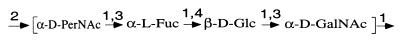Abstract
The O157:H7 clone of Escherichia coli, which causes major, often prolonged outbreaks of gastroenteritis with hemolytic-uremic syndrome (HUS) such as those in Japan, Scotland, and the United States recently, is thought to be resident normally in cattle or other domestic animals. This clone is of major significance for public health and the food industry. We have developed a fast method for sequencing a given O antigen gene cluster and applied it to O157. The O157 O antigen gene cluster is 14 kb in length, comprising 12 genes and a remnant H-repeat unit. Based on sequence similarity, we have identified all the necessary O antigen genes, including five sugar biosynthetic pathway genes, four transferase genes, the O unit flippase gene, and the O antigen polymerase gene. By PCR testing against all 166 E. coli O serogroups and a range of gram-negative bacterial strains, including some that cross-react serologically with E. coli O157 antisera, we have found that certain O antigen genes are highly specific to O157 E. coli. This work provides the basis for a sensitive test for rapid detection of O157 E. coli. This is important both for decisions on patient care, since early treatment may reduce the risk of life-threatening complications, and for detection of sources of contamination. The method for fast sequencing of O antigen gene clusters plus an ability to predict which genes will be O antigen specific will enable PCR tests to be developed as needed for other clones of E. coli or, once flanking genes are identified, clones of any gram-negative bacterium.
Escherichia coli is a clonal species, with clones normally being identified by their combination of O and H (and sometimes K) antigens. Some O antigens, such as O157, are characteristically found in pathogenic clones, with the O157:H7 clone being particularly significant in human disease, since it has caused approximately two-thirds of all recent cases of hemolytic-uremic syndrome in North America and Europe (32). All enterohemorrhagic E. coli strains produce Shiga toxins (Stx), but Stx-producing E. coli strains possessing O antigen 157 are the most frequently isolated from humans and are the predominant cause of hemolytic-uremic syndrome (32). Because of the very low infective dose of this organism (14), bacteria entering the human food chain can still pose a health problem after undergoing enormous dilution. For example, in January 1993 there was an outbreak due to contamination by O157:H7 E. coli at a large meat-processing plant making more than 1 million hamburger patties per day. The affected hamburgers were sold through one retail chain in four states, and 477 people became ill, of whom 3 died (3, 7). The scale of the plant and its operation was such that there would have been great dilution of the contaminated meat, and the highest count of O157:H7 found in hamburger patties of the same production date was 15 organisms per g, although obviously the hamburgers which caused the infection were not tested.
The O antigen, which contains many repeats of an oligosaccharide unit (O unit), is part of the lipopolysaccharide present in the outer membrane of gram-negative bacteria. It contributes major antigenic variability to the cell surface, and on the basis of this variation E. coli has been divided into 166 O serogroups. The surface O antigen is subject to intense selection by the host immune system, which may account for the maintenance of many different O antigen forms within species such as E. coli. Characteristically, all genes specific to O antigen synthesis are clustered (27). Several O antigen gene clusters have been cloned and sequenced, and some have been studied further. However, it is time-consuming to clone an O antigen gene cluster, which is normally longer than 10 kb. We have used the JUMPstart sequence, which is a 39-bp element present upstream of many polysaccharide gene clusters (12), and the gnd sequence, which is present downstream of O antigen gene clusters of E. coli, Salmonella enterica, and related species (4, 29), to amplify the O157 O antigen gene cluster by doing long PCR. The PCR product was then subjected to random cleavage and the fragments were cloned before sequencing. By using this method, a given O antigen gene cluster can be sequenced within a few weeks.
The O157 O antigen contains N-acetyl-d-perosamine, l-fucose, d-glucose, and N-acetyl-d-galactose. Analysis of the O157 sequence revealed four genes of the GDP-l-fucose pathway (manB, manC, gmd, and fcl), the per gene of the GDP-perosamine pathway, a gene which probably encodes the acetyltransferase to make GDP–N-acetylperosamine, three presumptive sugar transferase genes for synthesis of the O unit, the O unit flippase gene, and the O antigen polymerase gene. By PCR testing against all 166 known E. coli serogroups, a range of gram-negative bacterial strains, and O157 E. coli strains not related to the strain used for sequencing, we found that the O157 O antigen transferase, flippase, and polymerase genes are O157 specific.
Great efforts have been made to develop a method for timely and accurate detection of the O157:H7 strain (see, e.g., references 8, 10, 14, and 22). PCR-based methods are ideal for rapid detection of organisms at low concentrations. PCR detection with probes based on the Stx and eaeA genes and a plasmid has been developed, but each probe gave cross-reaction with other E. coli strains even when only a small number of strains were tested (10).
MATERIALS AND METHODS
Bacterial strains.
Plasmids were maintained in E. coli K-12 strain JM109. E. coli O157:H7 (isolate C664-1992) was from The International Escherichia and Klebsiella Centre. Standard E. coli O group strains (17) were used (see Table 1). Other strains used are also listed in Table 1, together with the names of the suppliers.
TABLE 1.
Bacterial strains and PCR pools used for testing of E. coli O157 primers
| Pool no. | Strains whose chromosomal DNA is included in the pool | Source |
|---|---|---|
| 1 | E. coli type strains for O serotypes 1, 2, 3, 4, 10, 16, 18, and 39 | IMVSa |
| 2 | E. coli type strains for O serotypes 40, 41, 48, 49, 71, 73, 88, and 100 | IMVS |
| 3 | E. coli type strains for O serotypes 102, 109, 119, 120, 121, 125, 126, and 137 | IMVS |
| 4 | E. coli type strains for O serotypes 138, 139, 149, 7, 5, 6, 11, and 12 | IMVS |
| 5 | E. coli type strains for O serotypes 13, 14, 15, 17, 19ab, 20, 21, and 22 | IMVS |
| 6 | E. coli type strains for O serotypes 23, 24, 25, 26, 27, 28, 29, and 30 | IMVS |
| 7 | E. coli type strains for O serotypes 32, 33, 34, 35, 36, 37, 38, and 42 | IMVS |
| 8 | E. coli type strains for O serotypes 43, 44, 45, 46, 50, 51, 52, and 53 | IMVS |
| 9 | E. coli type strains for O serotypes 54, 55, 56, 57, 58, 59, 60, and 61 | IMVS |
| 10 | E. coli type strains for O serotypes 62, 63, 64, 65, 66, 68, 69, and 70 | IMVS |
| 11 | E. coli type strains for O serotypes 74, 75, 76, 77, 78, 79, 80, and 81 | IMVS |
| 12 | E. coli type strains for O serotypes 82, 83, 84, 85, 86, 87, 89, and 90 | IMVS |
| 13 | E. coli type strains for O serotypes 91, 92, 95, 96, 97, 98, 99, and 101 | IMVS |
| 14 | E. coli type strains for O serotypes 103, 104, 105, 106, 107, 108, and 110 | IMVS |
| 15 | E. coli type strains for O serotypes 112, 162, 113, 114, 115, 116, 117, and 118 | IMVS |
| 16 | E. coli type strains for O serotypes 123, 165, 166, 167, 168, 169, 170, and 171 | —b |
| 17 | E. coli type strains for O serotypes 172, 173, 127, 128, 129, 130, 131, and 132 | —c |
| 18 | E. coli type strains for O serotypes 133, 134, 135, 136, 140, 141, 142, and 143 | IMVS |
| 19 | E. coli type strains for O serotypes 144, 145, 146, 147, 148, 150, 151, and 152 | IMVS |
| 20 | E. coli type strains for O serotypes 153, 154, 155, 156, 158, 159, and 160 | IMVS |
| 21 | E. coli type strains for O serotypes 161, 163, 164, 8, 9, 111, and 124 | IMVS |
| 22 | As pool 20, plus E. coli O157 type strain A2 (O157:H19) | IMVS |
| 23 | As pool 20, plus E. coli O157:H16 strain C475-89 | —d |
| 24 | As pool 20, plus E. coli O157:H45 strain C727-89 | —d |
| 25 | As pool 20, plus E. coli O157:H2 strain C252-94 | —d |
| 26 | As pool 20, plus E. coli O157:H39 strain C258-94 | —d |
| 27 | As pool 20, plus E. coli O157:H26 | —e |
| 28 | As pool 20, plus S. enterica serovar Landau | —f |
| 29 | As pool 20, plus B. abortus | —g |
| 30 | As pool 20, plus Y. enterocolitica O9 | —h |
| 31 | Y. pseudotuberculosis strains of O groups IA, IIA, IIB, IIC, III, IVA, IVB, VA, VB, VI, and VII | —i |
| 32 | S. boydii strains of serogroups 1, 3, 4, 5, 6, 8, 9, 10, 11, 12, 14, and 15 | —j |
| 33 | S. enterica strains of serovars (each representing a different O group) typhi, montevideo, ferruch, jangwani, raus, hvittingfoss, waycross, dan, dugbe, basel 65:i:e,n,z15, and 52:d:e,n,x,z15 | IMVS |
| 34 | V. cholerae strains of O groups 1, 6, 10, 37, 41, 52, 74, 81, 83, 99, and 139 | —k |
IMVS, Institute of Medical and Veterinary Science, Adelaide, Australia.
123 from IMVS; the rest from Statens Serum Institut, Copenhagen, Denmark.
172 and 173 from Statens Serum Institut, Copenhagen, Denmark; the rest from IMVS.
O157 strains from Statens Serum Institut, Copenhagen, Denmark.
O157:H26 from R. Brown, Royal Children’s Hospital, Melbourne, Australia.
S. enterica serovar Landau from M. Popoff, Institut Pasteur, Paris, France.
B. abortus from the culture collection of The University of Sydney, Sydney, Australia.
Y. enterocolitica O9 from K. Bettelheim, Victorian Infectious Diseases Reference Laboratory, Victoria, Australia.
S. Aleksic, Institute of Hygiene, Germany.
J. Lefebvre, Bacterial Identification Section, Laboratoire de Santé Publique du Québec, Canada.
O1 and O139 from National Institute of Cholera and Enteric Diseases, India; the rest from Tohio Shimada, Department of Bacteriology, National Institute of Health, Tokyo, Japan.
Construction of a random DNase I bank.
Oligonucleotides 482 (5′-CACTGCCATACCGACGACGCCGATCTGTTGCTTGG) and 412 (5′-ATTGGTAGCTGTAAGCCAAGGGCGGTAGCGT) were used to PCR amplify the O antigen gene cluster. Long PCR was carried out with the Expand Long Template PCR system from Boehringer. The PCR cycles were as follows: denaturation at 94°C for 10 s, annealing at 64°C for 30 s, and extension at 68°C for 15 min. Two aliquots containing 150 ng of DNA of long-PCR product each were subjected to DNase I digestion with the Novagen DNase shotgun cleavage kit and a modified protocol as follows. Each aliquot was diluted into 45 μl of 0.05 M Tris-HCl (pH 7.5)–0.05 mg of bovine serum albumin per ml–10 mM MnCl2. A 5-μl volume of a 1:3,000 or 1:4,500 dilution of DNase I (2 U/μl) (Novagen no. 69164-1) in the same buffer was added to each tube (one dilution per aliquot), and 10 μl of stop buffer (100 mM EDTA, 30% glycerol, 0.5% orange G, 0.075% xylene [Novagen no. 69165-1]) was added after incubation at 15°C for 5 min. The contents of the two DNase I reaction tubes were then combined and fractionated on a 0.8% LMT agarose gel, and the gel segment with DNA of about 1 kb (about 1.5 ml of agarose gel) was excised. DNA was extracted from the agarose with the Promega Wizard PCR Preps DNA purification system and resuspended in 200 μl of water before being extracted once with phenol and twice with ether and then precipitated. The DNA was resuspended in 17.25 μl of water and subjected to T4 DNA polymerase repair and single dA tailing with the Novagen single dA tailing kit. The reaction product (85 μl containing about 8 ng of DNA) was then extracted with chloroform-isoamyl alcohol (24:1) once and ligated to 3 × 10−3 pmol of pGEM-T (Promega) in a total volume of 100 μl. Ligation was carried out overnight at 4°C, and the ligated DNA was precipitated and resuspended in 20 μl of water before being electroporated into E. coli JM109 and plated out on 5-bromo-4-chloro-3-indolyl-β-d-galactoside (BCIG)-isopropyl-β-d-thiogalactopyr anoside (IPTG) plates to give a bank.
Sequencing and analysis.
The DNA template for sequencing was prepared with a 96-well-format plasmid DNA miniprep kit (Advanced Genetic Technologies Corp.) by the procedure developed in the Institute for Genomic Research (TIGR) (34). Sequencing was performed with an Applied Biosystems 377 automated DNA sequencer. Sequence data were assembled and analyzed by using the Australian National Genomic Information Service (ANGIS), which incorporates several sets of programs (28). We used the algorithm described by Eisenberg et al. (9) to identify potential transmembrane segments from the amino acid sequence.
Specificity assay by PCR.
Chromosomal DNA was isolated with the Promega Genomic isolation kit and checked by gel electrophoresis. A total of 34 pools were made, with 6 to 12 samples of DNA per pool (Table 1). Chromosomal DNAs from six E. coli O157 strains, S. enterica serovar Landau, Brucella abortus, and Yersinia enterocolitica O9 were individually added to one pool containing another seven samples to give pools 22 to 30. PCR was carried out in a total volume of 25 μl, and then 10 μl was run on an agarose gel to check for amplified DNA.
Nucleotide sequence accession number.
The DNA sequence described here has been deposited in GenBank under accession no. AF061251.
RESULTS
General strategy for sequencing O antigen gene clusters of E. coli and related species.
Oligonucleotides which bind to the 5′ end of the gnd gene and the middle of the JUMPstart sequence were used for successful PCR amplification of O antigen gene clusters from all 10 randomly chosen serotypes of E. coli and S. enterica (unpublished data).
A PCR fragment of about 14 kb was obtained from E. coli O157:H7 isolate C664-1992, subjected to DNase I digestion, and cloned into pGEM-T to make a bank. To limit the effect of PCR errors, products of five individual PCRs were pooled before making the bank. A total of 112 clones were first sequenced from one end, and 16 of the 112 were then sequenced from the other end to obtain 85% double-strand coverage. Gaps and regions of inadequate coverage were then sequenced from specific PCR products amplified from chromosomal DNA. A sequence of 14,002 bases was obtained, which covers the DNA from the end of JUMPstart to the start of gnd.
O157 O antigen genes.
Twelve open reading frames were predicted from the sequence (Fig. 1); all have the same transcriptional direction from JUMPstart to gnd. The nucleotide and amino acid sequences were used to search available databases for indications of possible function.
FIG. 1.
O antigen gene cluster of E. coli O157. Both gene names and orf numbers are given.
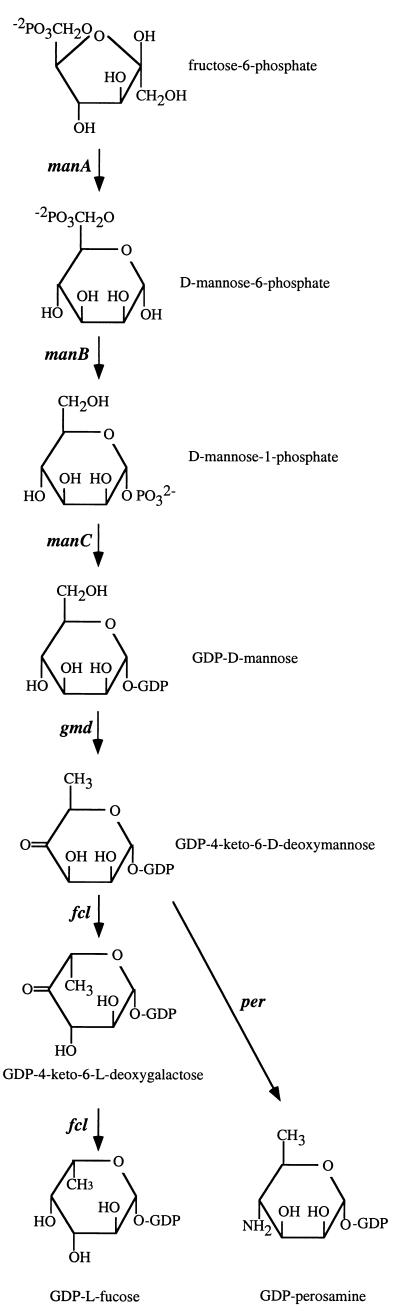
The structure of the O157 O unit is known (Fig. 2) (23), and we expect genes for GDP-l-fucose and GDP-N-acetylperosamine synthesis (Fig. 3). orf10 and orf11 showed similarity to the many published manC and manB genes, respectively, with the highest levels of identity at the amino acid level being 64 and 97% to manC of the E. coli O111 O antigen gene cluster (2) and manB of the E. coli colanic acid capsule gene cluster (30), respectively. orf10 and orf11 were named manC and manB, i.e., genes for synthesis of GDP-mannose. Orf7 showed 89% identity to Gmd encoded by the E. coli colanic acid capsule gene cluster which converts GDP-mannose to GDP-4-keto-6-d-deoxymannose (30), and the gene was named gmd. Orf8 showed 79 and 69% identity to WcaG of the E. coli colanic acid capsule gene cluster and to WbcJ (Orf14.8) of the Y. enterocolitica O8 O antigen gene cluster, respectively (42). Colanic acid and the Y. enterocolitica O8 O antigen both contain fucose, as does the O157 O antigen. Two enzymatic steps are required for GDP-l-fucose synthesis from GDP-4-keto-6-d-deoxymannose, the product of Gmd (11). It has been shown that the human FX protein carries out both reactions to convert GDP-4-keto-6-d-deoxymannose to GDP-l-fucose (33). WcaG is the same size as the human FX protein, and they have 29% amino acid sequence identity. We have recently shown that WcaG can also carry out both reactions (1) and have renamed the gene fcl (l-fucose). We identify orf8 as fcl on the basis of homology to fcl of the colanic acid cluster. In support of the one enzyme carrying out both reactions is the observation that there are no genes other than manB, manC, gmd, and fcl with a high level of similarity among the three sequenced bacterial gene clusters encoding fucose-containing structures (E. coli colanic acid, E. coli O157 O antigen, and Y. enterocolitica O8 O antigen [30, 42]). Orf5 is very similar to WbeE (RfbE) of Vibrio cholerae, which is thought to be the perosamine synthetase, which converts GDP-4-keto-6-d-deoxymannose to GDP-perosamine (31). V. cholerae O1 and E. coli O157 O antigens contain perosamine and N-acetylperosamine, respectively. The V. cholerae O1 manC, manB, gmd, and wbeE genes are the only genes of the V. cholerae O1 gene cluster with significant similarity to genes of the E. coli O157 gene cluster, and we believe that our observations both confirm the prediction made for the function of wbeE of V. cholerae and show that orf5 of the O157 gene cluster encodes GDP-perosamine synthetase. orf5 has therefore been named per. orf5 plus about 100 bp of upstream DNA was previously sequenced by Bilge et al. (5). orf12 includes 50 amino acids with high-level similarity to a conserved region of an acetyltransferase family (16), and we believe that it encodes the N-acetyltransferase which converts GDP-perosamine to GDP-N-acetylperosamine. orf12 has been named wbdR, but it could be renamed if the function were confirmed.
FIG. 2.
Structure of the O157 O antigen (23).
FIG. 3.
Biosynthetic pathway of GDP-l-fucose and GDP-perosamine with gene names. The pathway begins with fructose-6-phosphate, which is converted to d-mannose-6-phosphate by the phosphomannose isomerase encoded by manA. The manA gene is located outside the O antigen gene cluster (4). The GDP-l-fucose pathway was described by Ginsburg (11), and we have identified all the genes (reference 30 and unpublished data). The GDP-perosamine pathway is that proposed by Stroeher et al. (31), and the genes assigned to each step are based on sequence similarity described in the text.
We can also identify wzx (orf4, O unit flippase) and wzy (orf2, O antigen polymerase) genes. Most O antigens are synthesized with the Wzx-dependent system, in which repeat O units are synthesized on the inner face of cytoplasmic membrane before being transferred across the cytoplasmic membrane by Wzx and polymerized by Wzy (26, 35). orf4 is predicted to encode an integral inner membrane protein with 12 transmembrane segments. Orf4 shows similarity to many Wzx proteins and has the approximate 50-amino-acid segment found near the amino-terminal end of Wzx proteins with a conserved motif (30). The O antigen polymerases thus far identified are integral membrane proteins and have a similar predicted secondary structure (several transmembrane segments with a large periplasmic loop) (21) but little or no sequence similarity. orf2 encodes a protein with eight predicted membrane segments and a large loop on the predicted periplasmic face and fits the criteria to be named wzy.
The orf1 and orf3 gene products showed characteristics of the WcaA group of transferases: the members of this group have some similarity at the amino-terminal end, and about 35 amino acids at various positions are highly conserved within this region (30). orf1 and orf3 have been named wbdN and wbdO, respectively. The amino acid sequence encoded by orf6 showed the characteristics of another group of transferases (the WcaC group) (30), and in this case the conserved amino acids are located at various positions at the carboxy-terminal end. orf6 has been named wbdP. From the structure of the O157 O unit (Fig. 2), we expect four transferases, including one to add the first sugar to the carrier lipid undecaprenol phosphate (UndP), but have found only three presumptive transferase genes. Other than wzx and wzy, none of the genes in the cluster encode predicted integral membrane proteins, whereas the two proteins known to initiate O unit synthesis by transfer of a sugar phosphate to UndP, WecA (Rfe) and WbaP (RfbP), have several predicted transmembrane segments (13, 20). We therefore suggest that as proposed on quite good grounds for Y. enterocolitica (42), WecA transfers GalNAc phosphate to UndP to initiate O unit synthesis as well as initiating enterobacterial common-antigen synthesis by transfer of GlcNAc phosphate. We can thus account for all the required steps for synthesis of the O157 O antigen with 11 of the 12 genes of the cluster. orf9 shows high-level similarity (44% identity at amino acid level, same length) to the wcaH gene of the E. coli colanic acid capsule gene cluster. There is no homolog in the Y. enterocolitica O8 O antigen gene cluster, so a role in GDP-fucose synthesis is not indicated (see the discussion of fcl above). The function of this gene, named wbdQ, is unknown.
The DNA between manB and wbdR has strong sequence similarity to one of the H-repeat units of E. coli K-12 (43). The inverted-repeat sequences flanking this region are still recognizable, each with 2 of the 11 bases being changed. The H-repeat-associated open reading frame located within this region has a 267-bp deletion and a number of mutations (including frameshift mutations) at various positions.
Identification of O157-specific genes.
O antigen gene clusters generally contain about 8 to 20 genes, falling into three general classes: (i) genes for synthesis of nucleotide sugar precursors such as dTDP-rhamnose or GDP-N-acetylperosamine, (ii) genes for transfer of sugars to build the O unit, and (iii) genes which carry out specific assembly or processing steps in conversion of the O unit to the O antigen as part of the complete lipopolysaccharide (see the reviews by Reeves [25, 26] and Whitfield [35]). Genes of the first class are commonly present in many O antigen clusters, and sequence similarity is usually sufficient to identify these genes in database searches; they may therefore give cross-reactions in DNA-based assays. Genes of the second class are often group specific since they are specific for both sugars of the linkage, and, furthermore, transferase genes for a given linkage in different clusters may show negligible similarity (e.g., wbaW [rfbW] of S. enterica group C2 [18] and any of the presumptive α-mannose 1→2 mannose transferases of S. enterica group C1 [15]). Genes of the third class encode proteins such as the O antigen polymerase and the flippase: these are most easily identified on the basis of predicted transmembrane segments rather than sequence per se and may also be group specific.
We used 16 pairs of oligonucleotide primers (Table 2), based on the sequences of transferase, wzx, and wzy genes of O157, in PCR with 34 pools of DNA (Table 1) to test the specificity of these genes. The DNA in pools 1 to 21 includes DNA from strains representing the 165 other known E. coli serotypes, the DNA in pools 22 to 30 includes DNA from E. coli O157 strains or strains of other species (Brucella abortus, Y. enterocolitica O9, and S. enterica serovar Adelaide) which cross-react serologically with E. coli O157, and the DNA in pools 31 to 34 includes DNA from 11 Y. pseudotuberculosis, 12 Shigella boydii, 12 S. enterica, and 12 V. cholerae strains, respectively, with each strain having a different O antigen. Each of the 16 primer pairs produces a band of the predicted size from pools containing O157 DNA (pools 22 to 27) (Table 2). Several other pools gave bands of different sizes from that expected, which are attributed to nonspecific priming (Table 2). However, one primer pair produced two bands with Y. enterocolitica O9, one of which was the same size as that from the positive control: this pair of primers is based on the wzy sequence (Table 2). The predicted secondary structures of Wzy proteins are generally similar, although there is generally very low similarity at the amino acid or DNA level among the sequenced wzy genes. Thus, it is possible that Y. enterocolitica O9 has a wzy gene closely related to that of E. coli O157. It is also possible that this band is due to chance hybridization of another gene, since the other two wzy primer pairs did not produce any band with Y. enterocolitica O9. Pool 28 includes DNA of S. enterica serovar Landau (serogroup N), which has the same O antigen as E. coli O157 (23). Pools 29, 30, and 34 include DNA of B. abortus, Y. enterocolitica O9, and V. cholerae O1, respectively, all of which have perosamine or N-acetylperosamine in the O antigen, with the first two also cross-reacting serologically with E. coli O157 antisera (6). These results indicate that the genes tested are highly O157 specific, although one primer pair may have cross-reacted with Y. enterocolitica O9.
TABLE 2.
PCR testing of the specificity of O157 genes
| Gene | Base positions of gene | Base positions of forward/reverse primers | No. of pools (of pools 1–21 and 28–34) giving band of predicted sizea | Annealing temp (°C) of PCR |
|---|---|---|---|---|
| wbdN | 60–842 | 60–77/842–825 | 0* | 55 |
| 165–182/512–495 | 0* | 55 | ||
| 291–308/749–732 | 0 | 55 | ||
| wzy | 839–2023 | 839–856/2023–2006 | 1** | 50 |
| 1034–1051/1600–1583 | 0*** | 63 | ||
| 1259–1276/1894–1877 | 0 | 60 | ||
| wbdO | 1992–2738 | 1992–2009/2738–2721 | 0 | 50 |
| 2091–2108/2474–2457 | 0**** | 62 | ||
| 2286–2303/2663–2646 | 0 | 60 | ||
| wzx | 2725–4116 | 2725–2742/4116–4099 | 0 | 50 |
| 2923–2940/3609–3592 | 0***** | 63 | ||
| wbdP | 5238–6452 | 5238–5255/6452–6435 | 0 | 55 |
| 5421–5438/5954–5937 | 0* | 55 | ||
| 5688–5705/6212–6195 | 0 | 55 | ||
| wbdR | 13137–13802 | 13242–13259/13610–13593 | 0 | 55 |
| 13365–13382/13712–13695 | 0 | 60 |
Seven pairs of primers gave bands of quite different sizes from that predicted. These are indicated as follows: *, one band in one pool; **, two bands in pool 30 (one of the bands is of the correct size), three bands in 1 pool, one band in 21 pools; ***, two bands in one pool; ****, three bands in 11 pools and two bands in 14 pools; *****, two bands in 2 pools and one band in 7 pools.
Thus, PCR with primers based on genes wbdN, wzy, wbdO, wzx, wbdP, and wbdR is highly specific for E. coli O157, giving positive results with each of six unrelated O157 strains, while only one primer pair gave a band of the expected size with one of the three strains with O antigens known to cross-react serologically with E. coli O157.
DISCUSSION
We have sequenced the O157 O antigen gene cluster and identified, with various degrees of precision, all the genes required for the synthesis of the O antigen. All but one of the genes have a low G+C content (Fig. 1), as observed for other O antigen gene clusters, and this indicates that, as appears to be quite common, the O157 O antigen gene cluster was acquired by transfer from another species. The exception is manB, which closely resembles the colanic acid manB gene and has a 54% G+C content. This situation has been observed in the S. enterica C1 and E. coli O7 O antigen gene clusters (15, 19), and it has been suggested that in such cases the O antigen manB gene was derived from the colanic acid gene cluster by recombination (15).
A remnant H repeat, also with a low G+C content, is located upstream of wbdR. The deletion and mutation in the H-repeat unit indicate that it has been associated with this gene cluster for a long time since last undergoing transposition, perhaps having played a role in assembly of the gene cluster. It is possible that in the absence of wbdR, the predicted O-acetyltransferase gene, the O antigen would be synthesized with perosamine in place of N-acetylperosamine and that the gene is not essential for O antigen synthesis: one can speculate that the ancestral gene cluster contained perosamine and that the N-acetyltransferase gene was added by lateral transfer mediated by the H repeat, similar to the H-repeat-mediated gene transfer proposed for the S. enterica D2 O antigen cluster (41).
The currently accepted methods for the detection of Stx-positive O157 strains involve assays for the detection of Stx, either directly or by PCR, coupled with plating on selective media directly or after enrichment with O157-specific antibodies attached to paramagnetic particles, followed by serotyping (O157 O antigen determination). We have now identified genes highly specific to O157, since they were detected by PCR in each of the six unrelated O157 strains but not in any of the other strains tested, including representatives of the 165 other known E. coli O antigen forms, and a range of other gram-negative bacteria. Thus, we believe that the genes are suitable for use in a PCR-based method for identification of O157 strains to replace time-consuming plating and serotyping methods. We also showed that these genes were E. coli O157 specific in that they were not in general detectable by PCR in S. enterica serovar Landau, B. abortus, or Y. enterocolitica O9, which cross-react serologically with O157 antisera; this indicates that a PCR-based method would distinguish these strains from E. coli O157, whereas serotyping methods do not. For Y. enterocolitica O9, the choice of primer for the wzy gene appears to be important since one pair gave a band of the size expected for E. coli O157. The E. coli serotyping scheme is not yet fully comprehensive, and there are other, as yet unidentified, O antigens. For this reason, field strains and conditions must be tested to confirm the specificity, although we believe that all or most of these six genes will be specific to O157 strains. Further specificity can be gained by use of a combination of these genes, perhaps by PCR with primers binding to adjacent genes.
There are many O157 clones (38), and it is the O157:H7 clone for which there is major need for diagnostics. The use of an O157-specific test in screening for this organism is highly desirable, since in its serological form, it is the traditional test for identification of this clone. The O157 O antigen-specific test proposed would have to be combined with tests for other genes such as the stx gene. In this context, it has been shown that O157:H7 strains are in a small group which stands apart from most other E. coli strains (24, 36–40). There may well be sequences specific to this group which could also be useful to distinguish the O157:H7 clone from others. The use of O157-specific PCR allows one to use PCR tests for both stx (or other genes) and the O antigen, avoiding the need to include serological testing.
We used two or three pairs of oligonucleotides for each gene in the PCR test. Some of the primer pairs produced bands of the wrong size in some or all of the sample pools. We believe that this is due to chance priming elsewhere on the chromosome. This problem can be avoided by using other primer pairs for those genes.
About 20 O antigen gene clusters have been sequenced, 7 of them in this laboratory. Each involved laborious procedures for identification and cloning of the gene cluster before sequencing. For the O157 sequence, we have combined long PCR with approaches used for genome sequencing, involving DNase I shotgun library construction and 96-well tray plasmid preparation, to expedite the sequencing. These methods can be applied to any O antigen gene cluster for which the flanking sequence is known.
This study demonstrated that certain classes of O antigen genes can be highly specific, and now that a given O antigen gene cluster can be fully sequenced within a few weeks, it is possible to quickly sequence an O antigen gene cluster and determine specific probes for PCR-based detection of the O antigen for any new strain which emerges as a serious pathogen.
ACKNOWLEDGMENTS
We thank all the people and institutes listed in Table 1 for kindly supplying strains, and we thank Heather Curd for excellent technical assistance.
This investigation was supported by Bioproperties (Australia) Pty Ltd.
REFERENCES
- 1.Andrianopoulos K, Wang L, Reeves P R. Identification of the fucose synthetase gene in the colanic acid gene cluster of Escherichia coli K-12. J Bacteriol. 1998;180:998–1001. doi: 10.1128/jb.180.4.998-1001.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bastin D A, Reeves P R. Sequence and analysis of the O antigen gene (rfb) cluster of Escherichia coli O111. Gene. 1995;164:17–23. doi: 10.1016/0378-1119(95)00459-j. [DOI] [PubMed] [Google Scholar]
- 3.Bell B P, Goldoft M, Griffin P M, Davis M A, Gordon D C, Tarr P I, Bartleson C A, Lewis J H, Barrett T J, Wells J G, Baron R, Kobayashi J. A multistate outbreak of Escherichia coli O157:H7-associated bloody diarrhea and hemolytic uremic syndrome from hamburgers. The Washington experience. JAMA. 1994;272:1349–1353. [PubMed] [Google Scholar]
- 4.Berlyn M K B, Low K B, Rudd K E, Singer M. Linkage map of Escherichia coli K-12. In: Neidhardt F D, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C: American Society for Microbiology; 1996. pp. 1715–1902. [Google Scholar]
- 5.Bilge S S, Vary J C, Dowell S F, Tarr P I. Role of the Escherichia coli O157-H7 O side chain in adherence and analysis of an rfb locus. Infect Immun. 1996;64:4795–4801. doi: 10.1128/iai.64.11.4795-4801.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caroff M, Bundle D R, Perry M B. Structure of the O-chain of the phenol-phase soluble cellular lipopolysaccharide of Yersinia enterocolitica serotype O:9. Eur J Biochem. 1984;139:195–200. doi: 10.1111/j.1432-1033.1984.tb07994.x. [DOI] [PubMed] [Google Scholar]
- 7.Centers for Disease Control. Update. Multistate outbreak of Escherichia coli O157:H7 infections from hamburgers—western United States, 1992–1993. Morbid Mortal Weekly Rep. 1993;42:258–263. [PubMed] [Google Scholar]
- 8.Cubbon M D, Coia J E, Hanson M F, Thomson-Carter F M. A comparison of immunomagnetic separation, direct culture and polymerase chain reaction for the detection of verocytotoxin-producing Escherichia coli O157 in human faeces. J Med Microbiol. 1996;44:219–222. doi: 10.1099/00222615-44-3-219. [DOI] [PubMed] [Google Scholar]
- 9.Eisenberg D, Schwarz E, Komaromy M, Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J Mol Biol. 1984;179:125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- 10.Fratamico P M, Sackitey S K, Wiedmann M, Deng M Y. Detection of Escherichia coli O157:H7 by multiplex PCR. J Clin Microbiol. 1995;33:2188–2191. doi: 10.1128/jcm.33.8.2188-2191.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ginsburg V. Studies of the biosynthesis of guanosine diphosphate l-fucose. J Biol Chem. 1961;236:2389–2393. [PubMed] [Google Scholar]
- 12.Hobbs M, Reeves P R. The JUMPstart sequence: a 39 bp element common to several polysaccharide gene clusters. Mol Microbiol. 1994;12:855–856. doi: 10.1111/j.1365-2958.1994.tb01071.x. [DOI] [PubMed] [Google Scholar]
- 13.Jiang X M, Neal B, Santiago F, Lee S J, Romana L K, Reeves P R. Structure and sequence of the rfb (O antigen) gene cluster of Salmonella serovar typhimurium (strain LT2) Mol Microbiol. 1991;5:695–713. doi: 10.1111/j.1365-2958.1991.tb00741.x. [DOI] [PubMed] [Google Scholar]
- 14.Keene W E, McAnulty J M, Hoesly F C, Williams L P, Hedberg K, Oxman G L, Barrett T J, Pfaller M A, Fleming D W. A swimming-associated outbreak of hemorrhagic colitis caused by Escherichia coli O157:H7 and Shigella sonnei. N Engl J Med. 1994;331:579–584. doi: 10.1056/NEJM199409013310904. [DOI] [PubMed] [Google Scholar]
- 15.Lee S J, Romana L K, Reeves P R. Sequence and structural analysis of the rfb (O antigen) gene cluster from a group C1 Salmonella enterica strain. J Gen Microbiol. 1992;138:1843–1855. doi: 10.1099/00221287-138-9-1843. [DOI] [PubMed] [Google Scholar]
- 16.Lin W, Cunneen T, Lee C. Sequence analysis and molecular characterization of genes required for the biosynthesis of type 1 capsular polysaccharide in Staphylococcus aureus. J Bacteriol. 1994;176:7005–7016. doi: 10.1128/jb.176.22.7005-7016.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lior H. Classification of Escherichia coli. In: Gyles C L, editor. Escherichia coli in domestic animals and humans. Wallingford, United Kingdom: CAB International; 1994. pp. 31–72. [Google Scholar]
- 18.Liu D, Lindquist L, Reeves P R. Transferases of O-antigen biosynthesis in Salmonella enterica: dideoxhexosyl transferases of groups B and C2 and acetyltransferase of group C2. J Bacteriol. 1995;177:4084–4088. doi: 10.1128/jb.177.14.4084-4088.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marolda C L, Valvano M A. Identification, expression, and DNA sequence of the GDP-mannose biosynthesis genes encoded by the O7 rfb gene cluster of strain VW187 (Escherichia coli O7:K1) J Bacteriol. 1993;175:148–158. doi: 10.1128/jb.175.1.148-158.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meier-Dieter U, Barr K, Starman R, Hatch L, Rick P D. Nucleotide sequence of the Escherichia coli rfe gene involved in the synthesis of enterobacterial common antigen. J Biol Chem. 1992;267:746–753. [PubMed] [Google Scholar]
- 21.Morona R, Mavris M, Fallarino A, Manning P A. Characterization of the rfc region of Shigella flexneri. J Bacteriol. 1994;176:733–747. doi: 10.1128/jb.176.3.733-747.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park C H, Vandel N M, Hixon D L. Rapid immunoassay for detection of Escherichia coli O157 directly from stool specimens. J Clin Microbiol. 1996;34:988–990. doi: 10.1128/jcm.34.4.988-990.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perry M B, MacLean L, Griffith D W. Structure of the O-chain polysaccharide of the phenol-phase soluble lipopolysaccharide of Escherichia coli O157:H7. Biochem Cell Biol. 1986;64:21–28. doi: 10.1139/o86-004. [DOI] [PubMed] [Google Scholar]
- 24.Pupo G M, Karaolis D K R, Lan R, Reeves P R. Evolutionary relationships among pathogenic and nonpathogenic Escherichia coli strains inferred from multilocus enzyme electrophoresis and mdh sequence studies. Infect Immun. 1997;65:2685–2692. doi: 10.1128/iai.65.7.2685-2692.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reeves P R. Evolution of Salmonella O antigen variation by interspecific gene transfer on a large scale. Trends Genet. 1993;9:17–22. doi: 10.1016/0168-9525(93)90067-R. [DOI] [PubMed] [Google Scholar]
- 26.Reeves P R. Biosynthesis and assembly of lipopolysaccharide. New Compr Biochem. 1994;27:281–314. [Google Scholar]
- 27.Reeves P R, Hobbs M, Valvano M, Skurnik M, Whitfield C, Coplin D, Kido N, Klena J, Maskell D, Raetz C, Rick P. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 1996;4:495–503. doi: 10.1016/s0966-842x(97)82912-5. [DOI] [PubMed] [Google Scholar]
- 28.Reisner A H, Bucholtz C A, Smelt J, McNeil S. Proceedings of the Twenty-Sixth Annual Hawaii International Conference on Systems Science. Vol. 1. 1993. Australia’s National Genomic Information Service; pp. 595–602. [Google Scholar]
- 29.Sanderson K E, Hessel A, Rudd K E. Genetic map of Salmonella typhimurium, edition VIII. Microbiol Rev. 1995;59:241–303. doi: 10.1128/mr.59.2.241-303.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stevenson G, Andrianopoulos K, Hobbs H, Reeves P R. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J Bacteriol. 1996;178:4885–4893. doi: 10.1128/jb.178.16.4885-4893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stroeher U H, Karageorgos L E, Brown M H, Morona R, Manning P A. A putative pathway for perosamine biosynthesis is the first function encoded within the rfb region of Vibrio cholerae O1. Gene. 1995;166:33–42. doi: 10.1016/0378-1119(95)00589-0. [DOI] [PubMed] [Google Scholar]
- 32.Tarr P I. Escherichia coli O157:H7: clinical, diagnostic, and epidemiological aspects of human infection. Clin Infect Dis. 1995;20:1–8. doi: 10.1093/clinids/20.1.1. [DOI] [PubMed] [Google Scholar]
- 33.Tonetti M, Sturla L, Bisso A, Benatti U, De Flora A. Synthesis of GDP-l-fucose by the human FX protein. J Biol Chem. 1996;271:27274–27279. doi: 10.1074/jbc.271.44.27274. [DOI] [PubMed] [Google Scholar]
- 34.Utterback T R, McDonald L A, Fuldner R A. A reliable, efficient protocol for 96-well plasmid DNA miniprep with rapid DNA quantification for high-throughput automated DNA sequencing. Genome Sci Technol. 1995;1:1–8. [Google Scholar]
- 35.Whitfield C. Biosynthesis of lipopolysaccharide O antigens. Trends Microbiol. 1995;3:178–185. doi: 10.1016/s0966-842x(00)88917-9. [DOI] [PubMed] [Google Scholar]
- 36.Whittam T S, Ochman H, Selander R K. Multilocus genetic structure in natural populations of Escherichia coli. Proc Natl Acad Sci USA. 1983;80:1751–1755. doi: 10.1073/pnas.80.6.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whittam T S, Wachsmuth I K, Wilson R A. Genetic evidence of clonal descent of Escherichia coli O157:H7 associated with hemorrhagic colitis and hemolytic uremic syndrome. J Infect Dis. 1988;157:1124–1133. doi: 10.1093/infdis/157.6.1124. [DOI] [PubMed] [Google Scholar]
- 38.Whittam T S, Wilson R A. Genetic relationships among pathogenic Escherichia coli of serogroup O157. Infect Immun. 1988;56:2467–2473. doi: 10.1128/iai.56.9.2467-2473.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whittam T S, Wilson R A. Genetic relationships among pathogenic strains of avian Escherichia coli. Infect Immun. 1988;56:2458–2466. doi: 10.1128/iai.56.9.2458-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whittam T S, Wolfe M L, Wachsmuth I K, Ørskov F, Ørskov I, Wilson R A. Clonal relationships among Escherichia coli strains that cause hemorrhagic colitis and infantile diarrhea. Infect Immun. 1993;61:1619–1629. doi: 10.1128/iai.61.5.1619-1629.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiang S H, Hobbs M, Reeves P R. Molecular analysis of the rfb gene cluster of a group D2 Salmonella enterica strain: evidence for its origin from an insertion sequence-mediated recombination event between group E and D1 strains. J Bacteriol. 1994;176:4357–4365. doi: 10.1128/jb.176.14.4357-4365.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang L, Radziejewska-Lebrecht J, Krajewska-Pietrasik D, Toivanen P, Skurnik M. Molecular and chemical characterization of the lipopolysaccharide O-antigen and its role in the virulence of Yersinia enterocolitica serotype O8. Mol Microbiol. 1997;23:63–76. doi: 10.1046/j.1365-2958.1997.1871558.x. [DOI] [PubMed] [Google Scholar]
- 43.Zhao S, Sandt C H, Feulner G, Vlazny D A, Gray J A, Hill C W. rhs elements of Escherichia coli K-12: complex composites of shared and unique components that have different evolutionary histories. J Bacteriol. 1993;175:2799–2808. doi: 10.1128/jb.175.10.2799-2808.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]