Abstract
Recent progress in biological clock research has facilitated genetic analysis of circadian rhythm sleep disorders, such as delayed sleep phase syndrome (DSPS) and non-24-h sleep–wake syndrome (N-24). We analyzed the human period3 (hPer3) gene, one of the human homologs of the Drosophila clock-gene period (Per), as a possible candidate for rhythm disorder susceptibility. All of the coding exons in the hPer3 gene were screened for polymorphisms by a PCR-based strategy using genomic DNA samples from sleep disorder patients and control subjects. We identified six sequence variations with amino acid changes, of which five were common and predicted four haplotypes of the hPer3 gene. One of the haplotypes was significantly associated with DSPS (Bonferroni’s corrected P = 0.037; odds ratio = 7.79; 95% CI 1.59–38.3) in our study population. Our results suggest that structural polymorphisms in the hPer3 gene may be implicated in the pathogenesis of DSPS.
INTRODUCTION
Most living organisms exhibit a circadian (∼24 h) rhythm in their physiology and behavior, which is generated by an endogenous biological oscillator (Hastings, 1997). Genetic mutational studies using Drosophila led to the identification of the period (Per) gene, the first essential component of the circadian oscillator, which was followed by the isolation of various clock-relevant genes not only from Drosophila, but also from various species including mammals (Dunlap, 1999).
In mammals, clock-relevant genes such as Per1/2/3, CLOCK, Cry1/2, BMAL1 and Casein Kinase I epsilon (CK I ɛ) are expressed in the hypothalamic suprachiasmatic nuclei (SCN), the site of the principal circadian oscillator, and are thought to play a role in the clock mechanism (Dunlap, 1999; Lowrey et al., 2000). Mutations in Per2, CLOCK, Cry1/2 and CK I ɛ genes are known to modulate circadian rhythms in hamsters and mice (King and Takahashi, 2000; Lowrey et al., 2000); therefore, it is likely that human clock-relevant genes with mutations could also cause abnormal circadian rhythms.
In humans, sleep, plasma melatonin and the body core temperature show circadian rhythmicity, which is adjusted to 24 h by the environmental light–dark cycle and other stimuli such as social time cues (Hastings, 1997), whereas patients with circadian rhythm sleep disorders can not synchronize their sleep–wake cycle to the desired time schedule and have difficulties in their social lives (Campbell et al., 1999). Delayed sleep phase syndrome (DSPS) patients exhibit sleep-onset insomnia with an inability to wake up at a conventional time for school or work. Cases with non-24-h sleep–wake syndrome (N-24) show a daily delay of the sleep phase, resulting in cyclic episodes of insomnia. Recently, genetic contributions to the pathogenesis of DSPS and advanced sleep phase syndrome (ASPS), in which patients wake up and fall asleep too early relative to the desired time schedule, have been suggested by clinical genetic studies (Fink and Ancoli-Israel, 1997; Jones et al., 1999).
These results indicate that mutations in the clock-relevant genes may be involved in the susceptibility to circadian rhythm sleep disorders. To test this hypothesis, in this study we screened polymorphisms in the human Per3 (hPer3) gene, one of the three human homologs of the Drosophila clock-gene Per (Takumi et al., 1998; Zylka et al., 1998), and identified 20 sequence variations, of which six predicted amino acid changes. We characterized the polymorphisms and investigated their distribution in circadian rhythm sleep disorder patients and controls.
RESULTS
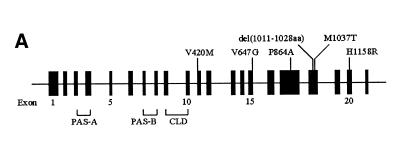
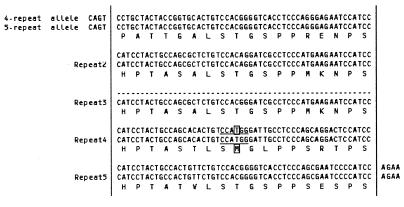
hPer3 cDNA was isolated from a size-fractionated cDNA library of the human brain (DDBJ/EMBL/GenBank accession No. AB047686). Sequence analysis of the cDNA revealed that the human chromosome 1 contig with accession No. Z98884 contained all of the coding sequences of hPer3 gene, as was suggested previously (Zylka et al., 1998). Table I summarizes the exon structure of the hPer3 gene, which consisted of 21 exons spanning >50 kbp. Based on the exon organization of the gene, polymerase chain reaction–single-strand conformation polymorphism (PCR-SSCP) analysis was performed, and genomic DNA from 36 patients (23 cases with DSPS and 13 with N-24) was screened for sequence variations in the hPer3 gene. Polymorphisms in DNA samples displaying aberrant SSCP migration patterns were identified by direct sequence analysis. A total of 20 sequence variations were detected: 13 single-nucleotide polymorphisms (SNPs) and a polymorphic repeat region with four or five copies of a 54 bp repetitive sequence in the exons, and four SNPs and two insertion/deletions in the portion of the introns examined (Table II). Five of the SNPs in the exons predicted amino acid substitutions: V420M, V647G, P864A, M1037T and H1158R. Exon 18 contained a polymorphic repeat domain, which harbored four or five copies of a tandemly repeated 54 bp sequence encoding 18 amino acids, in addition to the M1037T polymorphism (Figure 1A, 2). The prevalence of the six variations with putative amino acid changes was investigated in a total of 78 patients (48 with DSPS and 30 with N-24) and 100 controls by PCR-SSCP and subsequent direct sequence analysis. For exon 18, PCR–restriction fragment length polymorphism (RFLP) analysis was also used to confirm the number of repetitive sequences and the presence of M1037T polymorphism (data not shown). M1037T polymorphism was observed exclusively on a 4-repeat allele (Figure 2). Because V420M polymorphism was found only in one allele in one DSPS case out of the total study population, it was not included in further analyses. Genotype frequencies for the other five polymorphisms (V647G, P864A, 4-repeat/5-repeat, M1037T and H1158R) were not significantly different from those expected by Hardy–Weinberg equilibrium, both in the control population and the total study population. There was a marked linkage disequilibrium among the five polymorphisms. The V647G polymorphism was in complete concordance with H1158R. M1037T was always accompanied either by P864A or [V647G, H1158R]. On the basis of an expectation-maximization algorithm, we predicted four haplotypes, [V647, P864, 4-repeat, M1037, H1158] (designated as the H1 haplotype), [V647, P864, 5-repeat, M1037, H1158] (H2), [V647, A864, 4-repeat, T1037, H1158] (H3) and [G647, P864, 4-repeat, T1037, R1158] (H4), with a cumulative frequency of 100%. We could not identify a single recombinant event within these haplotypes in any of the patients or control individuals, who were unrelated except for one of the DSPS patients and one of the N-24 patients, who were siblings.
Table I. Exon structure of the hPer3 gene.
| Exon | Amino acids | Nucleotides (in Z98884) |
|---|---|---|
| 1 | 1–43 | (–224)–128 (61873–62224) |
| 2 | 43–92 | 129–274 (62660–62805) |
| 3 | 92–130 | 275–390 (63940–64055) |
| 4 | 131–198 | 391–592 (65264–65465) |
| 5 | 198–215 | 593–644 (71176–71227) |
| 6 | 215–265 | 645–793 (75744–75892) |
| 7 | 265–291 | 794–872 (78332–78410) |
| 8 | 291–327 | 873–979 (80266–80372) |
| 9 | 327–379 | 980–1136 (80911–81067) |
| 10 | 379–414 | 1137–1242 (86112–86217) |
| 11 | 415–457 | 1243–1371 (87112–87240) |
| 12 | 458–508 | 1372–1522 (87657–87807) |
| 13 | 508–553 | 1523–1658 (96504–96639) |
| 14 | 553–595 | 1659–1783 (97334–97458) |
| 15 | 595–653 | 1784–1957 (97686–97859) |
| 16 | 653–730 | 1958–2188 (103699–103929) |
| 17 | 730–962 | 2189–2886 (104337–105034) |
| 18 | 963–1072 | 2887–3214 (107053–107380) |
| 19 | 1072–1133 | 3215–3398 (112981–113164) |
| 20 | 1133–1183 | 3399–3549 (114218–114368) |
| 21 | 1184–1210 | 3550–3630 (119891–119971) |
In parentheses are the nucleotide positions in the sequence of accession No. Z98884.
Table II. Sequence variations detected in the hPer3 gene.
| Region | Nucleotide | Amino acid |
|---|---|---|
| change | change | |
| Intron 1 | 128 + 22(G→A) | none |
| Exon 2 | 195(C→T) | none |
| Intron 6 | 794–20insT | none |
| Intron 6 | 794–15(T→G) | none |
| Intron 7 | 872 + 53(T→C) | none |
| Intron 8 | 980–16delTTA | none |
| Exon 11 | 1258(G→A) | V420M |
| Exon 11 | 1338(T→C) | none |
| Exon 15 | 1881(G→A) | none |
| Exon 15 | 1940(T→G) | V647G |
| Exon 17 | 2259(G→A) | none |
| Exon 17 | 2415(C→T) | none |
| Exon 17 | 2484(A→G) | none |
| Exon 17 | 2590(C→G) | P864A |
| Exon 17 | 2616(G→A) | none |
| Exon 18 | 2934(C→T) | none |
| Exon 18 | del(3031–3084 nt) | del(1011–1028 aa) |
| Exon 18 | 3110(T→C) | M1037T |
| Exon 20 | 3473(A→G) | H1158R |
| Intron 20 | 3549 + 19(G→A) | none |
To avoid complications, nucleotide and amino acid positions are numbered according to the full-length hPer3 cDNA sequence containing five copies of a 54 bp repeat (AB047686). The 3110(T→C) [M1037T] and the 3473(A→G)[H1158R] polymorphisms are estimated to be exclusively on a 4-repeat allele; therefore, actual nucleotide (nt) and amino acid (aa) positions relative to the putative initiation site are 3056 nt (1019 aa) and 3419 nt (1141 aa), respectively.
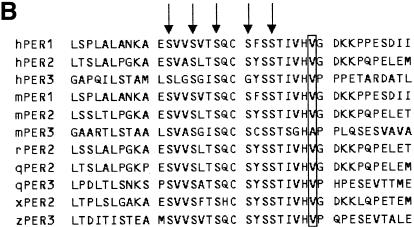
Fig. 1. (A) Schematic diagram of the hPer3 gene illustrating the locations of six polymorphisms with putative amino acid changes. Exons 1–21 are depicted as closed boxes and are numbered intermittently for clarity. Sizes of the introns and exons are not to scale. Polymorphisms with putative amino acid changes are shown above the gene. At the bottom are summarized the locations of the coding sequences for domains of hPER3. CLD, cytoplasmic localization domain. (B) Alignment of the amino acid sequences of vertebrate PER homologs. Only the region adjacent to the V647G polymorphism is shown. Amino acid residues that correspond to V647 in hPER3 are boxed. Arrows indicate the putative target residues of CK I ɛ (Toh et al., 2001). hPER1/2/3, human PER1/2/3; mPER1/2/3, mouse PER1/2/3; rPER2, rat PER2; qPER2/3, Japanese quail PER2/3; xPER2, Xenopus PER2; zPER3, zebrafish PER3.
Fig. 2. Nucleotide and amino acid sequences of each allele in the polymorphic repeat region of the hPer3 gene. The 4-repeat and 5-repeat alleles are aligned for optimal homology. The dotted line indicates a gap in the sequence created for optimal alignment. The nucleotide and amino acid that are subject to 3110(T→C) [M1037T] polymorphism, which was exclusively observed on a 4-repeat allele, are boxed. The NcoI restriction site used in RFLP analysis is underlined.
Carrier and estimated haplotype frequencies are reported in Table III. The frequency of the H4 haplotype was substantially higher in DSPS patients [7 (7.3%) of 96] than in controls [2 (1.0%) of 200], giving an odds ratio of 7.79 (95% CI 1.59–38.3; P = 0.0062). This difference remained significant after Bonferroni’s correction for two disorders and four haplotypes (P = 0.037). These results indicate that the H4 haplotype in the hPer3 gene is associated with increased susceptibility to DSPS. None of the other haplotypes showed any significant association with the disorders. Neither member of the sibling pair carried the H4 haplotype.
Table III. Carrier and estimated haplotype frequencies of hPer3 polymorphisms in patients and controls.
| Haplotype | Carrier frequency (%) |
Haplotype frequency (%) |
|||||
|---|---|---|---|---|---|---|---|
| DSPS | N-24 | Controls | DSPS | N-24 | Controls | ||
| H1[V647,P864,4-repeat,M1037,H1158] | 40/48 (83.3) | 28/30 (93.3) | 89/100 (89.0) | 62/96 (64.6) | 42/60 (70.0) | 137/200 (68.5) | |
| H2[V647,P864,5-repeat,M1037,H1158] | 16/48 (33.3) | 7/30 (23.3) | 34/100 (34.0) | 19/96 (19.8) | 7/60 (11.7) | 39/200 (19.5) | |
| H3[V647,A864,4-repeat,T1037,H1158] | 7/48 (14.6) | 10/30 (33.3) | 19/100 (19.0) | 8/96 (8.3) | 10/60 (16.7) | 22/200 (11.0) | |
| H4[G647,P864,4-repeat,T1037,R1158] | 7/48 (14.6)a | 1/30 (3.3) | 2/100 (2.0) | 7/96 (7.3)b | 1/60 (1.7) | 2/200 (1.0) | |
Underlined are minor alleles in each polymorphic locus.
Statistical comparisons were performed using a two-tailed Fisher’s exact test.
aP = 0.0055 versus controls, Bonferroni’s corrected P = 0.033.
bP = 0.0062 versus controls, odds ratio = 7.79, 95% CI 1.59–38.3, Bonferroni’s corrected P = 0.037.
DISCUSSION
There are two possibilities that might explain the association between the H4 haplotype of the hPer3 gene and DSPS. First, the H4 haplotype may be in linkage disequilibrium with functional mutations elsewhere within the hPer3 gene or a susceptibility locus nearby. Secondly, the polymorphisms on the H4 haplotype may directly contribute towards the increased susceptibility to DSPS. Out of the five polymorphisms on the H4 haplotype, G647 and R1158 are exclusively observed on the H4 haplotype, and G647 is a substitute of V647, which is conserved among most of the vertebrate PER homologs in the database (Table III; Figure 1B). Therefore, it is likely that G647 may be the causative variant. Of note, G647 is located within the region of amino acid similarity to the CK I ɛ binding domain of hPER1 and hPER2 (Vielhaber et al., 2000; Toh et al., 2001). Moreover, G647 is situated only five residues C-terminal relative to the region that corresponds to the putative phosphorylation site for CK I ɛ in hPER2 (Toh et al., 2001) (Figure 1B). Although CK I ɛ-dependent phosphorylation of PER3 protein has not been reported yet, structural conservation of the putative CK I ɛ phosphorylation site among PER1/2/3 indicates that PER3 is another substrate of CK I ɛ. In addition, substrate recognition of kinases can be affected by amino acid residues surrounding the consensus sequence (Pinna and Ruzzene, 1996). Taken together, we speculate that G647 polymorphism in the H4 haplotype may alter the CK I ɛ-dependent phosphorylation of hPER3. In our study population, two of the H4 haplotype carriers were controls (Table III); therefore, we can not exclude the possibility that an additional unknown polymorphism, such as one in CK I ɛ, might be necessary to exhibit an apparent alteration in phosphorylation of hPER3 and cause DSPS.
Phosphorylation of PER1 and PER2 by CK I ɛ seems to play an essential role in the circadian system (Keesler et al., 2000; Vielhaber et al., 2000; Toh et al., 2001). However, at the present time, it is difficult to elucidate how the G647-induced alteration of hPER3 phosphorylation, if any, could lead to the susceptibility to DSPS. While it is reported that a targeted disruption of the Per3 gene induced a subtle but significant alteration in the circadian cycle length in mice, the specific functional role of the Per3 gene in the clock system is not yet clearly understood (Shearman et al., 2000). The PER3 protein forms heterodimers with PER1/2 and CRY1/2, respectively, and enters into the nucleus, resulting in inhibition of CLOCK/BMAL1-mediated transcription. (Kume et al., 1999; Yagita et al., 2000). A possible contribution of PER3 in an output pathway of circadian oscillation is also proposed (Shearman et al., 2000). Altered phosphorylation of hPER3 might affect these functions. Alternatively, PER3 might act as a competitor for CK I ɛ binding against PER1/2 proteins. It is assumed that DSPS may be caused by subsensitivity to light, a prolonged endogenous circadian period, or abnormal coupling of the sleep–wake cycle to the circadian rhythm (Ozaki et al., 1996; Campbell et al., 1999). Functional analysis of the predicted form of the mutated hPER3 protein, in addition to the search for another polymorphism in CK I ɛ, may reveal the specific function of PER3 and the underlying physiological mechanism of developing DSPS.
No significant association was observed between the H4 haplotype and N-24, which often shows symptoms of DSPS during the course of the illness and is supposed to be caused by common underlying mechanisms (Weitzman et al., 1981; Campbell et al., 1999). In our study, 41 (85%) of the 48 DSPS patients did not carry the H4 haplotype (Table III); therefore, it is likely that DSPS is genetically heterogeneous with residual unknown genetic factors, of which some confer susceptibility to both DSPS and N-24.
A human CLOCK (hCLOCK) gene polymorphism in the 3′-untranslated region was reported to be associated with morningness–eveningness tendencies (individual differences in preferred timing of behavior) (Katzenberg et al., 1998). A recent study has revealed that a missense mutation in hPER2 causes familial ASPS (Toh et al., 2001). It is possible that the polymorphisms identified in this study, in combination with additional ones in other clock-relevant genes such as CLOCK and hPER2, may lead to physiological or pathological variations in circadian rhythmicity among individuals.
To our knowledge, this is the first study analyzing the hPer3 gene for polymorphisms in circadian rhythm sleep disorders. Our findings suggest that the H4 haplotype of the hPer3 gene may confer susceptibility to DSPS in ∼15% of affected people. Verification using other populations or familial based internal controls allowing for a linkage disequilibrium study is needed. Identification of a genetic factor will provide valuable insights into the pathogenesis of DSPS and in developing therapeutic strategies to treat patients with the disorder.
METHODS
Subjects
Cases and healthy control subjects, all of whom were sighted, were recruited at the participating institutes on mainland Japan. Cases comprised 48 patients with DSPS and 30 with N-24, and satisfied the International Classification of Sleep Disorders (ICSD1990) criteria. Cases with DSPS consisted of 29 males and 19 females (mean age 28.4 ± 10.0 years), and those with N-24 consisted of 22 males and eight females (mean age 26.4 ± 8.2 years). A total of 100 control subjects were recruited, 47 of whom were males and 53 were females (mean age 33.2 ± 7.9 years). All control subjects with a history of sleep disorders or psychoses were excluded. A part of the study population was reported previously (Ebisawa et al., 1999). Blood was aspirated and genomic DNA was prepared from white blood cells with a QIAamp DNA Blood Maxi Kit (Qiagen, Hilden) for use in the genetic analysis. The design of the study was approved by the ethics committees of the Saitama Medical School and the respective institutes from which blood samples were obtained. Each individual signed an informed consent form.
Isolation and sequencing of hPer3 cDNA
The hPer2 nucleotide sequence (DDBJ/EMBL/GenBank accession No. AB002345) was used to perform a BLAST search against expressed sequence tags (ESTs) of ∼10 000 cDNA clones isolated from size-fractionated human brain cDNA libraries (Ohara et al., 1997). Several clones showed high sequence similarity to the open reading frame (ORF) of hPer2. Sequence analysis of the full-length cDNAs suggested that they comprised two groups, of which one was made up of hPer1 cDNAs. cDNA clones in the second group showed 66% identity with mouse Per3 cDNA at the amino acid level, and >99% identity at the nucleotide level, to the corresponding sequences in a human chromosome 1 genomic contig, Z98884, which contains the sequence of the hPer3 gene (Zylka et al., 1998). Thus, we identified the second group clones as hPer3 cDNAs. The nucleotide sequence of a hPer3 cDNA with the longest 5′ region contained the first in-frame ATG preceded by a stop codon and encoded an ORF of 1210 amino acids, suggesting that the cDNA clone (AB047686) comprises a whole coding region of the hPer3 gene. The deduced amino acid sequence of the ORF showed 66, 39 and 42% identity with that of mouse PER3, human PER1 and human PER2 protein, respectively. From the comparison with the nucleotide sequence of Z98884, four discrepancies [1338(T→C), 2504(T→C)[L835P], 2852(A→G)[Q951R] and 3057(A→G)] were identified. After we had finished analyzing the sequence of the isolated clone, an hPer3 cDNA sequence assembled from public domain ESTs was independently released into the DDBJ/EMBL/GenBank database with the accession No. AL157954. The nucleotide sequence of AL157954 was >99% identical to that of AB047686.
SSCP and sequencing
Fluorescein-labeled oligonucleotide primers for SSCP analysis were synthesized based on the genomic sequence of the hPer3 gene to amplify all of the coding exons and the flanking exon–intron boundaries, generating PCR products of 209–414 bp (primer sequences are given in the Supplementary data, available at EMBO reports Online). SSCP analysis was performed as described previously (Ebisawa et al., 1999). Nucleotide sequences for the polymorphic repeat region and the insertion/deletions were confirmed by direct sequencing of the DNA samples homozygous for each variation.
RFLP analysis of the repeat domain
Exon 18 and adjacent introns were amplified using forward (5′-CAAAATTTTATGACACTACCAGAATGGCTGAC-3′) and reverse (5′-AACCTTGTACTTCCACATCAGTGCCTGG-3′) primers, the resultant PCR products separated on 1.5% agarose gels after digestion with NcoI, and visualized using SYBR Green I Stain (FMC BioProducts, Rockland, ME).
Statistical analysis
The calculations of Hardy–Weinberg equilibrium, pairwise linkage disequilibrium and haplotype frequencies were computed using the Arlequin software package (obtained from http://lgb.unige.ch/arlequin/software/). Fisher’s exact test was performed using Prism software (Graphpad Software Inc., San Diego, CA).
DDBJ/EMBL/GenBank accession Nos
The novel sequence data reported in this manuscript have been submitted to the DDBJ/EMBL/GenBank databases under accession Nos AB047520–AB047539 and AB047686.
Supplementary data
Primers used for SSCP analysis of the hPer3 gene are available as Supplementary data at EMBO reports Online.
Supplementary Material
Acknowledgments
ACKNOWLEDGEMENTS
Technical contributions by Eiichi Yamada, Kyoko Ohnishi and Masakazu Kinoshita are gratefully acknowledged. This work was supported by research grants from the Ministry of Health and Welfare, and the Ministry of Education, Science, Sports, and Culture (10557089, 11233206, 12470198) and Saitama Medical School.
REFERENCES
- Campbell S.S., Murphy, P.J., van den Heuvel, C.J., Roberts, M.L. and Stauble, T.N. (1999) Etiology and treatment of intrinsic circadian rhythm sleep disorders. Sleep Med. Rev., 3, 179–200. [DOI] [PubMed] [Google Scholar]
- Dunlap J.C. (1999) Molecular basis for circadian clocks. Cell, 96, 271–290. [DOI] [PubMed] [Google Scholar]
- Ebisawa T. et al. (1999) Allelic variants of human melatonin 1a receptor: function and prevalence in subjects with circadian rhythm sleep disorders. Biochem. Biophys. Res. Commun., 262, 832–837. [DOI] [PubMed] [Google Scholar]
- Fink R. and Ancoli-Israel, S. (1997) Pedigree of one family with delayed sleep phase syndrome. Sleep Res., 26, 713. [DOI] [PubMed] [Google Scholar]
- Hastings M.H. (1997) Central clocking. Trends Neurosci., 20, 459–464. [DOI] [PubMed] [Google Scholar]
- Jones C.R., Campbell, S.S., Zone, S.E., Cooper, F., DeSano, A., Murphy, P.J., Jones, B., Czajkowski, L. and Ptacek, L.J. (1999) Familial advanced sleep-phase syndrome: a short-period circadian rhythm variant in humans. Nature Med., 5, 1062–1065. [DOI] [PubMed] [Google Scholar]
- Katzenberg D., Young, T., Finn, L., Lin, L., King, D.P., Takahashi, J.S. and Mignot, E. (1998) A CLOCK polymorphism associated with human diurnal preference. Sleep, 21, 569–576. [DOI] [PubMed] [Google Scholar]
- Keesler G.A., Camacho, F., Guo, Y., Virshup, D., Mondadori, C. and Yao, Z. (2000) Phosphorylation and destabilization of human period1 clock protein by human casein kinase I ɛ. Neuroreport, 11, 951–955. [DOI] [PubMed] [Google Scholar]
- King D.P. and Takahashi, J.S. (2000) Molecular genetics of circadian rhythms in mammals. Annu. Rev. Neurosci., 23, 713–742. [DOI] [PubMed] [Google Scholar]
- Kume K., Zylka, M.J., Sriram, S., Shearman, L.P., Weaver, D.R., Jin, X., Maywood, E.S., Hastings, M.H. and Reppert, S.M. (1999) mCry1 and mCry2 are essential components of the negative limb of the circadian clock feedback loop. Cell, 98, 193–205. [DOI] [PubMed] [Google Scholar]
- Lowrey P.L., Shimomura, K., Antoch, M.P., Yamazaki, S., Zemenides, P.D., Ralph, M.R., Menaker, M. and Takahashi, J.S. (2000) Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science, 288, 483–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohara O., Nagase, T., Ishikawa, K.-I., Nakajima, D., Ohira, M., Seki, N. and Nomura, N. (1997) Construction and characterization of human brain cDNA libraries suitable for analysis of cDNA clones encoding relatively large proteins. DNA Res., 4, 53–59. [DOI] [PubMed] [Google Scholar]
- Ozaki S., Uchiyama, M., Shirakawa, S. and Okawa, M. (1996) Prolonged interval from body temperature nadir to sleep offset in patients with delayed sleep phase syndrome. Sleep, 19, 36–40. [PubMed] [Google Scholar]
- Pinna L.A. and Ruzzene, M. (1996) How do protein kinases recognize their substrates? Biochim. Biophys. Acta, 1314, 191–225. [DOI] [PubMed] [Google Scholar]
- Shearman L.P., Jin, X., Lee, C., Reppert, S.M. and Weaver, D.R. (2000) Targeted disruption of the mPer3 gene: subtle effects on circadian clock function. Mol. Cell. Biol., 20, 6269–6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takumi T. et al. (1998) A light-independent oscillatory gene mPer3 in mouse SCN and OVLT. EMBO J., 17, 4753–4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh K.L., Jones, C.R., He, Y., Eide, E.J., Hinz, W.A., Virshup, D.M., Ptacek, L.J. and Fu, Y.-H. (2001) An hPer2 phosphorylation site mutation in familial advanced sleep-phase syndrome. Science online, 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- Vielhaber E., Eide, E., Rivers, A., Gao, Z.-H. and Virshup, D.M. (2000) Nuclear entry of the circadian regulator mPER1 is controlled by mammalian casein kinase I ɛ. Mol. Cell. Biol., 20, 4888–4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzman E.D., Czeisler, C.A., Coleman, R.M., Spielman, A.J., Zimmerman, J.C. and Dement, W. (1981) Delayed sleep phase syndrome. Arch. Gen. Psychiatr., 38, 737–746. [DOI] [PubMed] [Google Scholar]
- Yagita K., Yamaguchi, S., Tamanini, F., van der Horst, G.T.J., Hoeijmakers, J.H.J., Yasui, A., Loros, J.J., Dunlap, J.C. and Okamura, H. (2000) Dimerization and nuclear entry of mPER proteins in mammalian cells. Genes Dev., 14, 1353–1363. [PMC free article] [PubMed] [Google Scholar]
- Zylka M.J., Shearman, L.P., Weaver, D.R. and Reppert, S.M. (1998) Three period homologs in mammals: differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron, 20, 1103–1110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.