Abstract
The type II secretion machinery allows most Gram-negative bacteria to deliver virulence factors into their surroundings. We report that in Erwinia chrysanthemi, GspE (the putative NTPase), GspF, GspL and GspM constitute a complex in the inner membrane that is presumably used as a platform for assembling other parts of the secretion machinery. The GspE–GspF–GspL–GspM complex was demonstrated by two methods: (i) co-immunoprecipitation of GspE–GspF–GspL with antibodies raised against either GspE or GspF; (ii) interactions in the yeast two-hybrid system between GspF and GspE, GspF and GspL, GspL and GspM. GspL was found to have an essential role in complex formation. We propose a model in which the GspE–GspF–GspL–GspM proteins constitute a building block within the secretion machinery on top of which another building block, referred to as a pseudopilus, assembles. By analogy, we predict that a similar platform is required for the biogenesis of the type IV pilus.
INTRODUCTION
A number of virulence determinants, in plant, animal or human pathogens, are secreted. Among the dedicated pathways for the secretion of virulence determinants, the type II secretory pathway is the most widely distributed in Gram-negative bacteria, including the pathogens Vibrio cholerae, Aeromonas hydrophila, Pseudomonas aeruginosa, Xanthomonas campestris, Erwinia chrysanthemi and Erwinia carotovora (Nunn, 1999). This pathway, also called the general secretory pathway (GSP), allows the secretion of a large variety of degradative enzymes (cellulases, pectinases, proteases) and toxins (aerolysin, cholera toxin). Type II protein secretion is thought to be a stepwise process: secreted proteins are first translocated by a Sec-dependent mechanism through the inner membrane (reviewed in Driessen et al., 1998), released into the periplasm, and transported across the outer membrane by the so-called Gsp proteins (for review see Filloux et al., 1998; Russel, 1998; Nunn, 1999).
Topological analysis of the Gsp proteins revealed a surprising feature since, despite being essential for crossing the outer membrane, 12 out of the 14 Gsp proteins are associated with the inner membrane (Filloux et al., 1998; Russel, 1998; Nunn, 1999). Interestingly, a subset of Gsp proteins is homologous to the proteins required for the biogenesis of type IV pili (Hobbs and Mattick, 1993). As a consequence, Gsp proteins were proposed to form a pseudopilus extending from the cytoplasmic side of the inner membrane up to the outer membrane through the periplasm. Recently, this point received support by the finding that GspG can form pilus-like structures (Sauvonnet et al., 2000).
Understanding how the 14 Gsp proteins interact and assemble, to allow protein secretion, has been a challenge in the last decade. Use of the classic biochemical approach as well as the yeast two-hybrid system has revealed only binary interactions, namely GspD–GspS, GspD–GspC, GspE–GspL and GspL–GspM (Nunn, 1999; Py et al., 1999; Possot et al., 2000). In the present study, the E. chrysanthemi GSP machinery was investigated. We report for the first time the existence of a heteromeric complex containing GspE–GspF–GspL–GspM proteins. We discuss a model in which these proteins constitute a platform in the inner membrane, used for the assembly of other parts of the secretion machinery.
RESULTS
In this study we used the E. chrysanthemi Out secretion machinery as a model. For the sake of simplicity, we decided to use the GSP nomenclature, instead of Out, throughout this report.
GspF interacts with GspE and GspL
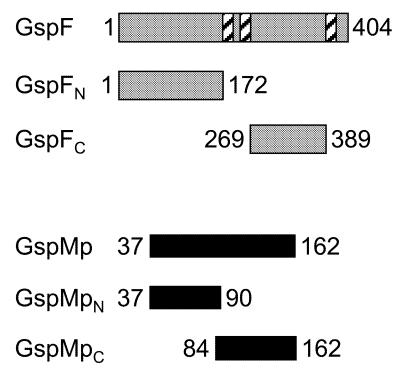
GspF is a polytopic, integral, cytoplasmic membrane protein with an N-terminal cytoplasmic domain followed by a small periplasmic loop and a large C-terminal cytoplasmic loop (Figure 1). Such a topology was deduced from hydropathy profile analysis, and by analysis of GspF–BlaM hybrids in Escherichia coli and E. carotovora (Thomas et al., 1997). Using the yeast two-hybrid system, we investigated whether GspF interacted with GspE and GspL, since these are thought to form a complex on the cytoplasmic side of the inner membrane. The two large cytoplasmic regions of GspF, referred to as GspFN and GspFC (Figure 1), were fused to LexA to produce hybrid proteins with DNA binding activity. The cytoplasmic domain of GspL, referred to as GspLc, and GspE were each fused to the transcriptional activator motif B42 (Py et al., 1999). Co-expression in yeast cells of GspFN with either GspE or GspLc led to transcriptional activation of lacZ (Table I). In contrast, no β-galactosidase activity was found in yeast cells expressing GspFC instead of GspFN (Table I). These results indicated that the first N-terminal 172 residues of GspF are able to interact with both GspE and the cytoplasmic domain of GspL.
Fig. 1. Schematic representation of the regions of GspF and GspM analysed in the yeast two-hybrid system. Schematic representation of the regions of GspF and GspM that have been fused at their N-terminus either to LexA or to the B42 activation domain. The number next to the boxes indicates the position of the residues that defined the domain studied. Hatched box, transmembrane region.
Table I. Interaction of GspF, GspE, GspL and GspM in the yeast two-hybrid system.
| LexA hybrid protein | B42 hybrid protein | β-galactosidase activity (units) |
|---|---|---|
| GspFN | – | 0 |
| GspFN | GspE | 720 ± 214 |
| GspFN | GspLc | 353 ± 20 |
| GspFC | – | 0 |
| GspFC | GspE | 0 |
| GspFC | GspLc | 0 |
| GspMp | – | 4 ± 4 |
| _ | GspLp | 0 |
| GspMp | GspLp | 345 ± 54 |
| GspMpC | GspLp | 378 ± 73 |
| GspMpN | GspLp | 0 |
| – | GspMp | 4 ± 4 |
| GspLp | – | 0 |
| GspLp | GspMp | 355 ± 68 |
| GspLp | GspMpC | 490 ± 89 |
| GspLp | GspMpN | 0 |
| GspMp | GspMp | 82 ± 23 |
| GspMpC | GspMpC | 128 ± 45 |
| GspMpN | GspMpN | 0 |
Mating was carried out between appropriate yeast strains in order to produce the pair of hybrid proteins indicated. β-galactosidase activity was assayed from diploid clarified cell lysates. A minimum of two independent assays were performed; the average of the β-galactosidase activity and the standard error are indicated. From the results of immunoblots using anti-HA or anti-LexA antibodies, we concluded that all the hybrid proteins were produced in yeast.
GspF, GspE and GspL form a stable complex in vivo
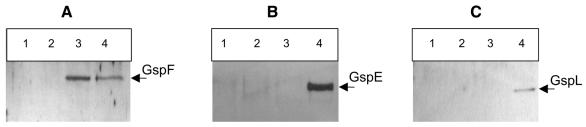
In order to test whether GspF formed a complex with GspE and GspL, co-immunoprecipitation experiments were performed. Anti-GspF antibodies coupled to protein A–Sepharose beads were used. GspE and GspL were tagged with the haemagglutinin (HA) and the vesicular stomatitis virus glycoprotein (VSV) epitope, respectively, allowing the use of the cognate monoclonal antibodies. GspF was produced from the cosmid pCPP2215, which carries the entire gsp cluster deleted for gspE (Lindeberg et al., 1996), to prevent potential competition between the native and the tagged GspE. Solubilized extracts were subjected to immunoprecipitation using anti-GspF antibodies.The presence of GspF, GspE or GspL in the immunoprecipitated material was analysed by immunoblotting with anti-GspF, anti-HA and anti-VSV antibodies, respectively. GspE, GspL and GspF were co-immunoprecipitated from strain DH5α(pCPP2215/pK-EL) (Figure 2, lane 4). Control experiments showed that GspE and GspL were not immunoprecipitated with anti-GspF antibodies in the absence of GspF (Figure 2, lane 2). Together, these results demonstrated that GspE, GspF and GspL form a stable complex in vivo.
Fig. 2. Co-immunoprecipitation of GspF, GspE and GspL with anti-GspF antibodies. Cells were solubilized in Triton X-100 and subjected to immunoprecipitation using anti-GspF antibodies. Immunoprecipitated material was analysed. Strains used were as follows: lane 1, DH5α; lane 2, DH5α(pK-EL); lane 3, DH5α(pCPP2215/pBADIK); lane 4, DH5α(pCPP2215/pK-EL). GspF, GspE and GspL were detected by immunoblotting with anti-GspF (A), anti-HA (B) or anti-VSV (C) antibodies, respectively. Visualization was by chemiluminescence (Amersham). GspF, GspE and GspL are indicated by an arrow.
In vivo biogenesis of the GspE–GspF–GspL complex
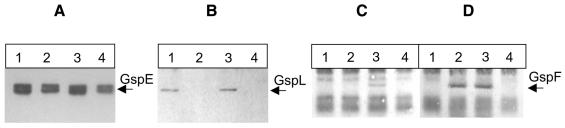
We tested whether GspL was necessary for the GspE–GspF complex to form. Co-immunoprecipitation experiments were performed using anti-GspE antibodies coupled to protein A–Sepharose beads, and strains containing the cosmid pCPP2222, which carries the entire gsp cluster deleted of gspL (Lindeberg et al., 1996). Triton X-100-solubilized extracts of strain DH5α(pCPP2222/pK-EL), which produces GspE, GspL and GspF, and of strain DH5α(pCPP2222/pK-E), which produces GspE and GspF, were used. The presence of GspE, GspL and GspF in the immunoprecipitated material was checked by immunoblotting. First, GspF, GspL and GspE were co-immunoprecipitated from strain DH5α(pCPP2222/pK-EL) by anti-GspE antibodies (Figure 3A, B and C, lane 3). This result gave further evidence for the existence of a GspE–GspF–GspL complex in vivo (see above). Secondly, as expected, GspE was immunoprecipited from strain DH5α(pCPP2222/pK-E) by anti-GspE antibodies, but GspF did not co-immunoprecipitate with GspE (Figure 3A and C, lane 2). As a control, we showed using this strain that GspF was immunoprecipitated by anti-GspF antibodies (Figure 3D, lane 2). These results demonstrated that GspL is required for the formation of the GspE–GspF complex.
Fig. 3. Analysis of the GspE–GspF–GspL complex in the absence of GspF or GspL. A Triton X-100-soluble cell extract was subjected to immunoprecipitation using antibodies raised against either GspE (A–C) or GspF (D). GspE, GspL and GspF were detected in the immunoprecipitated material by immunoblotting using anti-HA (A), anti-VSV (B) or anti-GspF (C and D) antibodies, respectively. Strains used were as follows: lane 1, DH5α(pK-EL); lane 2, DH5α(pCPP2222/pK-E); lane 3, DH5α(pCPP2222/pK-EL); lane 4, DH5α(pK-E). Visualization was either by chemiluminescence (Amersham) (A and B) or colorimetry (C and D).
Conversely, we tested whether the GspE–GspL complex forms in the absence of GspF. Triton X-100-solubilized extract from the strain that produces GspE and GspL (DH5α/pK-EL) was used and subjected to immunoprecipitation using anti-GspE antibodies. GspL and GspE were co-immunoprecipitated (Figure 3A and B, lane 1), indicating that GspF is not required for the formation of the GspE–GspL complex.
GspL interacts with GspM
Through its periplasmic domain, GspL could recruit other Gsp proteins into the GspE–GspF–GspL complex. According to its topology, GspM, an inner membrane protein with one transmembrane segment and a large periplasmic domain, was one of the potential candidates to interact with GspL on the periplasmic side of the membrane. We used the yeast two-hybrid system to test whether there is an interaction between the periplasmic domains of GspM and GspL, referred to as GspMp and GspLp, respectively. Each domain was fused either to LexA or to the transcriptional activator motif B42. Co-expression of hybrid proteins, containing GspLp or GspMp, in yeast cells led to transcriptional activation of lacZ (Table I), indicating that GspL and GspM interacted via their periplasmic domain. We delineated the region in GspM required for the interaction with GspL by using truncated forms of GspM, GspMpN and GspMpC (Figure 1). Yeast cells producing GspMpC and GspLp had a β-galactosidase activity similar to that detected in cells producing GspMp and GspLp. In contrast, no β-galactosidase activity was found in yeast cells expressing GspMpN and GspLp (Table I). These data indicated that GspM, via its last 79 residues, interacts with GspL.
Moreover, yeast cells producing the two fusion proteins containing the same region of GspM, GspMp or GspMpC, had similar β-galactosidase activities. In contrast, no β-galactosidase activity was found in yeast cells expressing the two fusion proteins containing GspMpN (Table I). Together, these data indicated that GspM forms homodimers, at least, via its last 79 residues.
DISCUSSION
Results obtained in this study allow us to propose that GspE, GspF, GspL and GspM form a complex within the Gsp machinery. Co-immunoprecipitation experiments allowed us to identify a complex containing GspE–GspF–GspL. The existence of such a complex is supported by results obtained in the yeast two-hybrid system, which revealed two new types of binary interactions on the cytoplasmic side of the inner membrane, namely GspF–GspE and GspF–GspL. This finding is of particular significance since no partner for GspF had ever been reported. That GspM also belongs to this complex is strongly supported both by our results from the yeast two-hybrid assay, showing an interaction GspL–GspM, and by studies in other systems homologous to Gsp (Michel et al., 1998; Sandkvist et al., 1999; Possot et al., 2000). The GspE–GspF–GspL–GspM complex will probably rank amongst the largest macromolecular complexes described in the cytoplasmic membrane. Indeed, we previously reported that GspE and GspL were able to form homomultimers (Py et al., 1999). We showed in the present work that GspM is also capable of homomultimerization. Furthermore, two GspE homologues, one involved in conjugative transfer of RP4 and the other found in the cag pathogenicity island of Helicobacter pylori, were reported to form homohexamers (Krause et al., 2000). Hence, the size of the GspE–GspF–GspL–GspM complex is expected to be 500 kDa, at least.
A major question relates to the biogenesis of the GspE–GspF–GspL–GspM complex. Our results suggested that GspE and GspL form a complex that subsequently associates with GspF. Indeed, in vivo, GspE–GspL complex formation is independent of GspF, while formation of the GspE–GspF complex is dependent on the presence of GspL. The step at which GspM is integrated into the complex remains to be determined. However, since we previously showed that the GspE–GspL interaction drives a conformational change of GspL (Py et al., 1999), we favour the idea that GspM joins the complex after association of GspE and GspL. In addition, a dynamic interaction between the hetero/homomeric protein forms should take place in the biogenesis of this complex, since attempts to pinpoint regions involved in partnership interactions revealed that the same region of GspM was found to be involved in both the GspM–GspM and GspM–GspL interactions.
Recently, it was shown that the major pseudopilin component of the type II secretory pathway can be assembled into pilus-like structures (Sauvonnet et al., 2000). It is because of their homologies with the structural subunit of the type IV pili, namely a prepilin peptidase cleavage site, that a subset of Gsp proteins has been referred to as pseudopilins. Moreover, the type II secretory apparatus can also assemble PpdD, the major E. coli K-12 type IV pilin, into a pilus (Sauvonnet et al., 2000). Proteins GspE, GspF, GspL and GspM have been shown to be required for pilus-like formation (Sauvonnet et al., 2000). Hence, a possibility is that the GspE–GspF–GspL–GspM complex helps to anchor the pilus-like structure in the inner membrane.
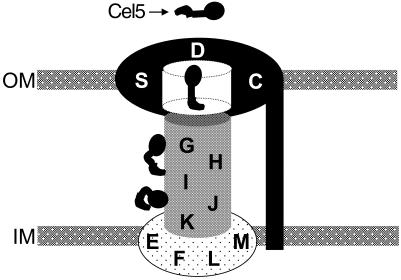
The assembly of the type II secretion apparatus appears to be very complex, since it involves 14 proteins that locate throughout both the membrane and the periplasm. We propose a modular view of the Gsp machinery permitting the delineation of three building blocks: (i) the GspE–GspF–GspL–GspM proteins, which form a large platform in the inner membrane; (ii) the pseudopilus constituted by the GspG, GspH, GspI, GspJ and GspK proteins; and (iii) the GspD–GspS–GspC proteins able to form a pore in the outer membrane (Figure 4). Because GspO processes and modifies two classes of substrate, the pseudopilins of the Gsp machinery and the pilins of the type IV pili, it is likely that GspO has no structural role in the Gsp machinery and should be considered on its own. Recent studies gave some support for contact between these building blocks. First, a contact between the GspG–GspH–GspI–GspJ–GspK pilus-like structure and the GspD–GspC–GspS pore is consistent with the reasonable supposition that the pilus spans the outer membrane inside the pore structure (Sauvonnet et al., 2000). Secondly, a contact between the GspE–GspF–GspL–GspM complex and the GspG–GspH–GspI–GspJ–GspK pilus-like structure is supported by the fact that in P. aeruginosa it has been shown that a mutation in gspE(xcpR) suppresses a mutation in gspG(xcpT) (Kagami et al., 1998). The GspE–GspF–GspL–GspM inner membrane platform could help in assembling and anchoring the pilus-like structure.
Fig. 4. Model for the type II secretion machinery. The secretion machinery can be depicted as comprising three building blocks: the inner membrane platform (GspE–GspF–GspL–GspM, light stippling); the pseudopilus (GspG–GspH–GspI–GspJ–GspK, heavy stippling); and the gated outer membrane pore (GspC–GspD–GspS, black area). Cel5 is shown as an example of a secreted protein ‘en route’ to the cell exterior.
SPECULATION
At some point, the protein to be secreted must interact with the secretion apparatus. Results from Shevchik et al. (1997) suggested a direct contact between the secreted protein and GspD. However, contradictory results were obtained by Guilvout et al. (1999). It was reported that secreted protein ‘en route’ to the cell exterior could exhibit a transient secretion-specific fold (McIver et al., 1995; Braun et al., 1996; Chapon et al., 2000). This led us to envisage a chaperoning role for some element of the secretion machinery. Hence, as a working hypothesis, we propose that the secreted proteins residing in the periplasm interact first with the pseudopilus, which helps them to adopt a conformation allowing their recognition by the proteins of the GspD–GspS–GspC pore (Figure 4). Moreover, the presence of the secreted protein could permit stable connection between the building blocks.
METHODS
Strains and media. The E.coli strain (DH5α) and the Saccharomyces cerevisiae strains (EGY48 and RF206), and rich and minimal media used, are described in Py et al. (1999).
Plasmids. Proteins fused either to the transcriptional activation motif (B42), which also includes the HA epitope, or to the DNA-binding protein (LexA) were produced from vector pJG4-5 and pEG202 (Golemis et al., 1994), respectively. Parts of the gspF and gspM genes were cloned as EcoRI–XhoI PCR fragments. DNA inserts were obtained by PCR using pCPP2006 (He et al., 1991) as a template. The sequence of the oligonucleotides used is available on request. Plasmids allowing the production of hybrid proteins with GspE or GspL parts are described in Py et al. (1999). All the cloned inserts were sequenced. During the course of these studies, we found that the published nucleotide sequences of gspF and gspM contained errors. The changes at the amino acid level are as follows. In GspM: (i) it is A43 and A64 instead of V43 and P64; (ii) between residues R66 and Q89, the sequence is LPPPEGARRQIAGRDISLTVLVP. In GspF: (i) there is insertion of an S and A residue after I65 and V80, respectively; (ii) it is Q161 and Q162 instead of H161 and E162; (iii) between A340 and D355, the sequence is ASGERSGELDGMLTRAA. The corrected sequences exhibit a higher degree of similarity with their homologues in E. carotovora.
Immunization. Antibodies against GspF and GspE were raised in rabbit by using an acrylamide band, obtained after SDS–PAGE, containing GspFN(His)6 or GspE(His)6, respectively. The acrylamide bands were excised, crushed and mixed with Freund’s adjuvant according to the method of Harlow and Lane (1988). GspFN(His)6 was obtained after purification on a Hitrap column (Pharmacia). GspE(His)6 was partially purified from inclusion bodies, in denaturating conditions.
Immunoprecipitation. Triton X-100-soluble cell extracts were prepared from strains that were grown in supplemented M9 medium at 30°C. Overnight cultures were diluted to 1/2 with fresh M9 medium supplemented with l-arabinose (0.02% final concentration) and incubated at 30°C for 3 h. Preparation of Triton X-100-soluble cell extracts and immunoprecipitation of this sample were performed essentially as described in Sandkvist et al. (1999). We used either anti-GspF or anti-GspE serum mixed with protein A–Sepharose beads. Samples were analysed by SDS–PAGE and immunoblotting as described in Py et al. (1999).
Yeast two-hybrid system. The yeast two-hybrid assay and β-galactosidase activity from diploid cells were performed as described in Py et al. (1999), using the method recommended by Golemis et al. (1994). Units of specific activity for β-galactosidase were nanomoles of o-nitrophenyl-β-d-galactoside hydrolysed per minute per milligram of protein (with ɛ = 4.5 × 103 M–1). Systematic controls for the production of the hybrid proteins and for the specificity of interaction were as described in Py et al. (1999).
Acknowledgments
ACKNOWLEDGEMENTS
We thank A. Collmer, M. Lindeberg for cosmids pCPP2215 and pCPP2222, R. Brent for material for the two-hybrid system, F. Denizot and J. Busuttil for DNA sequencing, and all the members of the Erwinia group for helpful discussions. This work was supported by grants from the CNRS, and the University of Aix-Marseille II.
REFERENCES
- Braun P., Tommassen, J. and Filloux, A. (1996) Role of the propeptide in folding and secretion of elastase of Pseudomonas aeruginosa. Mol. Microbiol., 19, 297–306. [DOI] [PubMed] [Google Scholar]
- Chapon V., Simpson, H.D., Morelli, X., Brun, E. and Barras, F. (2000) Alteration of a single tryptophan residue of the cellulose-binding domain blocks secretion of the Erwinia chrysanthemi Cel5 cellulase (ex-EGZ) via the type II system. J. Mol. Biol., 303, 117–123. [DOI] [PubMed] [Google Scholar]
- Driessen A.J.M., Fekkes, P. and van der Wolk, J.P.W. (1998) The Sec system. Curr. Opin. Microbiol., 1, 216–222. [DOI] [PubMed] [Google Scholar]
- Filloux A., Michel, G. and Bally, M. (1998) GSP-dependent protein secretion in Gram-negative bacteria: the Xcp system of Pseudomonas aeruginosa. FEMS Microbiol. Rev., 22, 177–198. [DOI] [PubMed] [Google Scholar]
- Golemis E.A., Gyuris, J. and Brent, R. (1994) Interaction trap/two-hybrid system to identify interacting proteins. In Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A. and Struhl, K. (eds), Current Protocols in Molecular Biology. Unit 13.14, John Wiley and Sons, NY.
- Guilvout I., Hardie, K.R., Sauvonnet, N. and Pugsley, A.P. (1999) Genetic dissection of the outer membrane secretin PulD: are there distinct domains for multimerization and secretion specificity? J. Bacteriol., 181, 7212–7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E. and Lane, D. (1988) Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- He S.Y., Lindeberg, M., Chatterjee, A.K. and Collmer, A. (1991) Cloned Erwinia chrysanthemi out genes enable Escherichia coli to selectively secrete a diverse family of heterologous proteins to its milieu. Proc. Natl Acad. Sci. USA, 88, 1079–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs M. and Mattick, J.S. (1993) Common components in the assembly of type IV fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol. Microbiol., 10, 233–243. [DOI] [PubMed] [Google Scholar]
- Kagami Y., Ratliff, M., Surber, M., Martinez, A. and Nunn, D.N. (1998) Type II protein secretion by Pseudomonas aeruginosa: genetic suppression of a conditional mutation in the pilin-like component XcpT by the cytoplasmic component XcpR. Mol. Microbiol., 27, 221–233. [DOI] [PubMed] [Google Scholar]
- Krause S., Barcena, M., Pansegrau, W., Lurz, R., Carazo, J.M. and Lanka, E. (2000) Sequence-related protein export NTPases encoded by the conjugative transfer region of RP4 and by the cag pathogenicity island of Helicobacter pylori share similar hexameric ring structures. Proc. Natl Acad. Sci. USA, 97, 3067–3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeberg M., Salmond, G.P. and Collmer, A. (1996) Complementation of deletion mutations in a cloned functional cluster of Erwinia chrysanthemi out genes with Erwinia carotovora out homologues reveals OutC and OutD as candidate gatekeepers of species-specific secretion of proteins via the type II pathway. Mol. Microbiol., 20, 175–190. [DOI] [PubMed] [Google Scholar]
- McIver K.S., Kessler, E., Olson, J.C. and Ohman, D.E. (1995) The elastase propeptide functions as an intramolecular chaperone required for elastase activity and secretion in Pseudomonas aeruginosa. Mol. Microbiol., 18, 877–889. [DOI] [PubMed] [Google Scholar]
- Michel G., Bleves, S., Ball, G., Lazdunski, A. and Filloux, A. (1998) Mutual stabilization of the XcpZ and XcpY components of the secretory apparatus in Pseudomonas aeruginosa. Microbiology, 144, 3379–3386. [DOI] [PubMed] [Google Scholar]
- Nunn D.N. (1999) Bacterial type II protein export and pilus biogenesis: more than just homologies? Trends Cell Biol., 9, 402–408. [DOI] [PubMed] [Google Scholar]
- Possot O., Vignon, G., Bomchil, N., Ebel, F. and Pugsley, A.P. (2000) Multiple interactions between pullulanase secreton components involved in stabilization and cytoplasmic membrane association of PulE. J. Bacteriol., 182, 2142–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Py B., Loiseau, L. and Barras, F. (1999) Assembly of the type II secretion machinery of Erwinia chrysanthemi: direct interaction and associated conformational change between OutE, the putative ATP-binding component and the membrane protein OutL. J. Mol. Biol., 289, 659–670. [DOI] [PubMed] [Google Scholar]
- Russel M. (1998) Macromolecular assembly and secretion across the bacterial cell envelope: Type II protein secretion systems. J. Mol. Biol., 279, 485–499. [DOI] [PubMed] [Google Scholar]
- Sandkvist M., Hough, L.P., Bagdasarian, M.M. and Bagdasarian, M. (1999) Direct interaction of the EpsL and EpsM proteins of the general secretion apparatus in Vibrio cholerae. J. Bacteriol., 181, 3129–3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvonnet N., Vignon, G., Pugsley, A.P. and Gounon, P. (2000) Pilus formation and protein secretion by the same machinery in Escherichia coli. EMBO J., 19, 2221–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchik V.E., Robert-Baudouy, J. and Condemine, G. (1997) Specific interaction between OutD, an Erwinia chrysanthemi outer membrane protein of the general secretory pathway, and secreted proteins. EMBO J., 16, 3007–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J.D., Reeves, P.J. and Salmond, G.P. (1997) The general secretion pathway of Erwinia carotovora subsp. carotovora: analysis of the membrane topology of OutC and OutF. Microbiology, 143, 713–720. [DOI] [PubMed] [Google Scholar]