Abstract
To detect specific partners of the small Golgi-localized GTPase rab1b we generated rab1b mutants and used them as bait proteins in yeast two-hybrid screens. We isolated several specifically interacting clones. Two of them encode large protein fragments highly homologous to rat GM130 and to human Golgin95. The full-length human GM130 cDNA was cloned and its interaction with rab1b was characterized in detail by yeast two-hybrid and in vitro binding assays. Here we report for the first time that the rab1b protein interacts specificially with GM130 in a GTP-dependent manner and therefore needs the hypervariable regions of the N- and C-termini. We mapped the rab1b binding site of GM130 and provide evidence that it is different to the previously described p115 and Grasp65 binding sites of the GM130 protein.
INTRODUCTION
Rab proteins are involved in regulating membrane traffic between various intracellular compartments. They can be detected in an inactive or an active conformation depending on the nucleotide-bound status, and are considered as switches, which cycle between an active, membrane-associated and an inactive cytosolic status (Olkkonen and Stenmark, 1997). Several approaches have been taken to trace out rab effector proteins acting downstream of or alongside the rab proteins. Using a biochemical approach, Zerial and co-workers recently identified >20 proteins interacting with the activated conformation of the rab5 protein (Christoforidis et al., 1999); therefore, it has been suggested that rab GTPases may be involved in the tethering of membranes by recruiting effectors to vesicle and target membranes. This step is thought to represent the initial step of the vesicle docking event (Waters and Pfeffer, 1999).
Mammalian rab1a and rab1b GTPases are localized at the endoplasmic reticulum (ER)–Golgi compartments (Tisdale et al., 1992; Saraste et al., 1995) and both rab1 isoforms are involved in regulating the anterograde traffic between ER and Golgi membranes (Nuoffer et al., 1994; Pind et al., 1994). It has been shown that the rab1 homologous yeast GTPase Ypt1p interacts genetically with Uso1p (Sapperstein et al., 1996; Cao et al., 1998). The mammalian homolog of Uso1p, p115, was identified as a protein required for intra-Golgi transport and as a component of transcytotic vesicles (Waters et al., 1992; Barroso et al., 1995). It is involved in the regrowth of Golgi cisternae from mitotic vesicles and stacking of cisternae during the post-mitotic formation of Golgi stacks in vitro (Rabouille et al., 1995, 1998; Shorter and Warren, 1999) and binds to the cis-Golgi matrix protein GM130 and Giantin (Nakamura et al., 1997; Sönnichsen et al., 1998; Dirac-Svejstrup et al., 2000). Recently, it was shown that p115 interacts in a GTP-dependent manner with rab1, suggesting that p115 is an effector protein of the rab1 GTPase (Allan et al., 2000). Here we describe a second effector protein of the rab1 GTPase. Using the yeast two-hybrid approach we surprisingly identified GM130 as a protein that interacts directly and in a GTP-dependent manner with rab1b. Using a set of GM130 deletion mutants we also demonstrate that the rab1b binding is independent of the N- and C-termini of GM130.
RESULTS
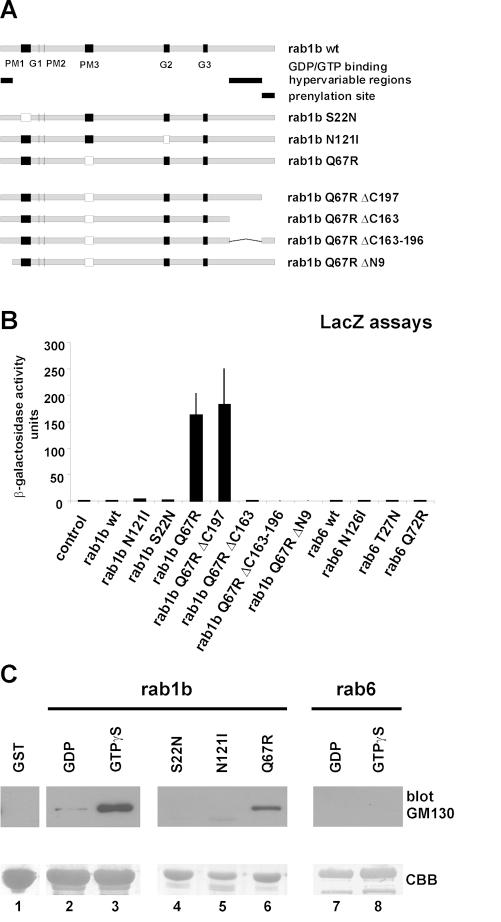
We generated mutants that present the inactive (GDP-bound) and active (GTP-bound) form of rab1b and used the GTPase-deficient rab1bQ67R mutant in yeast two-hybrid screening experiments to identify interacting proteins. The reporter yeast strain Y190 was transformed with a bait plasmid encoding the rab1bQ67R mutant fused to gal4BD and transformed with a plasmid cDNA library from human placenta. We screened up to 1 × 107 clones and isolated clones K9 and K18 (with the same 2.7 bp insert), which caused a strong activation of reporter genes in the presence of gal4BD–rab1bQ67R fusion proteins. No activation of the reporter genes could be observed when rab1b wt, the inactive rab1bS22N and rab1bN121I or rab6 wt and rab6 mutants (T27N, Q72R and N126I) were used (Figure 1). In addition, co-transformations with various members of the rab family, like rab2, rab3A, rab7ΔCC, rab9ΔCC or Ha-ras wild type and rhoQ61L, were performed to verify the specificity of the rab1b–K9/K18 interaction. Again, only the rab1bQ67R mutant displayed a specific interaction (not shown). To test whether the N- and C-terminal hypervariable regions and/or the prenylation site are necessary for the interaction with K9/K18, we constructed four rab1bQ67R deletion mutants (Figure 1A). The first double mutant, lacking the prenylation site (rab1bQ67R ΔC197), displayed the strongest activation of the reporter genes in the yeast co-transformation assay (Figure 1B). The two mutants lacking the C-terminal hypervariable regions, but including the prenylation site (rab1bQ67R ΔC163–197), or lacking the whole C-terminus (rab1bQ67R ΔC163) were unable to activate the reporter genes. The rab1bQ67R ΔN9 mutant also failed to activate the reporter genes, suggesting that the interaction with clone K9/K18 depends on the N-terminal hypervariable region of the rab1b protein. Sequence analysis of the K9/K18 cDNA insert showed that both inserts encode a truncated protein fragment of the human GM130 protein, lacking the first 224 amino acids (aa). The previously described Golgin95 amino acid sequence consists of the final 620 aa of the human GM130, suggesting that this protein is a deletion mutant of GM130, lacking the first 370 aa (Fritzler et al., 1993). We generated the full-length GM130 cDNA by a PCR-based strategy using a cDNA preparation from human placenta as a template for the 5′ amplification of the GM130 cDNA. The sequence data of the human GM130 cDNA have been submitted to DDBJ/EMBL/GenBank under accession No. AF248953. Human GM130 is a 990 aa protein with a predicted molecular weight of 111.59 kDa and a pI of 4.97. Further analysis of the amino acid sequence using the Lupas algorithm (Lupas, 1996) with a window of 28 and a minimum probability of 50% predicted that the human GM130 protein consists of six coiled-coiled (cc) regions: (cc1: aa 141–211; cc2: aa 214–437; cc3: aa 447–683; cc4: aa 716–766; cc5: aa 783–833; cc6: aa 836–886), suggesting that human GM130 displays a similar cc structure to the GM130 protein from rat (Nakamura et al., 1997). To confirm the data we obtained in the yeast two-hybrid assays, we performed an in vitro binding assay and expressed glutathione S-transferase (GST) rab1b wt fusion proteins and pre-loaded them with GDP or GTPγS, respectively. In addition, GST fusion proteins of rab1bS22N, rab1bN121I and the rab1bQ67R mutant were expressed. For the binding assays, cytosolic extracts of N-terminal hemagglutinin (HA)-tagged human GM130-overexpressing BHK cells were incubated with GST rab1b or rab6 fusion proteins. After a washing procedure, the bound proteins were separated by SDS–PAGE and analyzed by western blotting using antibodies against the HA tag. As shown in Figure 1C, only GST rab1b proteins, pre-loaded with GTPγS, and the GTPase-deficient rab1bQ67R mutant were able to recover significantly HA-tagged GM130 from the cytosol of transfected BHK cells (Figure 1C, lanes 3 and 6). No or only weak background signals were detectable in the controls (lanes 1, 2, 4, 5, 7 and 8).
Fig. 1. Specific interaction of rab1b with GM130. (A) We performed co-transformations with various rab1b mutants and a set of different rab control bait plasmids. (B) The β-galactosidase activities of the liquid lacZ assays are presented as mean ± SEM of three to six independent transformants. For the control co-transformations, the empty bait vector (pAS2-1) was used. (C) Lysates of BHK cells expressing HA-tagged GM130 were first pre-cleared with GST beads (lane 1) and then incubated with GST rab1b wt fusion proteins, pre-loaded either with GDP (lane 2) or GTPγS (lane 3) or with beads, coupled to GST rab1bS22N, GST rab1bN121I and GST rab1Q67R fusion proteins (lanes 4–6). As a control, pre-loaded GST rab6 wt beads were used (lanes 7 and 8). HA-GM130 was detected by western blotting (top panel). The amount of GST fusion proteins in the independent pull-down assays was monitored by Coomassie Blue (CBB) staining of the blot membranes (bottom panel). PM1–3, G1–3, conserved nucleotide binding motifs.
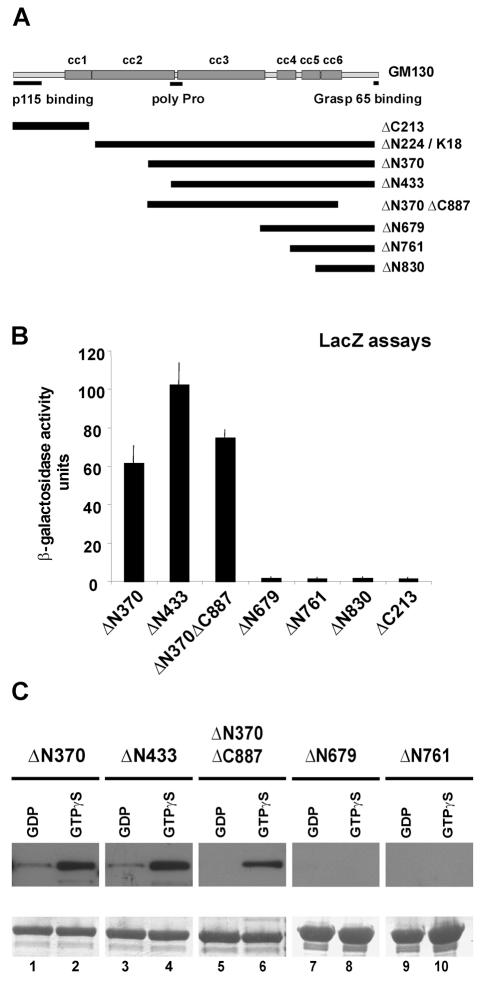
We mapped the rab1b binding site by using a set of GM130 deletion mutants (Figure 2A). The truncation mutant ΔN370 lacks the first 370 aa and encodes the previously decribed Golgin95. ΔN433 comprises the C-terminal part of GM130, containing the last four cc domains and the proline-rich site. The double truncated ΔN370Δ887 mutant contains the Golgin95 sequence lacking the last 104 aa of the C-terminus. The ΔN679, ΔN761 and ΔN830 truncation mutants encode the last three, two or the last cc domains, respectively. The truncation mutants were expressed as gal4AD fusion proteins in the yeast two-hybrid assays (Figure 2B) or as HA-tagged proteins in in vitro binding studies (Figure 2C). ΔC213 and ΔN224 (K9/18 protein fragment) were only tested in the yeast two-hybrid experiments. Every prey plasmid was co-transformed together with the rab1bQ67R bait construct (Figure 2B). Only deletion mutants ΔN224, ΔN370 and ΔN433 were able to activate the reporter genes, indicating that the p115 binding site of GM130 (aa 1–73) is not required for rab1b binding to GM130. This was confirmed by using mutant ΔC213, which has a p115 binding site but was unable to activate the reporter genes. Mutants ΔN679, ΔN761 and ΔN830 failed to interact with rab1bQ67R, whereas the double deletion mutant ΔN370ΔC887 led to a strong reporter gene activation (Figure 2B). To confirm these results, we performed in vitro binding assays (see Figure 1C) using the same series of deletion mutants. Again, only GST rab1b wt beads pre-loaded with GTPγS recovered the ΔN370, ΔN433 and ΔN370ΔC887 deletion mutants (Figure 2C, lanes 2, 4 and 6), whereas GST rab1b pre-loaded with GDP failed to bind (Figure 2C, lanes 1, 3 and 5). The ΔN679 and ΔN761 mutants were unable to bind to activated rab1b (Figure 2C, lanes 7–10). We therefore conclude that rab1b binding is independent of the N- and C-termini of GM130.
Fig. 2. Mapping of the rab1b binding site of GM130. (A) We generated GM130 truncation mutants. The numbers given correspond to the position of the first (ΔN series) or the last (ΔC mutants) amino acid residue in full-length human GM130. The mutants were expressed as gal4AD fusion proteins for yeast two-hybrid assays or as N-terminal HA-tagged proteins in BHK cells. (B) Liquid β-galactosidase assays with o-nitrophenyl-β-d-galactoside as a substrate were performed. The β-galactosidase activities are presented as mean ± SEM analyzing three independent transformants. (C) To confirm the yeast data, we performed in vitro binding assays. BHK lysates expressing HA-tagged GM130 mutants were pre-cleared with GST beads and then incubated with GST rab1b wt pre-loaded with GDP (lanes 1, 3, 5, 7 and 9) and GTPγS (lanes 2, 4, 6, 8 and 10), respectively. The recovery of HA-tagged GM130 mutants was determined by SDS–PAGE and western blotting (top panel). Equal amounts of each GST fusion protein in the pull-down assays were monitored by Coomassie Blue staining of the blot membranes (bottom panel).
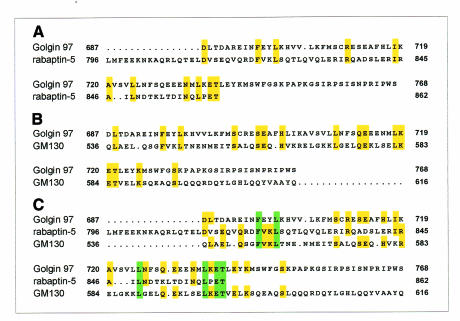
Unexpectedly, our results indicate that the rab1b binding site is localized at cc3. GM130/Golgin95 belongs to the family of Golgins (Fritzler et al., 1993; Nakamura et al., 1995). Recently, Barr (1999) identified Golgin97 and Golgin245 as effector proteins of activated rab6 and described a rab6 binding domain (R6BD) in the C-terminus of Golgin97/245. It was shown (Figure 3A) that this R6BD shares some similarities with the previously characterized rab5 binding domain (R5BD) of rabaptin-5 (Vitale et al., 1998; Barr, 1999). We performed an alignment of our GM130 minimal mutant (aa 370–887) and the R6BD of Golgin97 and found that aa 536–615 of GM130 display similarities to the R6BD of Golgin97 (Figure 3B). An additional alignment between the R5BD of rabaptin-5 (aa 796–862), the R6BD of Golgin97 (aa 687–768) and aa 536–615 of human GM130 revealed that the putative rab1b binding site also shows some similarities to the R5BD (Figure 3C). The results of the sequence alignments are consistent with our finding that rab1b specifically binds to the cc3 region of GM130 and lead to the suggestion that this aa pattern probably participates in the rab1b–GM130 interaction. However, although the ΔN679 deletion mutant of GM130 is unable to bind to rab1b, we could not exclude the possibility that the region between cc3 and cc4 as well as cc domains cc4–cc6 do cooperate with cc3 in rab1b binding.
Fig. 3. Sequence alignment using the PRETTY program (GCG package) between rab binding regions of different rab effector proteins. (A) Alignment between the rab5 binding domain (R5BD aa 796–862) of rabaptin-5 and the rab6 binding domain (R6BD aa 687–768) of Golgin97 (Barr, 1999). (B) Part of the alignment performed with the R6BD of Golgin97 and aa 370–887 from human GM130. (C) Alignment between the R5BD, the R6BD and aa 536–616 of human GM130. Yellow, identical in two sequences; green, identical in three sequences.
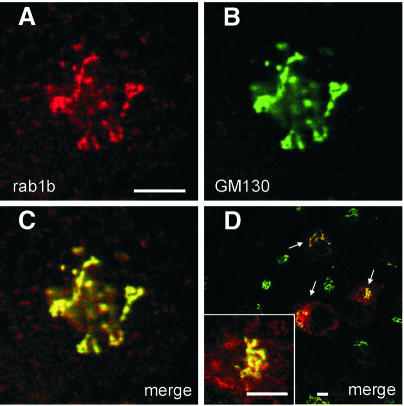
We performed immunolocalization studies in HeLa cells using polyclonal antibodies against GM130 and a monoclonal antibody (mAb) (1E7) raised against the hypervariable region of rab1b. Endogenous GM130 displays a high degree of co-localization with rab1b (Figure 4C) and also with overexpressed myc-tagged rab1b (Figure 4D) at the Golgi membranes. Endogenous as well as overexpressed myc-tagged rab1b show an additional staining in non-identified structures (Figure 4A and D). In conclusion, the immunofluorescence data are consistent with previous localization studies carried out on GM130 or rab1b, suggesting that the interaction between both proteins occurs at the cis-Golgi membranes (Nakamura et al., 1995; Saraste et al., 1995; Nelson et al., 1998).
Fig. 4. Localization of GM130 and rab1b by confocal immunofluorescence microscopy. HeLa cells were fixed, permeabilized and double labeled with the mAb 1E7 directed against rab1b (A) and a polyclonal antibody against GM130 (B). The merged image of (A) and (B) is given in (C) and shows the co-localization of overexpressed myc-rab1b (detected with mAB 9E10) and endogenous GM130 (D). Secondary antibodies were coupled to Alexa 594 (red) or Alexa 488 (green). Bar, 5 µm.
DISCUSSION
Here we report that human GM130 binds specifically to the activated form of the rab1b GTPase. Screening a human placenta cDNA library using the rab1bQ67R mutant as the bait protein resulted in the isolation of two specifically interacting clones (K9/K18). The interaction between rab1b and the ΔN224 GM130 mutant (clone K9/K18) depends on the hypervariable regions of the N- and C-termini of rab1b. This is consistent with previous results, which demonstrated that the hypervariable regions of rab5 are essential for proper localization, membrane association and regulatory function (Stenmark et al., 1994). We confirmed our data by in vitro binding assays and demonstrate that only the GTPase-deficient mutant rab1bQ67R and the rab1b wild type, pre-loaded with GTPγS, were able to bind to GM130. Surprisingly, the deletion mutants of GM130 used in this study (Figure 2A) indicate that rab1b binds to the cc3 domain, suggesting that rab binding sites within effector molecules are not restricted to non-α-helical regions of effector proteins, as found for example in rabaptin-5, EEA-1 or Golgin97 (Simonsen et al., 1998; Vitale et al., 1998; Barr, 1999). An alignment between aa 796–862 of rabaptin-5, aa 687–768 of Golgin97 and aa 536–615 of GM130 supports this point of view, because we found some similarities between rab5, rab6 and a putative rab1b binding site within the cc3 domain.
The differences in the sequence motifs (see Figure 3C) are likely to reflect the diversity of individual rab binding domains that are necessary to distinguish between rab-specific effector proteins. However, we can not exclude the possibility that additional regions do cooperate in rab1b binding to GM130.
As shown so far, GM130 interacts directly with at least three proteins: Grasp65 (a protein involved in Golgi cisternae stacking), p115 and rab1b (Nakamura et al., 1997; Barr et al., 1998). However, the role of GM130 in ER–Golgi traffic is still unclear. Nevertheless, Seemann et al. (2000) could demonstrate that a GM130 mutant lacking the p115 binding site inhibited transport of the VSV-G protein. Allan et al. (2000) showed that rab1 is able to program budding COPII vesicles for fusion via recruitment of p115 into a cis-SNARE complex. Thus, both proteins, p115 and GM130, are identified as specific effectors of the activated rab1 GTPase. This implies that p115 in complex with GM130 forms a tether/velcro factor that is recruited to membranes in a rab1-dependent manner. Interestingly, this tether is mitotically regulated by p34cdc2 kinase and it is worth noting that rab1 is also a substrate for this kinase (Bailly et al., 1991; Lowe et al., 2000). Furthermore, this GM130–p115 complex would have two rab binding sites, similar to rabaptin-5 and EEA-1 (Simonsen et al., 1998; Vitale et al., 1998), suggesting that it acts as a cross-linker between p115-containing membranes of the vesicular tubular clusters (VTCs) and GM130-containing cis-Golgi membranes (Nakamura et al., 1995; Nelson et al., 1998; Alvarez et al., 1999). Therefore, GM130 probably has a function in late steps of ER–Golgi traffic due to its interaction with p115 and rab1b (Nakamura et al., 1995; Nelson et al., 1998; Seemann et al., 2000). In this model, only a p115–GM130 interaction is needed. On the other hand, it is postulated by Warren and co-workers (Sönnichsen et al., 1998; Dirac-Svejstrup et al., 2000) that p115 also mediates tethering between COPI vesicles and Golgi membranes through its simultaneous interaction with GM130 and Giantin. An additional possibility for rab1 function is that the formation of this complex could be supported and/or regulated by an activated rab1 GTPase. The finding that GM130 is an effector protein of rab1 strongly supports the idea that GM130 has a function in ER–Golgi traffic. Future work is needed to elucidate the exact role of GM130 and the rab1–GM130 interaction in membrane traffic.
METHODS
Plasmids. Rab1b and rab6 point mutants have been described earlier (Schiedel et al., 1995; Weide et al., 1999). Details of plasmids and primers used are available from A.B.
Yeast two-hybrid methods. The yeast reporter strain Y190 was transformed with the bait plasmid pAS rab1bQ67R. The bait yeast strain was transformed with a human placenta cDNA library in pACT2 (Clontech). For yeast co-transformation assays, Y190 yeast cells were simultaneously transformed with the corresponding bait and prey constructs and analyzed by quantitative β-galactosidase assays.
Cell culture and transient transfection. BHK and HeLa cells were grown on tissue culture plates in Dulbecco’s modified Eagles medium (10% fetal calf serum, 100 U/ml penicillin–streptomycin). Transient transfections were performed as described earlier (Weide et al., 1999).
Antibodies. The mAb 1E7 against rab1b has been described earlier (Weide et al., 1999). The mAbs 12CA5 (Roche, Mannheim, Germany) and 9E10 recognize HA and myc epitopes, respectively. The polyclonal antibody against GM130 was a generous gift from Dr E. Sztul. Peroxidase-coupled secondary antibodies were from Amersham-Pharmacia Life Science. Texas red-conjugated secondary antibodies were from Dianova (Hamburg, Germany). Secondary antibodies coupled with Alexa 488 and Alexa 594 were from Molecular Probes (Leiden, The Netherlands).
Recombinant proteins. BL21 Escherichia coli cells were transformed with pGEX constructs containing myc-tagged cDNA inserts of rab1b wt/mutants and rab6 wt. Escherichia coli cells were sedimented, resuspended in lysis buffer [phosphate-buffered saline (PBS) with 5 mM MgCl2, 5 mM β-mercaptoethanol, 200 µM GDP, 5 µg/ml DNase I, 5 µg/ml RNase A and complete™ (Roche, Basel, Switzerland)] or in PBS containing 1% Triton X-100 and complete™ and sonicated on ice. Lysates were centrifuged at 100 000 g. The supernatant was incubated with glutathione–Sepharose 4B beads.
In vitro binding assays. BHK cells were transiently transfected with pSVHA- expression plasmids and resuspended in lysis buffer [20 mM HEPES pH 7.5, 100 mM NaCl, 5 mM MgCl2, 1 mM dithiothreitol (DTT), complete™]. Cell lysates were homogenized and centrifuged at 4500 g for 1 h. Supernatants were centrifuged at 100 000 g for 1 h at 4°C, dialyzed against lysis buffer without protease inhibitors for 2 h and centrifuged at 100 000 g. The supernatant was pre-cleared with GST beads for 1 h at 4°C and incubated with GST rab1b wt and GST rab6 wt beads, pre-loaded with GDP or GTPγS, or with GST rab1bS22N, GST rab1bN121I and GST rab1bQ67R fusion proteins, respectively. Pre-loaded GST rab1b and GST rab6 beads were treated as described by Christoforidis and Zerial (2000) with three additional washing steps using 2× WP1 buffer (20 mM HEPES pH 7.5, 250 mM NaCl, 1 mM DTT), 3× WP2 buffer (20 mM HEPES pH 7.5, 250 mM NaCl, 1 mM DTT, 1.0% Triton X-100, 0.1% SDS) and 1× WP3 (20 mM HEPES pH 7.5, 100 mM NaCl, 1 mM DTT). GST rab1b mutants were washed with WP1 (2×), WP2 (3×) and WP3 (1×). After washing, proteins were eluted by adding sample buffer, subjected to SDS–PAGE and analyzed by western blotting using the 12CA5 antibody against the HA tag.
Immunofluorescence analysis. Transiently transfected HeLa cells were grown on coverslips (60–80% confluency), prepared for immunofluorescence as described earlier (Weide et al., 1999) and then analyzed by confocal laser scanning microscopy.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr I.G. Macara (University of Virginia, Charlottesville) and Dr S.R. Pfeffer (Stanford University, CA) for control bait plasmids and we are grateful to Dr E. Sztul (University of Birmingham, AL) for the anti-GM130 antibody. We thank Dr J. Kremerskothen for critically reading the manuscript. This work was supported by the DFG and an FCI grant to A.B. and is part of the PhD theses of M.B. and M.K.
REFERENCES
- Allan B.B., Moyer, B.D. and Balch, W.E. (2000) Rab1 recruitment of p115 into a cis-SNARE complex: programming budding COPII vesicles for fusion. Science, 289, 444–448. [DOI] [PubMed] [Google Scholar]
- Alvarez C., Hideaki, F., Hubbard, A. and Sztul, E. (1999) ER to Golgi transport: requirements at a pre-Golgi VTC stage. J. Cell Biol., 147, 1205–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly E., McCaffrey, M., Touchot, N., Zahraoui, A., Goud, B. and Bornens, M. (1991) Phosphorylation of two small GTP-binding proteins of the Rab family by p34cdc2. Nature, 350, 715–718. [DOI] [PubMed] [Google Scholar]
- Barr F.A. (1999) A novel Rab6-interacting domain defines a family of Golgi-targeted coiled-coil proteins. Curr. Biol., 9, 381–384. [DOI] [PubMed] [Google Scholar]
- Barr F.A., Nakamura, N. and Warren, G. (1998) Mapping the interaction between GRASP65 and GM130, components of a protein complex involved in the stacking of Golgi cisternae. EMBO J., 17, 3258–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroso M., Nelson, D.S. and Sztul, E. (1995) Transcytosis-associated protein (TAP)/p115 is a general fusion factor required for binding of vesicles to acceptor membranes. Proc. Natl Acad. Sci. USA, 92, 527–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X., Ballew, N. and Barlowe, C. (1998) Initial docking of ER-derived vesicles requires Uso1p and Ypt1p but is independent of SNARE proteins. EMBO J., 17, 2156–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoforidis S. and Zerial, M. (2000) Purification and identification of novel Rab effectors using affinity chromatography. Methods, 20, 403–410. [DOI] [PubMed] [Google Scholar]
- Christoforidis S., McBride, H.M., Burgoyne, R.D. and Zerial, M. (1999) The Rab5 effector EEA1 is a core component of endosome docking. Nature, 397, 621–625. [DOI] [PubMed] [Google Scholar]
- Dirac-Svejstrup A.B., Shorter, J., Waters, M.G. and Warren, G. (2000) Phosphorylation of the vesicle-tethering protein p115 by a casein kinase II-like enzyme is required for Golgi reassembly from isolated mitotic fragments. J. Cell Biol., 150, 475–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzler M.J., Hamel, J.C., Ochs, R.L. and Chan, E.K. (1993) Molecular characterization of two human autoantigens: unique cDNAs encoding 95- and 160-kD proteins of a putative family in the Golgi complex. J. Exp. Med., 178, 49–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe M., Gonatas, N.K. and Warren, G. (2000) The mitotic phosphorylation cycle of the cis-Golgi matrix protein GM130. J. Cell Biol., 149, 341–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A. (1996) Coiled-coils: new structures and new functions. Trends Biochem. Sci., 21, 375–382. [PubMed] [Google Scholar]
- Nakamura N., Rabouille, C., Watson, R., Nilsson, T., Hui, N., Slusarewicz, P., Kreis, T.E. and Warren, G. (1995) Characterization of a cis-Golgi matrix protein, GM130. J. Cell Biol., 131, 1715–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N., Lowe, M., Levine, T.P., Rabouille, C. and Warren, G. (1997) The vesicle docking protein p115 binds GM130, a cis-Golgi matrix protein, in a mitotically regulated manner. Cell, 89, 445–455. [DOI] [PubMed] [Google Scholar]
- Nelson D.S., Alvarez, C., Gao, Y.S., Garcia-Mata, R., Fialkowski, E. and Sztul, E. (1998) The membrane transport factor TAP/p115 cycles between the Golgi and earlier secretory compartments and contains distinct domains required for its localization and function. J. Cell Biol., 143, 319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuoffer C., Davidson, H.W., Matteson, J., Meinkoth, J. and Balch, W.E. (1994) A GDP-bound form of rab1 inhibits protein export from the endoplasmic reticulum and transport between Golgi compartments. J. Cell Biol., 125, 225–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olkkonen V.M. and Stenmark, H. (1997) Role of Rab GTPases in membrane traffic. Int. Rev. Cytol., 176, 1–85. [DOI] [PubMed] [Google Scholar]
- Pind S.N., Nuoffer, C., McCaffery, J.M., Plutner, H., Davidson, H.W., Farquar, M.G. and Balch, W.E. (1994) Rab1 and Ca2+ are required for the fusion of carrier vesicles mediating endoplasmic reticulum to Golgi transport. J. Cell Biol., 125, 239–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabouille C., Misteli, T., Watson, R. and Warren, G. (1995) Reassembly of Golgi stacks from mitotic Golgi fragments in a cell-free system. J. Cell Biol., 129, 605–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabouille C., Kondo, H., Newman, R., Hui, N., Freemont. P. and Warren, G. (1998) Syntaxin 5 is a common component of the NSF- and p97-mediated reassembly pathways of Golgi cisternae from mitotic Golgi fragments in vitro. Cell, 92, 603–610. [DOI] [PubMed] [Google Scholar]
- Sapperstein S.K., Lupashin, V.V., Schmitt, H.D. and Waters, M.G. (1996) Assembly of the ER to Golgi SNARE complex requires Uso1p. J. Cell Biol., 132, 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste J., Lahtinen, U. and Goud, B. (1995) Localization of the small GTP-binding protein rab1p to early compartments of the secretory pathway. J. Cell Sci., 108, 1541–1552. [DOI] [PubMed] [Google Scholar]
- Schiedel A.C., Barnekow, A. and Mayer, T. (1995) Nucleotide induced conformation determines posttranslational isoprenylation of the ras related rab6 protein in insect cells. FEBS Lett., 376, 113–119. [DOI] [PubMed] [Google Scholar]
- Seemann J., Jokitalo, E.J. and Warren, G. (2000) The role of the tethering proteins p115 and GM130 in transport through the Golgi apparatus in vivo. Mol. Biol. Cell, 11, 635–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J. and Warren, G. (1999) A role for the vesicle tethering protein, p115, in the post-mitotic stacking of reassembling Golgi cisternae in a cell-free system. J. Cell Biol., 146, 57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A. et al. (1998) EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature, 394, 494–498. [DOI] [PubMed] [Google Scholar]
- Sönnichsen B., Lowe, M., Levine, T., Jamsa, E., Dirac-Svejstrup, B. and Warren, G. (1998) A role for giantin in docking COPI vesicles to Golgi membranes. J. Cell Biol., 140, 1013–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H., Valencia, A., Martinez, O., Ullrich, O., Goud, B. and Zerial, M. (1994) Distinct structural elements of rab5 define its functional specificity. EMBO J., 13, 575–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisdale E.J., Bourne, J.R., Khosravi-Far, R., Der, C.J. and Balch, W.E. (1992) GTP-binding mutants of rab1 and rab2 are potent inhibitors of vesicular transport from the endoplasmic reticulum to the Golgi complex. J. Cell Biol., 119, 749–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale G., Rybin, V., Christoforidis, S., Thornqvist, P.-Ö., McCaffrey, M., Stenmark, H. and Zerial, M. (1998) Distinct Rab-binding domains mediate the interaction of Rabaptin-5 with GTP-bound Rab4 and Rab5. EMBO J., 17, 1941–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters M.G. and Pfeffer, S.R. (1999) Membrane tethering in intracellular transport. Curr. Opin. Cell Biol., 11, 453–459. [DOI] [PubMed] [Google Scholar]
- Waters M.G., Clary, D.O. and Rothman, J.E. (1992) A novel 115-kD peripheral membrane protein is required for intercisternal protein transport in the Golgi stack. J. Cell Biol., 118, 1015–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weide T., Köster, M. and Barnekow, A. (1999) Inactive and active mutants of rab1b are not tightly integrated into target membranes. Int. J. Oncol., 15, 727–736. [DOI] [PubMed] [Google Scholar]