Abstract
The small GTP-binding protein Arf6 coordinates membrane traffic at the plasma membrane with aspects of cytoskeleton organization. This function does not overlap with that of other members of the ADP-ribosylation factor (Arf) family, although their switch regions, which are their major sites of interaction with regulators and effectors, have virtually identical sequences. Here we report the crystal structure of full-length, non-myristoylated human Arf6 bound to GTPγS. Unlike their GDP-bound forms, the active forms of Arf6 and Arf1 are very similar. Thus, the switch regions are discriminatory elements between Arf isoforms in their inactive but not in their active forms, a property that may generalize to other families of small G proteins. This suggests that GTP-bound Arfs may establish specific interactions outside the switch regions and/or be recognized in their cellular context rather than as isolated proteins. The structure also allows further insight into the lack of spontaneous GTPase activity of Arf proteins.
INTRODUCTION
Most families of small GTP-binding proteins include closely related members, which, despite high sequence similarities, display distinct cellular functions. A well-documented case is that of the ADP-ribosylation factor (Arf) family. Its major isoforms, Arf1 and Arf6, despite having ∼70% sequence identity, differ from each other in their cellular properties. Arf1, the most abundant Arf protein, acts mainly at the Golgi complex where it controls the recruitment of the COPI coatomer on budding vesicles (reviewed in Chavrier and Goud, 1999). In contrast, Arf6 localizes at the plasma membrane where it coordinates endocytotic membrane traffic with aspects of cytoskeleton organization (D’Souza-Schorey et al., 1995; Chavrier and Goud, 1999 and references therein). These functions depend on the concerted trigger of appropriate guanine exchange factors (GEFs), which catalyse the release of the tightly bound GDP, and GTPase activating proteins (GAPs), which stimulate the hydrolysis of bound GTP (reviewed in Donaldson and Jackson, 2000). There is growing evidence that GEFs, through their membrane-binding domains, are in charge of addressing Arfs at the appropriate subcellular localization, and are thus key players in sorting their different functions. Interestingly, several GEFs or GAPs have been shown in vitro and/or in vivo to be either ineffective on or specific for Arf6, suggesting that specific structural recognition of individual Arf isoforms is required for their functions (Donaldson and Jackson, 2000).
The structural cycle of Arf proteins differs from that of other small GTP-binding proteins in that it couples the classical GDP/GTP nucleotide switch, mediated by the switch I and II regions, to a membrane/cytosol switch mediated by the N-terminal helix and the region that connects switch I and II (the interswitch). The N-terminal helix in Arf–GDP blocks the interswitch in a retracted conformation (Amor et al., 1994; Greasley et al., 1995; Menetrey et al., 2000), which is released by the interaction of the helix with membranes (Franco et al., 1995). Membranes are therefore essential for GEFs to catalyse the shift of the interswitch that precedes the completion of switch I and II reorganization upon binding of GTP (Goldberg, 1998; Béraud-Dufour et al., 1999). The switch II and, to a lesser extent, the switch I regions are involved in the interaction of Arfs with GEFs and GAPs (Goldberg, 1998, 1999). Given that the switch regions have virtually identical sequences among Arf proteins, this addresses the question of how they implement the specificity of their cellular interactions.
As a first step towards understanding this paradox, we showed recently that human Arf6 and Arf1 have different conformations in their GDP-bound form, which result from a small number of sequence differences that have coordinated structural effects (Menetrey et al., 2000). Here, we report the structure of full-length, non-myristoylated Arf6–GTPγS, which is the first structure of a full-length activated Arf protein. Arf6–GTPγS displays the general features observed for Arf1Δ17–GDPNP, a mutant of Arf1 that lacks the N-terminal helix, including the backwards translation of the interswitch. Thus, although their GDP-bound conformations are different, Arf1 and Arf6 reach similar active structures, notably at their switch regions. This suggests that Arf1 and Arf6 are poorly discriminated by their switch regions when they are in their active form. Together with the structures of GDP-bound Arfs, our structure provides a framework to analyse the recognition of Arf proteins by effectors and GAPs. Comparison of the structural GDP/GTP cycle of Arf6 to that of other small G proteins suggests that the amplitude and nature of structural changes at the switch regions provide a general basis for biological diversity, and adds new insights into the lack of spontaneous GTPase activity of Arf proteins.
RESULTS AND DISCUSSION
Structure of Arf6–GTPγS and comparison to Arf6–GDP
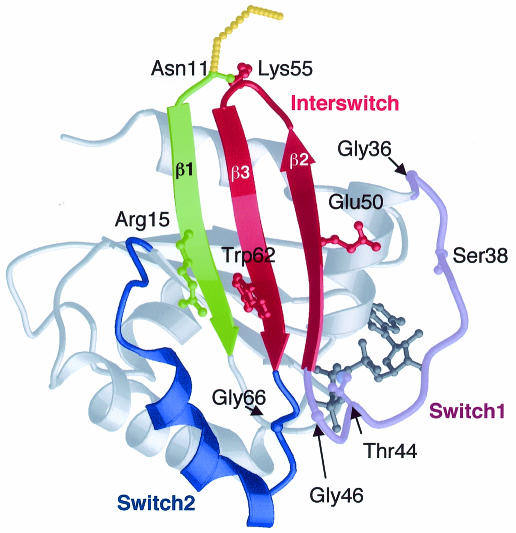
The crystal structure of full-length, non-myristoylated human Arf6 bound to the GTP analogue GTPγS was solved to 2.8 Å resolution by molecular replacement (Table I; Figure 1). The GTPγS nucleotide is clearly visible, as well as the Mg2+ ion and a small number of water molecules that form intramolecular interactions. The two switch regions and the interswitch are well defined, except for the interswitch loop where the B-factors are higher than average. The overall GDP-to-GTP conformational transition involves the disorganization of the N-terminal helix binding site and the release of the helix (residues 1–10) into the solvent, and the reorganization of a continuous 40 residue peptide (residues 36–76), which encompasses switch I, the interswitch and switch II. Thus full-length Arf6–GTPγS undergoes the dual switch as does truncated Arf1, as described below (Figure 2, top).
Table I. Statistics for X-ray structure determination.
| Measured reflections | 196 227 |
| Unique reflections | 8616 |
| Completeness (%) | 93.1 |
| Resolution range (Å) | 30.0–2.80 |
| Rsym (%) | 6.2 |
| R-factor (%) | 23.4 |
| Rfreea (%) | 27.7 |
| R.m.s.d. | |
| bond lengths (Å) | 0.008 |
| bond angles (°) | 1.4 |
| Average B-factor (Å2) | 50.2 |
aCalculated for a random set of 10% of the data excluded from the refinement.
Fig. 1. Structure of Arf6–GTPγS.
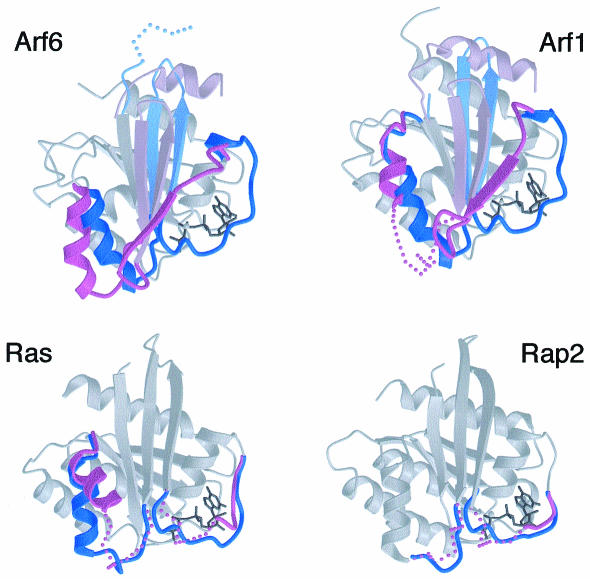
Fig. 2. Structural plasticity of the switch I and II regions is variable between related small GTP-binding proteins. Comparison of the GDP/GTP structural cycle between Arf6 and Arf1 (top) and between Ras and Rap2 (bottom) reveals variable GDP-bound conformations (in violet) and similar GTP-bound structures (in blue). Orientations are as in Figure 1. The switch I and II regions are in dark shades, other switch elements in Arf are in light shades. Flexible regions are shown by dotted lines. For clarity, only the GTP-bound form of each protein is shown except at the switch regions. From top to bottom and left to right: Arf6–GDP (PDB entry 1E0S) and Arf6–GTPγS; Arf1–GDP (1HUR) and Arf1 Δ17–GDPNP (Goldberg, 1998); Ras–GDP (4Q21) and Ras–GDPNP (5Q21); Rap2–GDP (1KAO) and Rap2–GTP (3RAP).
As expected from the structure of Arf1Δ17–GDPNP (Goldberg, 1998), the binding site for the N-terminal helix is destroyed by a rigid body translation by 6.5 Å of the interswitch. The N-terminal peptide is not visible in the electron density, which vanishes into the solvent before Asn11. Although this may be due to partial heterogeneity of the Arf6 sample (see Methods), it is likely that the N-terminus is not rigidly organized relative to the protein core, and may tether Arf6 rather than provide it with a definite orientation relative to membranes. This region forms a helix in Arf1–GDP (Amor et al., 1994; Greasley et al., 1995) and Arf6–GDP (Menetrey et al., 2000), and in the corresponding Arf1 peptide in the presence of membranes (Losonczi et al., 2000). This suggests that the N-terminus remains helical throughout the GDP/GTP cycle of Arf1, and probably in the cycle of Arf6 as well.
The displacement of the N-terminal peptide allows the translation of the interswitch to take place. This movement involves the unzipping of β-strands β2 and β3 from the switch I β-strand on one side, and from strand β1 on the other one. β1–β3 interactions reform in Arf6–GTPγS after a two-residue register shift of the interswitch, adding two more hydrogen bonds up to Asn11 as compared with Arf6–GDP. This releases a conformational strain at the main chain of Lys55 in the interswitch loop, whose unfavourable Φ and Ψ angles in Arf6–GDP return to low-energy values in Arf6–GTPγS. In addition, Lys55 protrudes from the side of Arf6 opposite to the nucleotide-binding site, where it may interact with the polar heads of membrane phospholipids and thereby contribute to stabilize and/or orient active Arf relative to membranes.
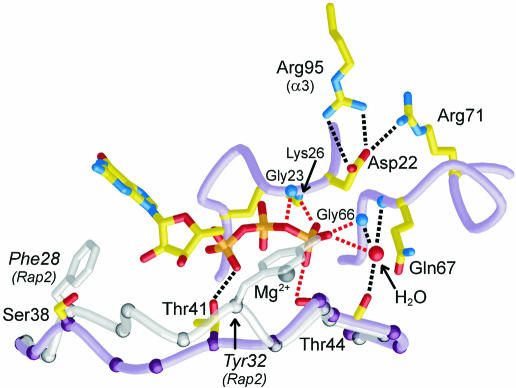
The GDP/GTP structural cycle is completed by the rearrangement of switch I and II in the vicinity of the γ-phosphate of GTP (Goldberg, 1998). Here we find that Arf6–GTPγS displays the hallmark interactions of GTP-bound small GTP-binding proteins (Figure 3). In switch I, Thr44 interacts with GTPγS and Mg2+ following a 14 Å movement from its position in Arf6–GDP. The unambiguous presence of Mg2+ contrasts with its absence in Arf6–GDP (Menetrey et al., 2000) and Arf1–GDP (Amor et al., 1994; Greasley et al., 1995), as well as in the related protein Arl3–GDP (Hillig et al., 2000). The switch II region, which forms a 310-helix interrupted by Pro72 in both Arf6–GDP and Arf6–GTPγS, has rotated by 25° with a rotation centre close to His76, thus positioning Gly66 and Gln67 from the DxxGQ motif near the γ-phosphate of GTPγS. The hinge regions for the movement of switch I and switch II are located near glycine residues at their ends (Gly36 and Gly46 in switch I, Gly66 in switch II), highlighting the role of glycines as key residues of the structural switches. The conformational change also breaks the Ser38–Glu50 hydrogen bond, which we showed to be responsible for the more rapid dissociation of GDP from Arf6 as compared with Arf1–GDP, where this hydrogen bond is not made as Ser38 is replaced by an isoleucine (Menetrey et al., 2000). In Arf6–GTPγS, we observe weak hydrogen bonds between Ser38 and the carbonyl of Thr157, and between Glu50 and the hydroxyl of Tyr31. A residue central to the switch motions is a conserved Arg (Arg15 in Arf6) that bridges the N-terminus to the interswitch and switch II, and accompanies their GDP/GTP structural switch. Whereas it contacts different residues in GDP-bound Arf1 (Gln83, Asn84) and Arf6 (Thr79, Gly80), its interactions are more similar in their GTP-bound forms, including a common aromatic hydrogen bond to Trp62 (Trp66 in Arf1) in the interswitch.
Fig. 3. The GTP-binding site of Arf6 and comparison with switch I of Rap2. Switch I (residues 36–47), switch II (64–72) and the P-loop (21–27) are shown in violet. Hydrogen bonds are shown by dotted lines. Switch I of Rap2–GTP (residues 26–37), representative of Ras, Rho and Ran, is shown in light grey after overall superposition of Rap2 onto Arf6–GTPγS. The switch I regions of Rap2/Arf6 superimpose at Gly26/Gly36, Pro34–Thr35/Pro43–Thr44 and Glu37/Phe47.
Implications for the interactions of Arf1 and Arf6 with regulatory and effector proteins
Our previous analysis of Arf6–GDP pointed to unexpected conformational differences with Arf1–GDP, which explained differences in nucleotide and Mg2+ affinities (Menetrey et al., 2000) and possibly functional properties (Al-Awar et al., 2000). Here we find that Arf1Δ17–GDPNP and Arf6–GTPγS converge to very similar structures (r.m.s.d. = 0.6 Å on 158 Cαs). The GDP-to-GTP conformational pathway is thus different in nature and amplitude for Arf1 and Arf6, most prominently at switch II. This region is essentially flexible in Arf1–GDP and undergoes a disorder-to-order transition upon binding of GTP (Figure 2, top right). In contrast, it is well organized in Arf6–GDP where it rotates as a rigid body to adopt the GTP-bound conformation (Figure 2, top left). We and others noticed earlier that G proteins display greater structural variations when bound to GDP than when bound to GTP (Berghuis et al., 1996; Cherfils et al., 1997). Arf1 and Arf6, together with the Ras family members H-ras and Rap2A, are the only pairs of related GTP-binding proteins whose full structural GDP/GTP cycle has been described for the uncomplexed proteins. Their pairwise comparison reveals strikingly similar features, including conversion of dissimilar GDP-bound structures to very similar GTP-bound forms (Figure 2). This suggests that modulations of the structural plasticity are likely to form a general basis for the specific properties of individual members within a family of small GTP-binding proteins.
Comparison of the structural GDP/GTP cycles of Arf1 and Arf6 provides further insight as to how they can be discriminated by regulators and effectors. It shows that their switch regions, which have virtually identical sequences, have different conformations when the proteins are bound to GDP, but adopt similar GTP-bound structures. Thus, the switch regions can be used efficiently by interacting partners to discriminate Arf1 from Arf6 in their inactive forms, but much less so in their active forms where the only sequence differences, located at the remote end of switch I (Gln37Ser38 in Arf6 and Glu41Ile42 in Arf1), do not result in differences in the main chain conformation. Yet, the GDP-to-GTP conformational change exposes these residues to a greater extent in GTP-bound Arfs than in their inactive forms, where they may contribute to direct interactions with specific effectors (Al-Awar et al., 2000). The remaining part of switch I and the entire switch II have identical sequences, so that they can signal that Arfs are in their active form, but do not carry structural information as to which isoform they belong. This suggests a non-exclusive alternative for the recognition of GTP-bound Arfs by effectors and GAPs: they might recognize regions other than the switch regions in which the sequences are different between Arf isoforms, or they might recognize Arf–GTP in its cellular context, possibly in complexes with other proteins. Both possibilities are supported by a recent report, which suggests that ArfGAP activity is enhanced by co-binding of the coatomer (Goldberg, 1999), as well as studies on Arf1–Arf6 chimeras that point to functional regions beyond the switch regions (Al-Awar et al., 2000). However, neither scenario escapes the requirement for specific structural recognition at some stage, most likely implemented by the GEFs, as they come first in the course of the GDP/GTP cycle. An attractive hypothesis would be that GEFs and downstream partners cross-talk through transient mixed complexes, possibly using coiled-coiled interactions, as this would conceptually provide a means to combine Arf proteins and their regulators/effectors into well-sorted pathways.
Why have Arf proteins no spontaneous GTPase?
A subtle difference between Arf proteins and their sister GTP-binding proteins is that they lack the weak spontaneous GTPase activity of Ras, Rho, Rab and Ran families (Weiss et al., 1989). Although it is unclear at the moment whether this underlines a functional requirement, it is interesting to scrutinize the structures of their GTP-binding sites for differences that might be related to the enzymic phosphate transfer reaction. Surprisingly, all classical interactions of the γ-phosphate are present in Arf6–GTPγS as well as in Arf1Δ17–GDPNP (Figure 3). Thus, the lack of hydrolysis must originate in other peculiarities of the Arf active site. First, we notice a difference in the register of switch I in Arf–GTP as compared with other GTP-binding proteins (Figure 3). In Arf1 and Arf6, it is one residue longer before the invariant Thr (Thr44 in Arf6) and one residue shorter after. As a consequence, Arfs have no equivalent to a conserved Tyr found in switch I in Ras, Rho and Ran families (Tyr32 in H-Ras), which caps the γ-phosphate of GTP. Its counterpart in Arf (Thr41 in Arf6) interacts with the α-phosphate instead. Secondly, Arfs feature an unusual Asp residue in the P-loop motif (22DGxxKT27 in Arf6), equivalent to Gly12 in Ras where its mutation to Asp impairs spontaneous GTP hydrolysis (Seeburg et al., 1984) (Figure 3). This Asp residue may result in similar impairment of the spontaneous GTPase of Arf. However, Asp22 holds the P-loop, switch II and helix α3 together in Arf6–GTPγS so that measurement of its effects on GTP hydrolysis in mutants may be compromised by a concomitant decrease in GTP binding (Kahn et al., 1995).
Despite these differences, we observe electron density for a well-ordered water molecule that interacts with the γ-phosphate, switch I (Thr44 carbonyl) and switch II (NH of Gly66 and Gln67), and is remarkably conserved in every structure of active forms of GTP-binding proteins, whether uncomplexed or interacting with an effector (Figure 3). This suggests that rather than being a nucleophilic water molecule awaiting activation, as it is generally described (reviewed in Maegley et al., 1996), this well-ordered water molecule may have a built-in inhibitory role in Arf as well as in other GTPases by preventing another water molecule binding with the in-line SN2 configuration that leads to hydrolysis of GTP. Arf may lack a mechanism to displace it, possibly because of the above differences in switch I and the P-loop. This also suggests that ArfGAPs as well as other GAPs must implement conformational and/or electrostatic changes to release this inhibitory interaction.
METHODS
Human full-length Arf6 was expressed in Escherichia coli and purified to homogeneity as described (Menetrey et al., 2000). Bound nucleotides, present as a mixture of GDP and GTP, were dissociated in the presence of 2.5 mM EDTA and 5 mM GTPγS, and hydrolysed by alkaline phosphatase, followed by addition of 3 mM MgCl2 to stop the exchange reaction. Crystals were grown by the vapour diffusion hanging drop method by mixing 2:1 vols of Arf6–GTPγS (8.6 mg/ml) and of the reservoir (17% PEG4000, 200 mM ammonium sulfate, 100 mM sodium acetate pH 4.6, 5 mM MgCl2, 1 mM dithiothreitol). Rod-shaped crystals (50 × 50 × 200 µm3) appeared within a few days, and were stabilized in the crystallization buffer complemented to 20% PEG4000 and 25% PEG400 before cryo-freezing in liquid ethane. Data were collected at the ESRF synchrotron facility (Grenoble, France) on beamline ID14-1 at wavelength 0.934 Å. Reflections were reduced and scaled with Denzo and Scalepack (Otwinowski, 1993). Crystals belong to space group P41212 with a = b = 72.9 Å, c = 131.5 Å, α = β = γ = 90°, and contain two molecules per asymmetric unit.
The structure was solved by molecular replacement with AMoRe (Navaza, 1994) using Arf1Δ17–GDPNP (Goldberg, 1998) as a search model. The structure was refined using CNS (Brunger et al., 1998) and graphical building was carried out with TURBO (http://afmb.cnrs-mrs.fr/subjects/turbo) using σ-weighted 2Fo – Fc and Fo – Fc maps (CCP4, 1994) and CNS composite omit maps. Refinement was carried out with non-crystallographic symmetry restraints applied to most of the structure except flexible loops. There is no density for residues 1–10, which were not modelled. The presence of the N-terminal helix was assessed by N-terminal Edman sequencing of the protein sample, showing a 10% contamination from Arf6 proteolysed of its five N-terminal residues. MALDI-TOF mass spectroscopy on dissolved crystals confirms the presence of a fraction of proteolysed Arf6, which may preclude observation of the N-terminal helix at the medium resolution of 2.8 Å. Statistics for structure determination are given in Table I. Coordinates have been deposited with the Protein Data Bank (PDB) with entry code 1HFV.
Acknowledgments
ACKNOWLEDGEMENTS
We thank the staff at the LURE and ESRF synchrotron facilities for making beamlines available to us. We are grateful to D. Lê (CNRS, Gif sur Yvette) for N-terminal sequences, and P. Decottignies and P. Le Maréchal (Université Paris-Sud, Orsay) for mass spectroscopy. This work was supported by grants from the Association pour la Recherche contre le Cancer and AstraZeneca. S.P. is supported by a grant from Institut Curie.
REFERENCES
- Al-Awar O., Radhakrishna, H., Powell, N.N. and Donaldson, J.G. (2000) Separation of membrane trafficking and actin remodeling functions of ARF6 with an effector domain mutant. Mol. Cell. Biol., 20, 5998–6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amor J.C., Harrison, D.H., Kahn, R.A. and Ringe, D. (1994) Structure of the human ADP-ribosylation factor 1 complexed with GDP. Nature, 372, 704–708. [DOI] [PubMed] [Google Scholar]
- Béraud-Dufour S., Paris, S., Chabre, M. and Antonny, B. (1999) Dual interaction of ADP ribosylation factor 1 with Sec7 domain and with lipid membranes during catalysis of guanine nucleotide exchange. J. Biol. Chem., 274, 37629–37636. [DOI] [PubMed] [Google Scholar]
- Berghuis A.M., Lee, E., Raw, A.S., Gilman, A.G. and Sprang, S.R. (1996) Structure of the GDP-Pi complex of Gly203→Ala giα1: a mimic of the ternary product complex of gα-catalyzed GTP hydrolysis. Structure, 4, 1277–1290. [DOI] [PubMed] [Google Scholar]
- Brunger A.T. et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: program for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Chavrier P. and Goud, B. (1999) The role of ARF and rab GTPases in membrane transport. Curr. Opin. Cell Biol., 11, 466–475. [DOI] [PubMed] [Google Scholar]
- Cherfils J., Menetrey, J., Le Bras, G., Janoueix-Lerosey, I., de Gunzburg, J., Garel, J.R. and Auzat, I. (1997) Crystal structures of the small G protein Rap2A in complex with its substrate GTP, with GDP and with GTPγS. EMBO J., 16, 5582–5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson J.G. and Jackson, C.L. (2000) Regulators and effectors of the ARF GTPases. Curr. Opin. Cell Biol., 12, 475–482. [DOI] [PubMed] [Google Scholar]
- D’Souza-Schorey C., Li, G., Colombo, M.I. and Stahl, P.D. (1995) A regulatory role for ARF6 in receptor-mediated endocytosis. Science, 267, 1175–1178. [DOI] [PubMed] [Google Scholar]
- Franco M., Chardin, P., Chabre, M. and Paris, S. (1995) Myristoylation of ADP-ribosylation factor 1 facilitates nucleotide exchange at physiological Mg2+ levels. J. Biol. Chem., 270, 1337–1341. [DOI] [PubMed] [Google Scholar]
- Goldberg J. (1998) Structural basis for activation of ARF GTPase: mechanisms of guanine nucleotide exchange and GTP-myristoyl switching. Cell, 95, 237–248. [DOI] [PubMed] [Google Scholar]
- Goldberg J. (1999) Structural and functional analysis of the ARF1–ARFGAP complex reveals a role for coatomer in GTP hydrolysis. Cell, 96, 893–902. [DOI] [PubMed] [Google Scholar]
- Greasley S.E., Jhoti, H., Teahan, C., Solari, R., Fensome, A., Thomas, G.M., Cockcroft, S. and Bax, B. (1995) The structure of rat ADP-ribosylation factor-1 (ARF-1) complexed to GDP determined from two different crystal forms. Nature Struct. Biol., 2, 797–806. [DOI] [PubMed] [Google Scholar]
- Hillig C.H., Hanzal-Bayer, M., Linari, M., Becker, J., Wittinghofer, A. and Renault, L. (2000) Structural and biochemical properties show ARL3–GDP as a distinct GTP-binding protein. Structure, 8, 1239–1245. [DOI] [PubMed] [Google Scholar]
- Kahn R.A., Clark, J., Rulka, C., Stearns, T., Zhang, C.J., Randazzo, P.A., Terui, T. and Cavenagh, M. (1995) Mutational analysis of Saccharomyces cerevisiae ARF1. J. Biol. Chem., 270, 143–150. [DOI] [PubMed] [Google Scholar]
- Losonczi J.A., Tian, F. and Prestegard, J.H. (2000) Nuclear magnetic resonance studies of the N-terminal fragment of adenosine diphosphate ribosylation factor 1 in micelles and bicelles: influence of N-myristoylation. Biochemistry, 39, 3804–3816. [DOI] [PubMed] [Google Scholar]
- Maegley K.A., Admiraal, S.J. and Herschlag, D. (1996) Ras-catalyzed hydrolysis of GTP: a new perspective from model studies. Proc. Natl Acad. Sci. USA, 93, 8160–8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menetrey J., Macia, E., Pasqualato, S., Franco, M. and Cherfils, J. (2000) Structure of Arf6–GDP suggests a basis for guanine nucleotide exchange factors specificity. Nature Struct. Biol., 7, 466–469. [DOI] [PubMed] [Google Scholar]
- Navaza J. (1994) AMoRe: an automated package for molecular replacement. Acta Crystallogr. A, 50, 157–163. [Google Scholar]
- Otwinowski Z. (1993) Oscillation data reduction program. In Sawyer, L., Isaacs, N. and Bailey, S. (eds), CCP4 Study Weekend: Data Collection and Processing. Daresbury Laboratory, Warrington, UK, pp. 56–62.
- Seeburg P.H., Colby, W.W., Capon, D.J., Goeddel, D.V. and Levinson, A.D. (1984) Biological properties of human c-Ha-ras1 genes mutated at codon 12. Nature, 312, 71–75. [DOI] [PubMed] [Google Scholar]
- Weiss O., Holden, J., Rulka, C. and Kahn, R.A. (1989) Nucleotide binding and cofactor activities of purified bovine brain and bacterially expressed ADP-ribosylation factor. J. Biol. Chem., 264, 21066–21072. [PubMed] [Google Scholar]