Abstract
A three-dimensional structural model of an influenza virus ribonucleoprotein particle reconstituted in vivo from recombinant proteins and a model genomic vRNA has been generated by electron microscopy. It shows a circular shape and contains nine nucleoprotein monomers, two of which are connected with the polymerase complex. The nucleoprotein monomers show a curvature that may be responsible for the formation of helical structures in the full-size viral ribonucleoproteins. The monomers show distinct contact boundaries at the two sides of the particle, suggesting that the genomic RNA may be located in association with the nucleoprotein at the base of the ribonucleoprotein complex. Sections of the three-dimensional model show a trilobular morphology in the polymerase complex that is consistent with the presence of its three subunits.
INTRODUCTION
The genome of influenza A virus is a set of eight single-stranded RNA segments of negative polarity. For them to be functional in transcription and replication, they have to be associated to nucleoprotein (NP) 55 kDa-molecules and to the polymerase in ribonucleoprotein particles (RNPs) (reviewed in Portela et al., 1999). Transcription and replication of viral RNPs take place in the nucleus of infected cells. Replication involves the generation of a full-length RNA copy of positive polarity that is encapsidated with NP molecules and the polymerase (cRNP). These cRNPs serve as intermediates for the synthesis of vRNA progeny molecules. Transcription is initiated with capped primers generated from cellular hnRNAs by a cap-stealing mechanism (reviewed in Lamb and Krug, 1996). Termination and polyadenylation involves the reiterative copying of an oligo(U) sequence located close to the 5′ terminus of the vRNAs, (Robertson et al., 1981; Poon et al., 1999). These processes require the interaction of the polymerase with the conserved 5′-terminal sequences of the template (Pritlove et al., 1999).
The virus polymerase is a heterotrimer formed by the PB1, PB2 and PA proteins (Digard et al., 1989). The PB2 subunit (83 kDa) is a cap-binding protein (Ulmanen et al., 1981) involved in the initiation of viral transcription (Bárcena et al., 1994). The PB1 protein (83 kDa) is responsible for the RNA polymerase activity (Biswas, 1998). The PA subunit (78 kDa) is a phosphoprotein (Sanz-Ezquerro et al., 1998) with protease activity (Sanz-Ezquerro et al., 1995; A. Nieto, unpublished results) whose activity correlates with the replication capacity of the polymerase (Perales et al., 2000). The most abundant component of the RNP is the NP, a basic protein that binds RNA without sequence specificity and protects its sugar-phosphate backbone from modification (Baudin et al., 1994; Albo et al., 1995).
Early studies of viral RNPs by electron microscopy (Compans et al., 1972; Heggeness et al., 1982) showed supercoiled structures with a terminal loop. The polymerase is present at one end of the supercoil, as shown by immunogold labelling (Murti et al., 1988) and helps in maintaining the RNA ends linked together (Klumpp et al., 1997). NP–vRNA complexes generated in vitro show structural and biochemical properties similar to natural RNPs (Yamanaka et al., 1990) and purified NP, essentially RNA-free, can also self-assemble into oligomers and coiled structures analogous to RNPs (Ruigrok and Baudin, 1995). The flexibility and heterogeneity of natural RNPs have precluded a better understanding of their structure. We recently used recombinant, more uniform populations of vRNPs to obtain structural information by electron microscopy and image processing that allowed for the first time a low-resolution visualization of the virus polymerase (Ortega et al., 2000). Here we have applied a similar approach to generate a three-dimensional (3D) reconstruction of a smaller recombinant RNP. The 3D structure obtained reveals the structural features of the NP monomers and their interaction with the polymerase complex, in which the presence of the three subunits is delineated.
RESULTS AND DISCUSSION
Improved structure analysis of a recombinant mini-RNP
In an earlier report, we described the in vivo reconstitution and analysis of the structure of a recombinant influenza RNP containing a vRNA-like genome of 313 nt (Ortega et al., 2000). Classification and averaging of the images obtained indicated the presence of a prominent RNP population with 11-NP monomers and minor size classes with 10 and 12 monomers. These RNP populations comprised circular and elliptic particles. In addition to the size and shape heterogeneity observed, the main limitation in these studies was the flexibility of the particles, suggested by the formation of circular and elliptic structures, and especially the flexibility of the polymerase complex in relation to the NP ring. Such structural plasticity led to the loss of information upon image averaging.
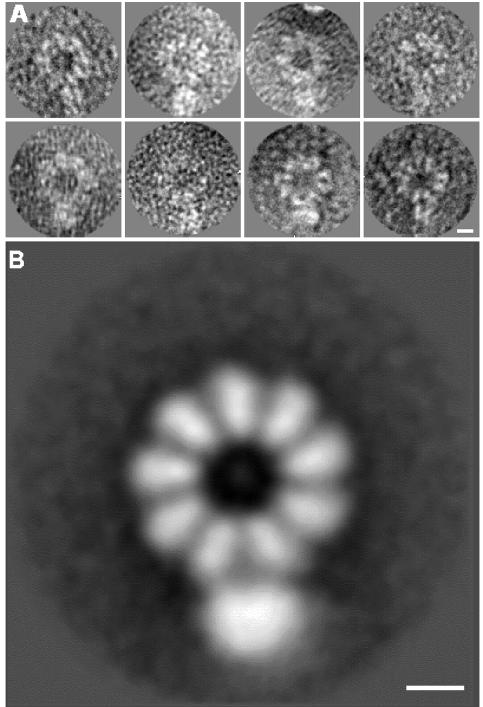
To overcome these problems, we took advantage of the availability of a series of ribozyme constructs that directed the reconstitution of virus RNPs of smaller size with higher efficiency of replication than before (Ortega et al., 2000). One such clone led to the generation of RNPs with a genome of only 248 nt. This length is sufficient for the RNP to incorporate eight NP monomers but in fact contained a mixture of particles with eight and nine NP molecules. We have shown that RNPs with an odd number of NP molecules replicate less efficiently than those containing an even number (Ortega et al., 2000). However, the reconstituted RNP 9-mers were used for structural studies because they gave more homogeneous particles and yielded a better average image than the 8-mers when analysed by electron microscopy. A gallery of these individual images and the average image obtained is presented in Figure 1. It is clear from these results that the quality of the average image was better than that obtained previously for the 11-mers (compare with Figure 4, Ortega et al., 2000) and the apparent size of the polymerase was in proportion to that of the NP. Two reasons may have contributed to the improvement in the resolution of the images: (i) the particle size is smaller and hence it may be more rigid; and (ii) the number of NP monomers in the 9-mers is probably at the upper limit of capacity of the RNA genome, leading to a somewhat strained, less flexible structure. In view of the enhanced rigidity of these RNPs we set out to determine their 3D structure by electron microscopy.
Fig. 1. Average image of an influenza virus mini-RNP. Individual images of the RNP complexes obtained after two-dimensional alignment (A) and 2D-average (B) of the population of influenza virus recombinant RNPs containing nine NP monomers. In this and subsequent Figures, the scale bar represents 50 Å.
3D reconstruction of a recombinant mini-RNP
The approach followed for 3D reconstruction involved the collection of images with a sample tilt of 35°, in order to apply the angular refinement algorithms provided by SPIDER (Frank et al., 1996). An initial set of 2040 images was classified according to size and shape yielding a uniform set of 890 images that were aligned and averaged. A first 3D crude model was obtained stacking 20 single planes of this 2D average. The model was improved by iterative refinement steps in which its projections were compared with the images in the set and those that showed the best fit were selected for further refinement. Finally, a collection of 190 images that showed correlation coefficients above 0.65 were selected, which allowed distinction between the top-on-carbon and bottom-on-carbon orientations. The iterative refinement was carried out until the correlation coefficients did not change.
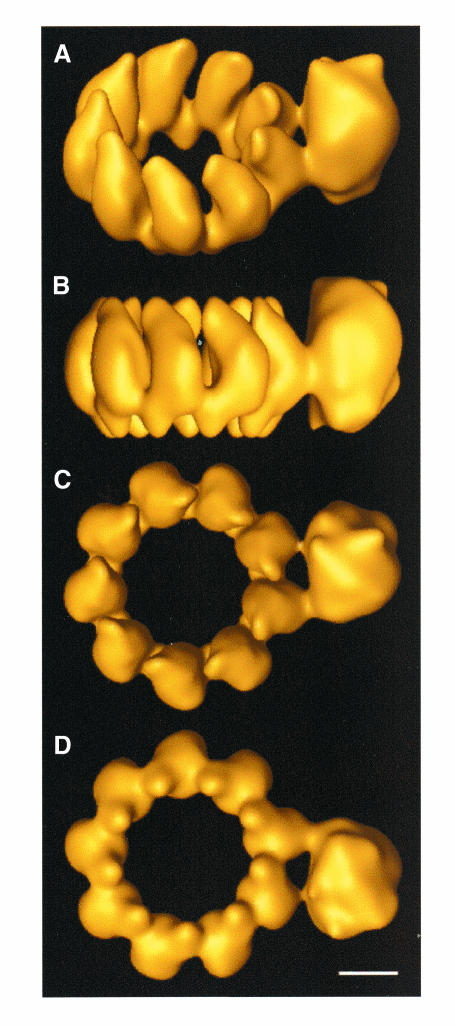
The model obtained is presented in Figure 2. The particle includes a ring of nine NP monomers, in which symmetry restrictions have been applied, and an additional volume that corresponds to the polymerase complex. The resolutions obtained were 27 and 36 Å, respectively, the difference being due to the imposition of symmetry in the NP ring. The mass in the NP ring is asymmetrically distributed along the vertical axis. A tight connection between the NP monomers is observed at the bottom of the model. In addition, the NP monomers present a clear curvature, especially apparent in the perspective and side views (Figure 2A and B). The polymerase complex appears as a rounded particle that is connected to the NP ring by two contacts involving adjacent NP monomers, one of which is thicker than the other. The fact that the polymerase complex is ∼15 Å taller than the NP ring supports the idea that most of the sample was contrasted by the staining reagent. Furthermore, the mass of the reconstructed volume represents ∼95% of its expected mass (∼820 kDa).
Fig. 2. Three-dimensional model of a recombinant influenza virus mini-RNP. (A) Perspective view. (B) Side view. (C) Top view. (D) Bottom view.
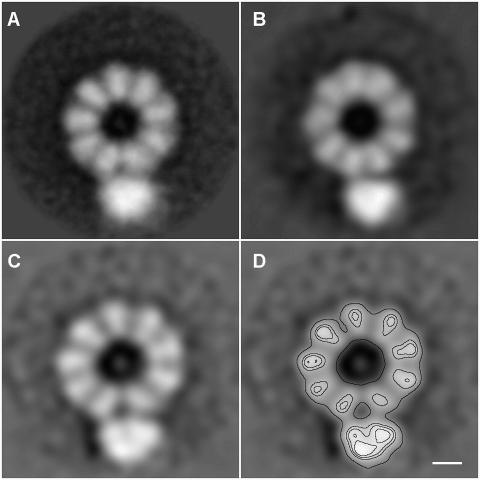
The correlation of the model obtained with the experimental data is illustrated in Figure 3. The average image obtained with the 190 images included in the final set (generated with a 35° sample tilt; Figure 3A) is compared with the 35° projection of the model presented in Figure 2 (Figure 3B), and with the central section of the model, in which no symmetry was imposed (Figure 3C and D). The mass distributions observed and predicted show a very good fit, in line with the correlation coefficients obtained. From the central section of the model (Figure 3C and D) it is apparent that the two adjacent NP monomers involved in the interaction with the polymerase are different from the rest. While the other NP monomers are placed with their axis radially oriented towards the centre of the NP ring, these two monomers are slightly displaced to accommodate the interaction with the polymerase. In addition, the internal density of the reconstructed polymerase is not uniform but rather shows three distinct areas, suggesting the presence of three closely connected subunits.
Fig. 3. Comparison of the predictions of the three-dimensional model of an influenza virus recombinant RNP with the experimental data. The average image of the 190 images in the final set for 3D reconstruction, obtained with a 35° sample tilt (A) is shown in comparison with the 35° projection of the model (B) and with the central section of the model without any symmetry imposed (C and D).
Biological implications derived from the 3D model
The model presented in Figures 2 and 3 represents the first 3D structural information available to date about an influenza virus RNP particle and, moreover, about any negative-stranded RNA virus RNP. Although the resolution obtained so far is limited, the structure presented here provides information that may have relevant biological consequences. Thus, the NP monomers show a higher degree of association at the bottom than at the top of the particle. This mass concentration may reflect the presence of the genomic RNA bound to the NP. The total length of the RNA present in the RNP particle (248 nt × ∼3.5 Å/nt ≈ 870 Å) is higher than the perimeter of the NP ring (∼400 Å), indicating that it must be wrapped around the NP monomers in some way. The sequences of NP mainly responsible for its association to RNA have been mapped to its N-terminal region by deletion mutagenesis (Albo et al., 1995). Other approaches have indicated that several amino acids along the entire NP molecule are implicated in RNA binding (Elton et al., 1999). Our model cannot distinguish between these possibilities but suggests that considerable packaging of the RNA around the NP molecule takes place. In addition, since the RNA is linked to the NP monomers and the NP monomers are interacting at the bottom of the 3D model, we suggest that the bottom area of the particle contain at least part of the RNA molecule.
The general structure of influenza virus RNPs is not circular but helical (Compans et al., 1972; Heggeness et al., 1982). In fact, the structure of the mini-RNPs depends on the length of the genomic RNA present in the particles. Thus, RNPs whose genome contains 248 nt are essentially circular (this report), while those containing 350 nt show a mixture of circular and elliptic structures (Ortega et al., 2000). On the other hand, RNPs with lengths of 400 nt and over showed only helical structures (Ortega et al., 2000). Similar results have been reported by Ruigrok (Ruigrok and Baudin, 1995) when studying the conformation of complexes containing only NP. These results indicate that the NP monomers contain the information necessary to yield a helical structure upon self-association and a circular or elliptic structure can only be detected when the length of the RNA present in the RNP particle is not sufficient to allow supercoiling. The 3D structure proposed in this report illustrates how this supercoiling may be obtained. The curvature of the NP monomer gives ground to interactions leading to helical assemblies. The presence of RNA is not required for this conformation and, moreover, if the RNA is short enough it can prevent helical coiling.
In addition to its role in the packaging of viral RNA, the NP is functionally important in the process of replication. This is evident from the existence of mutant viruses affected in the NP gene that show a replication-defective phenotype. Some of them are affected in the synthesis of vRNA at restrictive temperatures (reviewed in Mahy, 1983) whilst others show a defect in the synthesis of cRNA from a vRNA template (Mena et al., 1999). Such phenotypes might be the result of changes in the interactions of the NP with the polymerase complex, since there is no specificity in the NP–vRNA interaction (Baudin et al., 1994). In fact, interaction of NP with the PB1 and PB2 subunits has been reported (Biswas and Nayak, 1994). The 3D model presented here shows two distinct protein contacts between the polymerase complex and the adjacent NP monomers. One of them seems to be weaker than the other and might represent a NP–PB2 interaction, since it is more labile than the NP–PB1 interaction (Biswas and Nayak, 1994).
With regard to the polymerase complex, the resolution obtained to date is not high enough to derive many biological implications. However, it is worth mentioning that it has a very compact structure in spite of the fact that the inter-subunit interaction domains have been mapped to the N- and C-terminal regions of PB1, the C-terminal region of PA and the N-terminal region of PB2 (reviewed in Portela et al., 1999). Further work involving cryo-electron microscopy of a larger collection of RNP particles will be required to gain deeper insights into the structure of the viral polymerase.
METHODS
Biological materials. The vaccinia vTF7-3 virus (Fuerst et al., 1987) was a gift of B. Moss. Plasmids pGPB1, pGPB2, pGPA and pGNPpolyA have been described (Perales and Ortín, 1997). Plasmid pT7ΔNSRT clone 23, able to generate a 248 nt vRNA-like transcript, was described previously (Ortega et al., 2000).
Reconstitution and purification of recombinant RNPs. The generation of active recombinant RNPs was carried out essentially as described (Ortega et al., 2000). Cultures of COS-1 cells were infected with vTF7-3 virus and, after adsorption for 1 h at 37°C, were washed with DMEM and transfected with a mixture of plasmids containing 3 µg of pGPB1 and pGPB2, 0.6 µg of pGPA, and 12 µg of pGNPpolyA and pT7ΔNSRT clone 23. The DNA mixtures were diluted to 0.5 ml in DMEM. Cationic liposomes (1–2 µl/µg of DNA) were diluted to 0.5 ml in DMEM. Both dilutions were mixed, kept at room temperature for 15 min and added to the cultures containing 4 ml of DMEM. After 24 h of incubation at 37°C, the medium was replaced with 10 ml of DMEM containing 2% fetal bovine serum and incubated for a further 24 h. The cultures were collected 48 h after transfection and lysed for 2 h at 0°C in buffer A (10 mM Tris–HCl, 1 mM EDTA, 7.5 mM ammonium sulfate, 0.025% NP40, 1 mM DTT pH 7.9). After centrifugation at 10 000 g, the extract was centrifuged on a 20–35% glycerol gradient in TN buffer (150 mM NaCl, 50 mM Tris–HCl pH 7.8) for 17 h at 35 000 r.p.m. and 4°C in a SW41 rotor. The fractions were assayed for transcriptional activity as described (Ortega et al., 2000). Fractions containing active RNPs were centrifuged in a step glycerol gradient in TN buffer for 8 h at 55 000 r.p.m. and 4°C in a SW55 rotor. Active fractions were resedimented in a similar step gradient for 8 h at 55 000 r.p.m. and 4°C in a TL55 rotor. The active fractions were then used for further analyses.
Electron microscopy and image processing. Samples were applied to carbon-coated collodion grids, previously glow discharged at low air pressure, stained with 2% uranyl acetate and visualized in a JEOL 1200 EXII microscope at a nominal magnification of 4000×. Individual RNP images were recorded following a low-dose protocol using a GATAN slow scan CCD with a magnification factor of 19× (3.1 Å/pixel) and a defocus range of 3000–5000 Å. Images were 2D processed and aligned by cross-correlation free-pattern methods (Marco et al., 1996) using the XMIPP package (Marabini et al., 1998). Due to the heterogeneity of the particles, the images were subjected to Kohonen’s self-organizing feature maps (Marabini and Carazo, 1994). After this classification, homogeneous populations were obtained and averaged. For 3D reconstruction the sample stage was tilted 35°. As an initial model, a stacking of 20 single pixel planes identical to the average image was used. The model was refined using the angular refinement algorithms provided by SPIDER (Frank et al., 1996) and the reconstruction was performed using ART (Marabini et al., 1996). The final 3D model was obtained applying 9-fold symmetry to the entire NP ring but not to the outer area, due to the presence of the polymerase. The resolution of the model was estimated by Fourier ring correlation of two independent reconstructions and these values were used to low-pass filter the volume in the final model.
Acknowledgments
ACKNOWLEDGEMENTS
We are indebted to J.A. Melero, A. Nieto and A. Portela for their critical comments on the manuscript. The technical assistance of Y. Fernández and J. Fernández is gratefully acknowledged. We thank Carlos Oscar Sánchez for his help in 3D reconstruction. J.Ortega is a fellow of the Instituto de Estudios Turolenses. E.A. is a fellow of the Comunidad de Madrid. This work was supported by the Programa Sectorial de Promoción General del Conocimiento (grants PB97-1160 and PB96-0818).
REFERENCES
- Albo C., Valencia, A. and Portela, A. (1995) Identification of an RNA binding region within the N-terminal third of the influenza A virus NP polypeptide J. Virol., 69, 3799–3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bárcena J., Ochoa, M., de la Luna, S., Melero, J.A., Nieto, A., Ortín, J. and Portela, A. (1994) Monoclonal antibodies against influenza virus PB2 and NP polypeptides interfere with the initiation step of viral mRNA synthesis in vitro. J. Virol., 68, 6900–6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin F., Bach, C., Cusack, S. and Ruigrok, R.W. (1994) Structure of influenza virus RNP.I. Influenza virus nucleoprotein melts secondary structure in panhandle RNA and exposes the bases to the solvent. EMBO J., 13, 3158–3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S.K. and Nayak, D.P. (1994) Mutational analysis of the conserved motifs of influenza A virus polymerase basic protein 1. J. Virol., 68, 1819–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S.K., Boutz, P.L. and Nayak, D.P. (1998) Influenza virus nucleoprotein interacts with influenza virus polymerase proteins. J. Virol., 72, 5493–5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compans R.W., Content, J. and Duesberg, P.H. (1972) Structure of the ribonucleoprotein of influenza virus. J. Virol., 4, 795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digard P., Blok, V.C. and Inglis, S.C. (1989) Complex formation between influenza virus polymerase proteins expressed in Xenopus oocytes. Virology, 171, 162–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elton D., Medcalf, L., Bishop, K., Harrison, D. and Digard, P. (1999) Identification of amino acid residues of influenza virus nucleoprotein essential for RNA binding. J. Virol., 73, 7357–7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J., Radermacher, M., Penczek, P., Zhu, J., Li, Y., Ladjadj, M. and Leith, A. (1996) Spider and web: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol., 116, 190–199. [DOI] [PubMed] [Google Scholar]
- Fuerst T.R., Earl, P.L. and Moss, B. (1987) Use of a hybrid vaccinia virus-T7 RNA polymerase system for expression of target genes. Mol. Cell. Biol., 7, 2538–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heggeness M.H., Smith, P.R., Ulmanen, I., Krug, R.M. and Chopin, P.W. (1982) Studies on the helical nucleocapsid of influenza virus. Virology, 118, 466–470. [DOI] [PubMed] [Google Scholar]
- Klumpp K., Ruigrok, R.W. and Baudin, F. (1997) Roles of the influenza virus polymerase and nucleoprotein in forming a functional RNP structure. EMBO J., 16, 1248–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb R.A. and Krug, R.M. (1996) Orthomyxoviruses: the viruses and their replication. In Fields, B.N. (ed.), Virology. Lippincott-Raven Publishers, PA, pp. 1353–1395.
- Mahy B.W.J. (1983) Mutants of influenza virus. In Palese, P. and Kingsburg, D.W. (eds), Genetics of Influenza Viruses. Springer-Verlag, Vienna, Austria, pp. 192–253.
- Marabini R. and Carazo, J.M. (1994) Pattern recognition and classification of images of biological macromolecules using artificial neural networks. Biophys. J., 66, 1804–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marabini R., Masegosa, I.M., San Martín, C., Marco, S., Fernández, J.J., de la Fragra, L.G., Vaquerizo, C. and Carazo, J.M. (1996) X-mipp: an image processing package for electron microscopy. J. Struct. Biol., 116, 237–240. [DOI] [PubMed] [Google Scholar]
- Marabini R., Herman, G.T. and Carazo, J.M. (1998) 3D reconstruction in electron microscopy using ART with smooth spherically symetric volume elements (blobs). Ultramicroscopy, 72, 53–65. [DOI] [PubMed] [Google Scholar]
- Marco S., Chagoyen, M., de la Fraga, L.G., Carazo, J.M. and Carrascosa, J.L. (1996) A variant of the ‘random approximation’ of the reference-free alignment algorithm. Ultramicroscopy, 66, 5–10. [Google Scholar]
- Mena I., Jambrina, E., Albo, C., Perales, B., Ortin, J., Arrese, M., Vallejo, D. and Portela, A. (1999) Mutational analysis of influenza A virus nucleoprotein: identification of mutations that affect RNA replication. J. Virol., 73, 1186–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murti K.G., Webster, R.G. and Jones, I.M. (1988) Localization of RNA polymerases of influenza viral ribonucleoproteins by immunogold labeling. Virology, 164, 562–566. [DOI] [PubMed] [Google Scholar]
- Ortega J., Martín-Benito, J., Zürcher, T., Valpuesta, J.M., Carrascosa, J.L. and Ortín, J. (2000) Ultrastructural and functional analyses of recombinant influenza virus ribonucleoproteins suggest dimerization of nucleoprotein during virus amplification. J. Virol., 74, 156–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales B. and Ortín, J. (1997) The influenza A virus PB2 polymerase subunit is required for the replication of viral RNA. J. Virol., 71, 1381–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales B., Sanz-Ezquerro, J.J., Gastaminza, P., Ortega, J., Fernández-Santarén, J., Ortín, J. and Nieto, A. (2000) The replication activity of influenza virus polymerase is linked to the capacity of the PA subunit to induce proteolysis. J. Virol., 74, 1307–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon L.L.M., Pritlove, D.C., Fodor, E. and Brownlee, G.G. (1999) Direct evidence that the poly(A) tail of influenza A virus mRNA is synthesized by reiterative copying of a U track in the virion RNA template. J. Virol., 73, 3473–3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portela A., Zürcher, T., Nieto, A. and Ortín, J. (1999) Replication of orthomyxoviruses. Adv. Virus Res., 54, 319–348. [DOI] [PubMed] [Google Scholar]
- Pritlove D.C., Poon, L.L.M., Devenish, L.J., Leahy, M.B. and Brownlee, G.G. (1999) A hairpin loop at the 5′ end of influenza A virus virion RNA is required for synthesis of poly(A)+ mRNA in vitro. J. Virol., 73, 2109–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson J.S., Schubert, M. and Lazzarini, R.A. (1981) Polyadenylation sites for influenza mRNA. J. Virol., 38, 157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruigrok R.W. and Baudin, F. (1995) Structure of influenza virus ribonucleoprotein particles. II. Purified RNA-free influenza virus ribonucleoprotein forms structures that are indistinguishable from the intact influenza virus ribonucleoprotein particles. J. Gen. Virol., 76, 1009–1014. [DOI] [PubMed] [Google Scholar]
- Sanz-Ezquerro J.J., de la Luna, S., Ortín, J. and Nieto, A. (1995) Individual expression of influenza virus PA protein induces degradation of co-expressed proteins. J. Virol., 69, 2420–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz-Ezquerro J.J., Fernández Santarén, J., Sierra, T., Aragón, T., Ortega, J., Ortín, J., Smith, G.L. and Nieto, A. (1998) The PA influenza polymerase subunit is a phosphorylated protein. J. Gen. Virol., 79, 471–478. [DOI] [PubMed] [Google Scholar]
- Ulmanen I., Broni, B.A. and Krug, R.M. (1981) The role of two of the influenza virus core P proteins in recognizing cap 1 structures (m7GpppNm) on RNAs and in initiating viral RNA transcription. Proc. Natl Acad. Sci. USA, 78, 7355–7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka K., Ishihama, A. and Nagata, K. (1990) Reconstitution of influenza virus RNA-nucleoprotein complexes structurally resembling native viral ribonucleoprotein cores. J. Biol. Chem., 265, 11151–11155. [PubMed] [Google Scholar]