Abstract
Conditional gene inactivation using the Cre/loxP system is widely used, but the difficulty in properly regulating Cre expression remains one of the bottlenecks. One approach to regulate Cre activity utilizes a mutant estrogen hormone-binding domain (ERT) to keep Cre inactive unless the non-steroidal estrogen analog 4-hydroxytamoxifen (OHT) is present. Here we describe a mouse strain expressing Cre-ERT from the ubiquitously expressed ROSA26 (R26) locus. We demonstrate efficient temporal and spatial regulation of Cre recombination in vivo and in primary cells derived from these mice. We show the existence of marked differences in recombination frequencies between different substrates within the same cell. This has important consequences when concurrent switching of multiple alleles within the same cell is needed, and highlights one of the difficulties that may be encountered when using reporter mice as indicator strains.
INTRODUCTION
Methods that permit specific gene modifications in vivo in mice provide a powerful approach to assess gene function. However, germline modification often results in embryonic lethality or has pleiotropic effects interfering with the analysis of specific disease aspects. The use of the bacterial (Cre/loxP) and yeast-derived (FLP/Frt) site-specific DNA recombination systems permits the analysis of gene function in vivo in a cell-type restricted fashion (Gu et al., 1993; Dymecki, 1996; Shibata et al., 1997; Vooijs et al., 1998; Marino et al., 2000). These systems are based on the ability of Cre and FLP recombinases to catalyze the excision of DNA flanked by loxP or Frt recognition sequences (Sauer and Henderson, 1988; O’Gorman et al., 1991). LoxP or Frt sites can be inserted around one or more exons of the cognate gene via homologous recombination in embryonic stem (ES) cells. Conditional mutant mice can be crossed with transgenic mice expressing Cre- or FLP recombinase under the control of tissue-specific promoters. This strategy ensures normal development of the animal, fixes the window of gene inactivation to defined cell types and is now widely used in a broad range of biological settings (Rajewsky et al., 1996; Metzger and Feil, 1999).
The lack of control over the onset of Cre expression in classical transgenic mice has important consequences for the phenotypic outcome. If switching occurs early in a particular cell lineage, the phenotypic aberrations reflect the accumulation of defects from the earliest stage at which the gene is required, complicating the analysis of later developmental stages. Temporally controlled Cre-mediated recombination overcomes this and permits analysis of gene function at selected timepoints.
Spatial and temporal control of Cre-mediated recombination in vivo has been achieved through the use of adenoviruses expressing Cre (Rohlmann et al., 1996; Wang et al., 1996; Akagi et al., 1997; Shibata et al., 1997). However, the immune response elicited by these viruses compromises the persistence of infected cells and affects the general health status of mice (Akagi et al., 1997). Recently, temporal regulation of Cre recombinase in vivo has been accomplished using tetracycline-controlled gene expression (Utomo et al., 1999) and interferon-inducible expression (Kuhn et al., 1995).
An alternative approach utilizes engineered recombinases fused to the hormone-binding domain (HBD) of the mutated estrogen receptor (ERT). The fusion protein becomes active upon administration of the synthetic estrogen antagonist 4-hydroxytamoxifen (OHT), but not in the presence of the natural ligand 17β-estradiol (Feil et al., 1996). Transgenic mouse lines have been generated that express cre-ERT fusion genes controlled by tissue-specific promoters, which show ligand-dependent recombination (Brocard et al., 1997; Danielian et al., 1998; Schwenk et al., 1998; Vasioukhin et al., 1999). These approaches depend on properly regulated Cre expression from tissue-specific promoters in transgenic mice. However, the random integration of transgenes into the mouse genome often results in mosaic gene expression (Garrick et al., 1998; Henikoff, 1998) and offers little control over the level of gene-expression obtained. This requires the screening of a large number of independent transgenic lines to obtain one with appropriate expression characteristics.
To date, no mouse strains exist that permit highly efficient ubiquitous and inducible gene inactivation in vivo. To achieve this we used gene targeting to insert a single copy of a ligand responsive cre gene into a locus that is expressed ubiquitously throughout mouse development and in the adult.
RESULTS
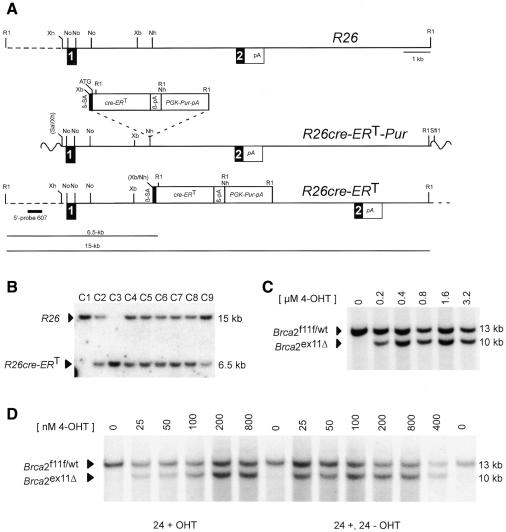
To express a ligand-dependent Cre recombinase we used a fusion gene encoding Cre and the mutated ligand binding domain (LBD) of the human estrogen receptor (G521R) (Schwenk et al., 1998). A targeting vector containing the cre-ERT fusion gene preceded by a β-globin splice acceptor site was inserted into the R26 locus (Figure 1A). This strategy is nearly identical to that reported by Soriano for the generation of the R26R mouse (Soriano, 1999). Mouse ES cells were electroporated and Southern analysis revealed that the majority of clones had undergone homologous recombination (Figure 1B). Clone C2 was used for the studies described below.
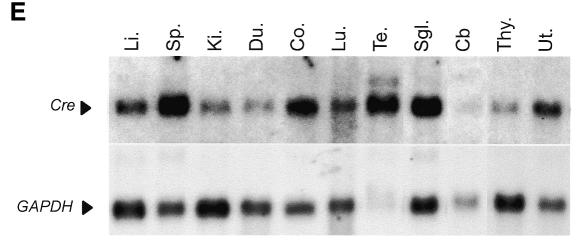
Fig. 1. Knock-in of cre-ERT fusion gene into the ROSA26 locus and ligand-inducibility in vitro. (A) Structure of the ROSA26 locus, the R26cre-ERT targeting vector and targeted locus. R1, EcoR; Xh, XhoI; No; NotI; Xb, XbaI; Nh, NheI. (B) Southern blot analysis with probe pHA607 (black box) on EcoRI digested DNA from puromycin resistant clones C1–C9. Wildtype (15 kb) and targeted (6.5 kb) R26 alleles are indicated. Clone C1 a non-targeted clone, C2 and C4–C9 heterozygous targeted clone, and C3 a homozygous targeted clone. (C) Ligand-inducible recombination of Brca2F11F in ES cells cultured for 60 h in the presence of OHT analyzed by Southern blot. Unrecombined Brca2F11F/wt (13 kb) and the recombined Brca2ex11 (10 kb) fragments are indicated. (D) Ligand-inducible recombination of Brca2F11F in primary keratinocytes analyzed as in (C), directly after removal of OHT (24+) or after culturing another 24 h without OHT (24+; 24–). (E) Northern blot analysis of poly(A)+ isolated RNA from R26cre-ERT mice. Upper panel shows hybridization to Cre probe and lower panel control hybridization to GAPDH. Tissues are Li (Liver), Sp (Spleen), Ki (Kidney), Du (Duodenum), Co (Colon), Lu (Lung), Te (Testis), Sgl (Salivary Gland), Cb (Cerebellum), Thy (Thymus) and Ut (Uterus).
We first determined whether introduction of cre-ERT into the R26 locus resulted in ligand-dependent recombination in ES cells. R26cre-ERT ES cells already harbored loxP sites surrounding exon 11 of breast cancer susceptibility gene-2 (Brca2)(e.g. Brca2F11F; J. Jonkers, R. Menwissen, H. van der Gulden, H. Peterse, M. van der Valk and A. Berns, in preparation). R26cre-ERT;Brca2F11F/wt ES cells, cultured in the absence of ligand showed no recombination. Addition of OHT to the medium for 60 h resulted in a dose-dependent recombination of Brca2F11F/wt reaching 100% at 400 nM (Figure 1C). Targeted ES cells were injected into blastocysts and heterozygous mice were obtained. R26cre-ERT;Brca2F11F/wt mice were healthy and fertile and showed no evidence for excision of Brca2F11F in the absence of ligand. This indicates that physiological levels of estrogens present in chimeric mice and subsequent offspring do not promote recombination of Brca2F11F.
Next, we tested if R26cre-ERT mice could be used as a source of primary cells in which conditional alleles could be efficiently switched in culture. Primary keratinocytes from newborn Brca2F11F/wt;R26cre-ERT mice were isolated and after 48 h of culture, OHT was added to the medium for 24 h followed by culturing in normal medium for an additional 24 h (24+, 24–). When cultured in the presence of 100 nM of OHT Brca2F11F could be switched with nearly 100% efficiency (Figure 1D). Analysis of recombination efficiencies directly after removal of OHT (24+) showed that recombination was incomplete with 100 nM suggesting residual Cre activity after removing ligand from the medium. As expected a further reduction in the concentration of OHT reduced the frequency of recombination on Brca2F11F.
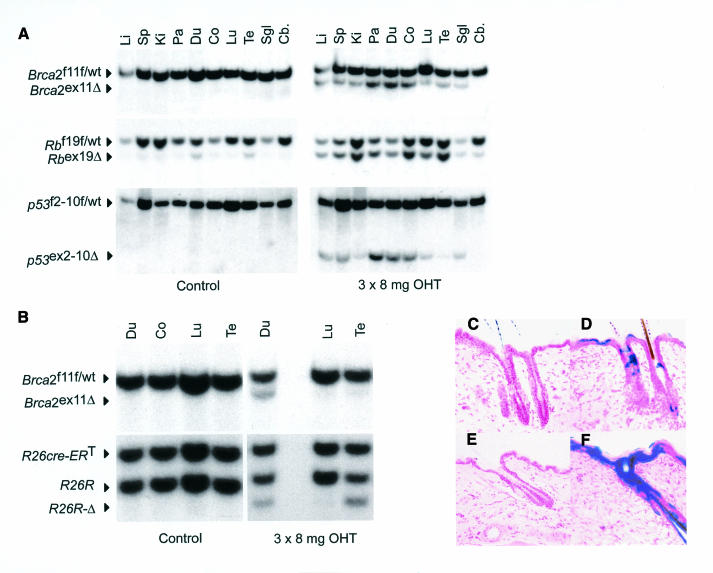
Conditional gene targeting permits the introduction of specific mutations in mice similar to those found in human diseases. Modeling of multifactorial disease in mice requires concurrent switching of floxed alleles in vivo. To date it is not known whether different conditional alleles within the same cell show a comparable switching efficiency. To investigate this we crossed R26cre-ERT mice with mice carrying conditional alleles for the retinoblastoma susceptibility gene (Rb) (RbF19F; Vooijs and Berns, 1999; Marino et al., 2000), Brca2F11F and p53 (Marino et al., 2000). RbF19F/wt;Brca2F11F/wt;p53F2-10F/wt;R26cre-ERT mice were injected for three days with 8 mg of OHT (i.p.) and tissue DNA was isolated 7 days later. Southern blot analysis showed that recombination of RbF19F approached 100% in most tissues, whereas moderate recombination frequencies were observed of Brca2F11F and p53F2-10F in the same samples (Figure 2A). A consistent low level of OHT-independent recombination in some tissues was seen on RbF19F but not on Brca2F11F or p53F2-10F.
Fig. 2. Differential recombination efficiencies in vivo between RbF19F, p53F2-10F Brca2F11F and R26R alleles. (A) Southern blot analysis of tissue DNA from OHT-treated RbF19F/wt;Brca2F11F/wt;p53F2-10F/wt;R26cre-ERT mice. Fragment sizes are 5.0 kb (RbF19F/wt), 4.5 kb (Rbex19Δ), 15 kb (p53F2-10F/wt) and 7 kb (p53ex2-10Δ). Tissues are as in Figure 1C. (B) Southern blot analysis of tissues from Brca2F11F;R26R;R26cre-ERT. Fragments are 4.8 kb (R26cre-ERT), 4.2 kb for the unrecombined and 3.8 kb for the recombined R26R allele. (C–F) Dose-dependent local Cre-mediated recombination in R26R;R26cre-ERT analyzed by X-Gal staining on frozen sections of treated (D, F) and non-treated (C, E) skin. Mice received 1× 0.5 mg OHT (C, D) or 3× 8 mg OHT (E, F) in DMSO. Counterstain is neutral red. Magnification 10×.
Since Cre reporter mice are widely used to monitor Cre activity (Akagi et al., 1997; Soriano, 1999), we considered it important to directly compare in vivo the behavior of a reporter allele with an independent conditional allele. R26R mice that express LacZ after Cre-mediated excision of a neo cassette (Soriano, 1999), were crossed with R26cre-ERT;Brca2F11F/wt mice to obtain R26R;Brca2F11F/wt;R26cre-ERT mice. These mice were readily obtained and we did not observe significant effects on their viability (Zambrowicz et al., 1997). Mice were injected i.p. with 3× 8 mg of OHT and tissue DNA was analyzed for recombination at both alleles. Whereas both loci behaved similarly in lung and duodenum, significant differences were seen in the testis where R26R recombined to a much greater extent than Brca2F11F (Figure 2B). To investigate whether these differences were related to tissue-specific differences in the expression of Cre-ERT we performed northern blot analysis on poly(A)+ RNA extracted from R26cre-ERT mice (Figure 1E). This analysis demonstrated that Cre-ERT is expressed in all tissues examined albeit at different levels. Most notably, expression in the cerebellum was found to be relatively low.
Having demonstrated that R26cre-ERT mice permit efficient deletion of floxed alleles in most tissues in vivo, we investigated whether local administration of OHT led to more restricted switching (Vasioukhin et al., 1999). We applied a single dose of 0.5 mg or three consecutive doses of 8 mg of OHT, respectively, to the shaved back of R26R;R26cre-ERT mice. Treated and untreated skin and various internal organs were processed for β-galactosidase activity 7 days after OHT application. At the lowest concentration (0.5 mg OHT) blue-stained cells were detected in cells of the follicular and interfollicular epidermis in the area where OHT was applied (Figure 2D). Increasing the topical dose to 3× 8 mg resulted in widespread staining throughout the treated area (Figure 2F). Under these conditions no appreciable staining was detected in non-treated adjacent skin nor in the underlying dermis (Figure 2C and E).
To investigate further the ligand-independent recombination seen in the absence of ligand at the cellular level we analyzed frozen-tissue sections from R26R;R26cre-ERT mice by staining for β-galactosidase activity. In most tissues analyzed (skeletal muscle, skin, liver, lungs, heart, kidney, spleen, stomach, eye, pituitary, testis, cerebellum and cerebrum) <0.1% of stained cells were identified. In contrast in duodenum and pancreas patches of stained cells were consistently found (1–5%, not shown).
DISCUSSION
The introduction of defined mutations in the mouse in a spatially and temporally controlled manner is considered an important next step in generating better mouse models of human diseases (Metzger and Feil, 1999; Porter, 1999; Vooijs and Berns, 1999). Our findings show that introduction of a ligand responsive cre gene (cre-ERT) under the transcriptional control of a broadly expressed gene fulfills this requirement.
Using R26cre-ERT mice in conjunction with Brca2, Rb, p53 and R26R conditional mutants, we show that Cre-mediated recombination in vivo can be efficiently regulated in a temporal, spatial and dose-dependent manner by OHT. The reduced activity observed in cerebellum may be explained by a lower or less uniform expression of R26-cre-ERT or less efficient activation of Cre-ERT due to a lower local concentration of OHT (Robinson et al., 1991). The ligand-independent Cre activity observed on the RbF19F and R26R alleles may be a consequence of inappropriate nuclear transport or proteolysis of the Cre-fusion protein, sufficient to catalyze a low level of recombination. We extended the use of R26cre-ERT mice by showing that restricted switching could be achieved with localized application of OHT on the skin. The short half-life of OHT in vivo (Robinson et al., 1991) should also permit localized switching at other tissue sites.
A comparison of recombination frequencies of different alleles in vivo showed marked differences. The distance between LoxP sites in the various targets tested may in part explain these differences. These distances are 0.45 kb for Rb, 2.5 kb for R26R, 6.6 kb for Brca2 and 7.9 kb for p53. However, the distance between loxP sites cannot solely explain the differences in recombination frequency as we also found a tissue-specific effect on recombination efficiency of the R26R allele compared to the Brca2F11F allele. Possibly these differences may be related to the expression level of the target gene or the local chromatin structure. R26 and Brca2 are both expressed in testes (Sharan and Bradley, 1997; Zambrowicz et al., 1997). However, whereas R26 is expressed ubiquitously, Brca2 is only expressed in proliferating cells (Blackshear et al., 1998). As these cells only constitute a fraction of the total number of cells used for the analysis, it is possible that recombination in Brca2-expressing cells is below the limit of our detection sensitivity. Further experiments are required to determine the relationship between target gene expression and Cre-mediated gene excision. Thus, not only are the expression characteristics an important variable in Cre-reporter strains, but also the susceptibility between different conditional alleles to be switched by Cre.
Finally, we have shown that in ES cells and primary keratinocytes complete control over gene switching can be exercised using R26cre-ERT. This system has significant advantages, since experimental and control cells only differ in their exposure to OHT. This difference (i) abrogates the effect of experimental variation between independent isolates, (ii) eliminates the effect of genetic differences between isolates from non-inbred strains, (iii) ensures rapid and controlled gene-excision without the need for selection, and (iv) permits gene-inactivation in both dividing and non-dividing cells.
Together, our results show that the introduction of cre-ERT in the R26 locus results in non-variegated OHT-inducible Cre activity, yielding a mouse strain that is of more general use than the currently available cre transgenic mice.
METHODS
ROSA26 targeting. A 13 kb genomic XhoI–EcoRI fragment from the ROSA26 locus was subcloned in the SalI–EcoRI sites of a modified pBR322 vector. A NheI site was used to introduce a 1.6 kb PGK-Puromycin selection cassette (pR26-MCS13-Pur). A 2 kb HindIII–KpnI fragment of pNPKCreER1c (Schwenk et al., 1998) containing the cre-ERT fusion gene was fitted with EcoRI sites and subcloned into the EcoRI site of a plasmid CAGGS containing the 3′ splice and polyadenylation signal from the rabbit β-globin gene (Niwa et al., 1991). A 2.2 kb XbaI–NheI fragment containing the 3′ splice site, the cre-ERT gene and the polyadenylation signal was inserted into the NheI site of pR26-MCS13-Pur to obtain the targeting vector R26cre-ERT-Pur. The resulting targeting vector was linearized with SfiI and purified by agarose gel electrophoresis.
Cell culture. All ES manipulations were performed as described previously (Clarke et al., 1992; Vooijs et al., 1998). E14 (129/Ola) ES cells were electroporated with R26cre-ERT-Pur and selected for 1 week on gelatin-coated dishes in BRL-conditioned medium containing 1.8 µg/ml of puromycin. Clones were expanded in triplicate and 1/3 was used for DNA analysis, the remainder was frozen in liquid N2. Homologous recombination at the R26 locus was confirmed by Southern blot analysis using a probe (pHA607) located outside of the targeting vector. One clone (C2) was karyotyped and injected into recipient blastocysts to obtain chimeras and heterozygous R26cre-ERT mice.
Primary mouse keratinocytes were isolated by trypsin flotation overnight at 4°C from newborn mice (Hennings et al., 1980; Missero et al., 1995). Cells were plated on mouse collagen type IV (1.5 µg/cm2, Becton Dickinson Labware, MA) coated plastic dishes in EMEM containing 4% chelexed FBS (Gibco-BRL), 10 ng/ml EGF (Upstate Biotechnology, NY) and 0.2 mM CaCl2. The next day cells were re-fed with the same medium containing only 0.045 mM CaCl2. Cells were cultured for another 24 h before adding OHT (Sigma H-6278, 30 mM stock solution in 100% EtOH) to the medium. DNA isolation from cultured cells was as described below for tissue DNA.
Genotyping. Mice were genotyped by PCR analysis on tail tip DNA (Laird et al., 1991). Genotyping of the Brca2F11F was performed using a primer in intron 11 int11F (5′-CTCATCATTTGTTGCCTCACTTC-3′) and int11R (5′-TGTTGGATACAAGGCATGTACAC-3′) yielding products of 529 and 450 bp for the floxed and wildtype alleles, respectively. Deletion of Brca2 was monitored using primers int10F (5′-GGCTGTCTTAGAACTTAGGCTG-3′) and int11R, yielding a 324 bp fragment. p53-int10-fwd (5′-AAGGGGTATGAGGGACAAGG-3′) and p53-int10-rev (5′-GAAGACAGAAAAGGGGAGGG-3′) primers were used to identify wildtype (391 bp) and p53 floxed alleles (461 bp). R26cre-ERT mice were screened with Cre1 (5′-GCACGTTCACCGGCATCAAC-3′) and Cre2 (5′-CGATGCAACGAGTGATGAGGTTC-3′). Genotyping of RbF19F mice (Vooijs et al., 1998) and of R26R Cre-reporter strain was performed by PCR as described (Soriano, 1999).
Ligand administration. For i.p. injection (300 µl) of mice, OHT was dispersed in sunflower seed oil by sonication. For topical administration OHT was dissolved in DMSO at 100 µg/µl and applied to the shaved back of mice.
Tissue DNA/RNA analysis. For Southern blot analysis of tissue DNA, tissues were isolated, minced and digested in lysis buffer (Laird et al., 1991). For the detection of Brca2 deletion, DNA was digested with SacI, separated by gel electrophoresis, transferred to Hybond N+ (Amersham) and hybridized with an exon-14 probe. p53 deletion was monitored using a BglII digest and the XbaI probe. Deletion of the Rb floxed and R26R alleles were monitored as described as described (Vooijs et al., 1998; Soriano, 1999). For northern blot analysis poly(A)+ RNA was isolated [Promega, poly(A)+-tract system] from total tissue RNA (Trizol, Gibco-BRL) separated by gel electrophoresis, transferred to Hybond N+ and hybridized to hGAPDH and Cre probes.
Probes. Southern analysis were performed for Brca2 with a 255 bp probe generated by PCR with ex14F (5′-GCTTCTGTCTAAAGGGCATC-3′) and ex14R (5′-TCTTCCCTGTCTCCATCTG-3′) primers and for p53 with a 700 nt XbaI fragment located ∼1 kb upstream of p53 exon-1. Deletion of the Rb floxed allele was detected with probe pHA153 (Clarke et al., 1992; Vooijs et al., 1998) and recombination at the R26 locus was analyzed using as a probe a PstI–SalI fragment subcloned in pGEM11 and amplified by PCR using T3 and T7 primers. Cre expression was analyzed using a Klenow-labeled probe comprising the complete Cre open reading frame.
β-galactosidase staining. Tissues were fixed in 2% paraformaldehyde/PBS, equilibrated overnight in 30% sucrose/PBS at 4°C, frozen in OCT compound (Miles Scientific) and stored at –80°C. Sections, 15 µM, were air-dried for 20 min, postfixed in 0.2% paraformaldehyde, washed in PBS and stained with X-gal reaction buffer according to standard procedures (Akagi et al., 1997).
Acknowledgments
ACKNOWLEDGEMENTS
We acknowledge Jan-Hermen Dannenberg and Hein te Riele (Division of Molecular Carcinogenesis, NKI, Amsterdam) for the ROSA26 genomic clone, Jurjen Bulthuis for technical assistance, Phil Soriano (Fred Hutchinson, Seattle, WA) for reagents and R26R mice, Francis Stewart (EMBL, Heidelberg) for pNPKCreER1c and Erik Danen (Division of Cell Biology, NKI, Amsterdam) for critically reading the manuscript. This work was supported by the organization for Medical Research (M.V.) and the Dutch Cancer Society (J.J.).
REFERENCES
- Akagi K., Sandig, V., Vooijs, M., van der Valk, M., Giovannini, M., Strauss, M. and Berns, A. (1997) Cre-mediated somatic site-specific recombination in mice. Nucleic Acids Res., 25, 1766–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackshear P.E. et al. (1998) Brca1 and Brca2 expression patterns in mitotic and meiotic cells of mice. Oncogene, 16, 61–68. [DOI] [PubMed] [Google Scholar]
- Brocard J., Warot, X., Wendling, O., Messaddeq, N., Vonesch, J.L., Chambon, P. and Metzger, D. (1997) Spatio-temporally controlled site-specific somatic mutagenesis in the mouse. Proc. Natl Acad. Sci. USA, 94, 14559–14563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke A.R., Robanus-Maandag, E., van Roon, M., van der Lugt, N.M., van der Valk, M., Hooper, M.L., Berns, A. and te Riele, H. (1992) Requirement for a functional Rb-1 gene in murine development. Nature, 359, 328–330. [DOI] [PubMed] [Google Scholar]
- Danielian P.S., Muccino, D., Rowitch, D.H., Michael, S.K. and McMahon, A.P. (1998) Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr. Biol., 8, 1323–1326. [DOI] [PubMed] [Google Scholar]
- Dymecki S.M. (1996) Flp recombinase promotes site-specific DNA recombination in embryonic stem cells and transgenic mice. Proc. Natl Acad. Sci. USA, 93, 6191–6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R., Brocard, J., Mascrez, B., LeMeur, M., Metzger, D. and Chambon, P. (1996) Ligand-activated site-specific recombination in mice. Proc. Natl Acad. Sci. USA, 93, 10887–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrick D., Fiering, S., Martin, D.I. and Whitelaw, E. (1998) Repeat-induced gene silencing in mammals. Nature Genet., 18, 56–59. [DOI] [PubMed] [Google Scholar]
- Gu H., Zou, Y.R. and Rajewsky, K. (1993) Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre-loxP-mediated gene targeting. Cell, 73, 1155–1164. [DOI] [PubMed] [Google Scholar]
- Henikoff S. (1998) Conspiracy of silence among repeated transgenes. BioEssays, 20, 532–535. [DOI] [PubMed] [Google Scholar]
- Hennings H., Michael, D., Cheng, C., Steinert, P., Holbrook, K. and Yuspa, S.H. (1980) Calcium regulation of growth and differentiation of mouse epidermal cells in culture. Cell, 19, 245–254. [DOI] [PubMed] [Google Scholar]
- Kuhn R., Schwenk, F., Aguet, M. and Rajewsky, K. (1995) Inducible gene targeting in mice. Science, 269, 1427–1429. [DOI] [PubMed] [Google Scholar]
- Laird P.W., Zijderveld, A., Linders, K., Rudnicki, M.A., Jaenisch, R. and Berns, A. (1991) Simplified mammalian DNA isolation procedure. Nucleic Acids Res., 19, 4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino S., Vooijs, M., van Der Gulden, H., Jonkers, J. and Berns, A. (2000) Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev., 14, 994–1004. [PMC free article] [PubMed] [Google Scholar]
- Metzger D. and Feil, R. (1999) Engineering the mouse genome by site-specific recombination. Curr. Opin. Biotechnol., 10, 470–476. [DOI] [PubMed] [Google Scholar]
- Missero C., Calautti, E., Eckner, R., Chin, J., Tsai, L.H., Livingston, D.M. and Dotto, G.P. (1995) Involvement of the cell-cycle inhibitor Cip1/WAF1 and the E1A-associated p300 protein in terminal differentiation. Proc. Natl Acad. Sci. USA, 92, 5451–5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H., Yamamura, K. and Miyazaki, J. (1991) Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene, 108, 193–199. [DOI] [PubMed] [Google Scholar]
- O’Gorman S., Fox, D.T. and Wahl, G.M. (1991) Recombinase-mediated gene activation and site-specific integration in mammalian cells. Science, 251, 1351–1355. [DOI] [PubMed] [Google Scholar]
- Porter A. (1999) Controlling your losses: conditional gene silencing in mammals. Trends Genet., 14, 73–79. [DOI] [PubMed] [Google Scholar]
- Rajewsky K., Gu, H., Kuhn, R., Betz, U.A., Muller, W., Roes, J. and Schwenk, F. (1996) Conditional gene targeting. J. Clin. Invest., 98, 600–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson S.P., Langan-Fahey, S.M., Johnson, D.A. and Jordan, V.C. (1991) Metabolites, pharmacodynamics, and pharmacokinetics of tamoxifen in rats and mice compared to the breast cancer patient. Drug Metab. Dispos., 19, 36–43. [PubMed] [Google Scholar]
- Rohlmann A., Gotthardt, M., Willnow, T.E., Hammer, R.E. and Herz, J. (1996) Sustained somatic gene inactivation by viral transfer of Cre recombinase. Nature Biotechnol., 14, 1562–1565. [DOI] [PubMed] [Google Scholar]
- Sauer B. and Henderson, N. (1988) Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc. Natl Acad. Sci. USA, 85, 5166–5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk F., Kuhn, R., Angrand, P.O., Rajewsky, K. and Stewart, A.F. (1998) Temporally and spatially regulated somatic mutagenesis in mice. Nucleic Acids Res., 26, 1427–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharan S.K. and Bradley, A. (1997) Murine Brca2: sequence, map position, and expression pattern. Genomics, 40, 234–241. [DOI] [PubMed] [Google Scholar]
- Shibata H. et al. (1997) Rapid colorectal adenoma formation initiated by conditional targeting of the Apc gene. Science, 278, 120–123. [DOI] [PubMed] [Google Scholar]
- Soriano P. (1999) Generalized lacZ expression with the ROSA26 Cre reporter strain. Nature Genet., 21, 70–71. [DOI] [PubMed] [Google Scholar]
- Utomo A.R., Nikitin, A.Y. and Lee, W.H. (1999) Temporal, spatial, and cell type-specific control of Cre-mediated DNA recombination in transgenic mice. Nature Biotechnol., 17, 1091–1096. [DOI] [PubMed] [Google Scholar]
- Vasioukhin V., Degenstein, L., Wise, B. and Fuchs, E. (1999) The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc. Natl Acad. Sci. USA, 96, 8551–8556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vooijs M. and Berns, A. (1999) Developmental defects and tumor predisposition in Rb mutant mice. Oncogene, 18, 5293–5303. [DOI] [PubMed] [Google Scholar]
- Vooijs M., van der Valk, M., te Riele, H. and Berns, A. (1998) Flp-mediated tissue-specific inactivation of the retinoblastoma tumor suppressor gene in the mouse. Oncogene, 17, 1–12. [DOI] [PubMed] [Google Scholar]
- Wang Y., Krushel, L.A. and Edelman, G.M. (1996) Targeted DNA recombination in vivo using an adenovirus carrying the cre recombinase gene. Proc. Natl Acad. Sci. USA, 93, 3932–3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrowicz B.P., Imamoto, A., Fiering, S., Herzenberg, L.A., Kerr, W.G. and Soriano, P. (1997) Disruption of overlapping transcripts in the ROSA beta geo 26 gene trap strain leads to widespread expression of β-galactosidase in mouse embryos and hematopoietic cells. Proc. Natl Acad. Sci. USA, 94, 3789–3794. [DOI] [PMC free article] [PubMed] [Google Scholar]