patterns. Chromosoma, 104, 84–91. [DOI] [PubMed] [Google Scholar]
- Hauptmann G. and Gerster, T. (1996) Multicolor whole mount in situ hybridisation to Drosophila embryos. Genes Dev. Evol., 206, 292–295. [DOI] [PubMed] [Google Scholar]
- Kennison J.A. (1995) The Polycomb and trithorax group proteins of Drosophila: trans-regulators of homeotic gene function. Annu. Rev. Genet., 29, 289–303. [DOI] [PubMed] [Google Scholar]
- Kitabayashi I., Yokoyama, A., Shimizu, K. and Ohki, M. (1998) Interactional and functional cooperation of the leukemia-associated factors AML1 and p300 in myeloid cell differentiation. EMBO J., 17, 2994–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M., O’Carroll, D., Rea, S., Mechtler, K. and Jenuwein, T. (2001) HP1 proteins recognize the Suv39h-dependent methylation mark in the histone H3 terminus. Nature, in press. [DOI] [PubMed] [Google Scholar]
- Linder B., Newman, R., Jones, L.K., Debernardi, S., Young, B.D., Freemont, C.P., Verrijzer, C.P. and Saha, V. (2000) Biochemical analyses of the AF10 protein: The extended LAP/PHD-finger mediates oligomerisation. J. Mol. Biol., 299, 369–378. [DOI] [PubMed] [Google Scholar]
- Locke J., Kotarski, M.A. and Tartof, K.D. (1988) Dosage-dependent modifiers of position effect variegation in Drosophila and a mass action model that explains their effect. Genetics, 120, 181–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Look A.T. (1997) Oncogenic transcription factors in the human acute leukemias. Science, 278, 1059–1064. [DOI] [PubMed] [Google Scholar]
- Murzina N., Verrault, A., Laue, E. and Stillman, B. (1999) Heterochromatin dynamics in mouse cells: interaction between chromatin assembly factor 1 and HP1 proteins. Mol. Cell. Biol., 4, 529–540. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C., Wieschaus, E. (1980) Mutations affecting segment number and polarity in Drosophila. Nature, 287, 795–801. [DOI] [PubMed] [Google Scholar]
- Paro R. (1995) Propagating memory of transcriptional states. Trends Genet., 11, 295–297. [DOI] [PubMed] [Google Scholar]
- Paro R. and Hogness, D.S. (1991) The polycomb protein shares a homologous domain with a heterochromatin-associated protein of Drosophila.Proc. Natl Acad. Sci. USA, 88, 263–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan R.F. et al. (1999) KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Kruppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol. Cell. Biol., 19, 4366–4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha V., Young, B.D. and Freemont, P.S. (1998) Translocations, fusion genes, and acute leukemia. J. Cell. Biochem., 3, 264–276. [DOI] [PubMed] [Google Scholar]
- Smothers J.F. and Henikoff, S. (2000) The HP1 chromo shadow domain binds a consensus peptide pentamer. Curr. Biol., 10, 27–30. [DOI] [PubMed] [Google Scholar]
- Spofford J.B. (1967) Position-effect variegation in Drosophila. In Ashburner, M. and Novitski, E. (eds), The Genetics and Biology of Drosophila. Vol 1c. Academic Press, London, UK, pp. 955–1018.
- Strasburger E.H. (1935) Drosophila melanogaster Meig. Eine Einführung in den Bau und die Entwicklung. Julius Springer-Verlag, Berlin.
- Ye Q. and Worman, H.J. (1996) Interaction between an integral protein of the nuclear envelope inner membrane and human chromodomain proteins homologous to Drosophila HP1. J. Biol. Chem., 271, 14653–14656. [DOI] [PubMed] [Google Scholar]
- Yu B.D., Hess, J.L., Horning, S.E., Brown, G.A. and Korsmeyer, S.J. (1995) Altered Hox expression and segmental identity in Mll-mutant mice. Nature, 378, 505–508. [DOI] [PubMed] [Google Scholar]