Abstract
The quality control mechanism in the endoplasmic reticulum (ER) discriminates correctly folded proteins from misfolded polypeptides and determines their fate. Terminally misfolded proteins are retrotranslocated from the ER and degraded by cytoplasmic proteasomes, a mechanism known as ER-associated degradation (ERAD). We report the cDNA cloning of Edem, a mouse gene encoding a putative type II ER transmembrane protein. Expression of Edem mRNA was induced by various types of ER stress. Although the luminal region of ER degradation enhancing α-mannosidase-like protein (EDEM) is similar to class I α1,2-mannosidases involved in N-glycan processing, EDEM did not have enzymatic activity. Overexpression of EDEM in human embryonic kidney 293 cells accelerated the degradation of misfolded α1-antitrypsin, and EDEM bound to this misfolded glycoprotein. The results suggest that EDEM is directly involved in ERAD, and targets misfolded glycoproteins for degradation in an N-glycan dependent manner.
INTRODUCTION
In eukaryotes, newly synthesized proteins in the endoplasmic reticulum (ER) are transported to their final destinations if they are correctly folded, while misfolded proteins are selectively degraded (Bonifacino and Weissman, 1998; Ellgaard et al., 1999). Recently, an increasing number of misfolded membrane and secreted proteins in the ER have been shown to be degraded by cytoplasmic proteasomes, which is known as ER-associated degradation (ERAD) (Kopito, 1997; Bonifacino and Weissman, 1998; Plemper and Wolf, 1999). In both human and yeast cells, trimming of mannose residues from N-linked oligosaccharides determines the fate of misfolded glycoproteins. If trimming of the Man9GlcNAc2 (Man9) oligosaccharide precursor is prevented by α1,2-mannosidase inhibitors or by gene disruption, misfolded glycoproteins are stabilized (Jakob et al., 1998, Liu et al., 1999). The presence of a Man8GlcNAc2 (Man8)-binding lectin, which targets misfolded glycoproteins to degradation, has been postulated (Jakob et al., 1998), but the molecular identity of the putative lectin, as well as other molecules involved in the ERAD pathway, remains unknown.
Since the cellular requirement for the molecules involved in ERAD would be expected to increase under stress, we searched for novel genes that are upregulated following ER stress. In this study, we cloned Edem (DDBJ/EMBL/GenBank accession No. AB042828), a mouse gene encoding a protein with homology to ER α1,2-mannosidase I (Gonzalez et al., 1999; Tremblay and Herscovics, 1999), but no α1,2-mannosidase enzymatic activity. Functional analysis suggests that ER degradation enhancing α-mannosidase-like protein (EDEM) is directly involved in ERAD, and that it accelerates the degradation of misfolded glycoproteins depending on the extent of trimming of mannose residues from N-linked oligosaccharides.
RESULTS
Edem mRNA is upregulated by ER stress
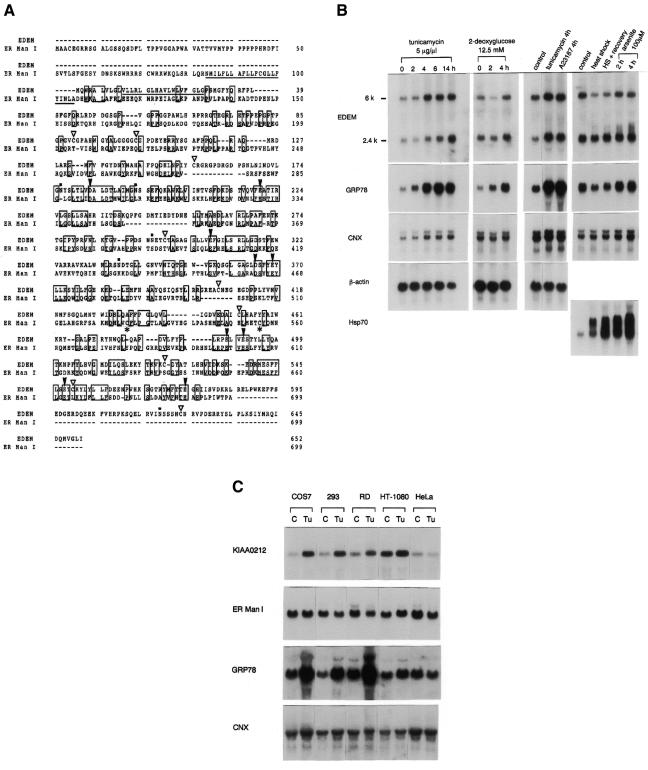
To search for ER stress inducible genes, we prepared RNA from mouse BALB/c 3T3 cells that had been treated with or without 5 µg/ml tunicamycin for 16 h. From a subtraction library generated using suppression subtractive hybridization, we focused on a 338-bp gene fragment whose deduced amino acid sequence is similar to that of class I α1,2-mannosidases. Nucleic acid data base (GenBank) searches revealed two EST clones (DDBJ/EMBL/GenBank accession No. AA260553 and AA726979) which have long 5′ extensions of this fragment. Using AA260553 as a probe, we obtained a 2.3 kb full-length cDNA from a mouse embryo cDNA library. This cDNA encodes a predicted type II transmembrane protein of 652 amino acids, containing a hydrophobic stretch at the N-terminus. We refer to this novel protein as EDEM. A search of the protein data base revealed a human homologue encoded by the KIAA0212 gene (Nagase et al., 1996) that is 92% identical to mouse EDEM. Homologous proteins were also identified in other species, including the Caenorhabditis elegans C47E12.3 gene product and the Saccharomyces cerevisiae hypothetical 91.2 kDa protein encoded by the gene YHR204W. The mouse Edem gene product shares 41 and 27% amino acid identity with the C. elegans and S. cerevisiae homologues, respectively. The human ER α1,2-mannosidase I (ER Man I) (Gonzalez et al., 1999; Tremblay and Herscovics, 1999), which converts Man9 oligosaccharide to Man8 isomer B, has the highest similarity to the mouse EDEM, among members of the α1,2-mannosidase family. Although the identity between human ER Man I and mouse EDEM is only ∼18%, EDEM contains signature motifs characteristic of class I α1,2-mannosidases including the conserved acidic amino acids (Figure 1A) (Lipari and Herscovics, 1999; Vallée et al., 2000).
Fig. 1. Peptide sequence of mouse EDEM and induction of Edem mRNA by ER stress. (A) Alignment of peptide sequences of mouse EDEM and human ER α1,2-mannosidase I (ER Man I). Identical residues are boxed. Two Cys residues conserved among processing α-mannosidases are shown by *, and Cys of EDEM are marked by open triangles. Conserved acidic amino acids are shown by arrowheads. Putative transmembrane regions are underlined, and possible N-glycosylation sites of mouse EDEM are dotted. (B) Northern blot analysis of mouse BALB/c 3T3 cells treated with various agents to induce ER stress (tunicamycin, 2-deoxyglucose, A23187) or cytoplasmic stress (heat shock, arsenite). Ten micrograms of total cellular RNA were hybridized with 32P-labelled cDNAs encoding EDEM, GRP78 (BiP), calnexin (CNX), β-actin, or Hsp70. (C) Northern blot analysis of various primate cell lines. Ten micrograms of total cellular RNA from control cells (C) or cells treated with tunicamycin for 4 h (Tu) were hybridized with 32P-labelled cDNAs encoding human homologue of EDEM (KIAA0212), ER Man I, GRP78 or CNX.
The mouse Edem gene was transcribed into two mRNA species of 2.4 kb and 6 kb (Figure 1B), both of which were detected in all mouse tissues examined (data not shown). The human homologue KIAA0212 mRNA is 6 kb in size, including a long 3′ untranslated region (UTR) of 4 kb (Nagase et al., 1996). 3′-rapid amplification of cDNA ends (3′-RACE) of Edem from mouse liver cDNA revealed another Edem cDNA carrying a 3.8 kb 3′ UTR, which was 70% identical to the entire KIAA0212 human gene, indicating that the two mRNA species transcribed from mouse Edem originate from different polyadenylation sites.
Edem expression after ER stress was examined by northern blot analysis (Figure 1B), and was quantified following normalization against β-actin mRNA (see Supplementary data). When mouse BALB/c 3T3 cells were exposed to ER or cytoplasmic stress, the expression of both forms of Edem mRNA (2.4 and 6 kb) increased by ∼3-fold in response to ER stress, whereas no increase was observed in response to cytoplasmic stress. We next examined whether the stress response was specific to Edem among various members of the class I α1,2-mannosidases. Several primate cultured cell lines were exposed to tunicamycin, and the expression profiles of various ER genes were compared (Figure 1C). In cells capable of responding to ER stress, the mRNA expression levels of GRP78 (BiP) and KIAA0212 (human homologue of Edem) increased by ∼4-fold and 3-fold, respectively. In contrast, neither ER Man I (Figure 1C) nor Golgi α1,2-mannosidase IB (Man IB; Herscovics et al., 1994) (data not shown) was increased by ER stress.
EDEM-HA localizes in the ER
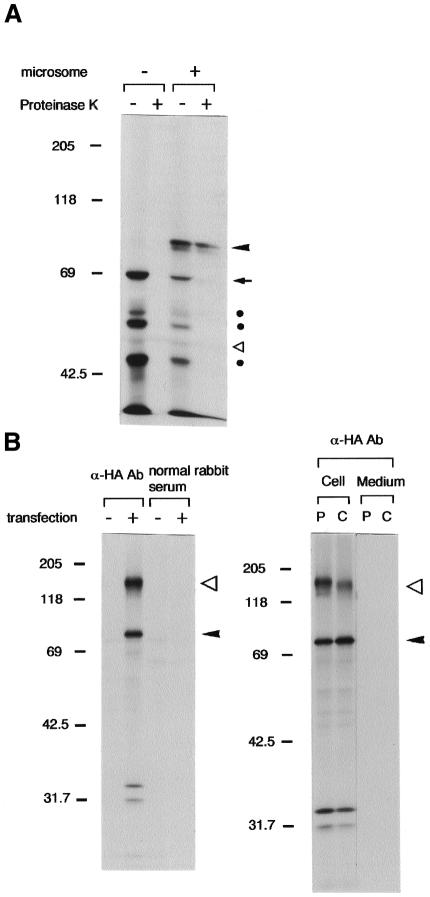
To examine the intracellular localization of EDEM, we expressed C-terminally HA-tagged EDEM (EDEM-HA) in COS-7 cells. The expressed protein was confined to the reticular structure around the nucleus, and colocalized with calnexin in the ER (data not shown). When EDEM-HA was translated in vitro in rabbit reticulocyte lysate, a 69 kDa protein was synthesized, and addition of canine pancreatic microsomes to the lysate produced a 78 kDa protein translocated into the microsomes that was resistant to proteinase K digestion (Figure 2A). Furthermore, this protein was recovered in the membrane fraction by the alkali-floatation method (Kutay et al., 1995) (data not shown). The same 78 kDa form was detected by immunoprecipitation of 35S-labelled COS-7 cells expressing EDEM-HA (Figure 2B), and was converted to the 69 kDa form by endoglycosidase H digestion (data not shown). No secretion into the medium was observed (Figure 2B). Thus, we conclude that EDEM-HA is a type II ER transmembrane protein that is inserted into the membrane as a 69 kDa form, and is then converted to a 78 kDa form by core glycosylation. A 150 kDa protein band was also found in the immunoprecipitates (Figure 2B), but the identity of this band is unknown, and its association with EDEM-HA is transient (Figures 2B and 3B).
Fig. 2. EDEM-HA localizes in the ER. (A) Autoradiogram of in vitro translated EDEM-HA in rabbit reticulocyte lysate metabolically labelled with [35S]methionine. In vitro transcribed Edem-HA RNA was used as template. EDEM-HA became resistant to proteinase K digestion when it was translocated into the microsomes. An arrowhead indicates the position of N-glycosylated EDEM-HA within the microsomes (78 kDa), and an arrow shows EDEM-HA without N-glycosylation (69 kDa). Open triangle shows the position of band detected without RNA template, and filled circles denote the degradation or immature products which were detected by α-HA antibody. Molecular mass standards (kDa) are shown on the left. (B) Autoradiogram of immunoprecipitation from metabolically labelled COS-7 cells. Arrowheads indicate the transfected EDEM-HA, and open triangles show the 150 kDa associated cellular protein that decreases during the chase period. P, pulse label for 45 min; C, chase for 45 min.
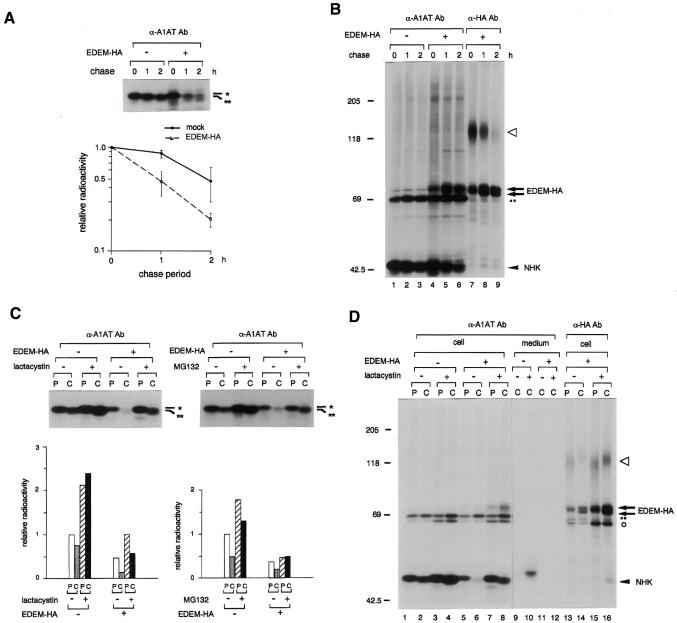
Fig. 3. EDEM accelerates the degradation of misfolded A1AT glycoprotein. (A) Autoradiogram following SDS–PAGE of immunoprecipitated material from HEK 293 cells cotransfected with NHK and EDEM-HA, or transfected with NHK and mock (indicated by –). The position of the predicted Man9 form of the A1AT variant (NHK) is shown by *, and that of the Man8 form by **. Cells were pulse-labelled for 15 min with [35S]methionine, and chased for the periods indicated. In this experiment only, lysates of cells cotransfected with EDEM-HA containing 1.6-fold more TCA-insoluble radioactivity than mock-transfected cells were used, since if equal amounts were used, the intensity of NHK signals in cells cotransfected with EDEM-HA was reduced to 42 ± 5% of the signal in mock-transfected cells at 0 h chase [see (C)]. The relative radioactivity in NHK at different times of chase was plotted on a semi-log scale relative to the intensity observed at 0 h chase. Error bars indicate standard deviations from the average of four (mock) or six (EDEM-HA) independent experiments. (B) Autoradiogram showing coimmunoprecipitation of NHK and EDEM-HA. The gel was exposed heavily to show coimmunoprecipitated bands. An arrowhead indicates the position of NHK, arrows show the transfected EDEM-HA, and the open triangle denotes the 150 kDa associated protein. The non-specific band detected in the immunoprecipitates using protein G-Sepharose is shown by dots (··). EDEM-HA appears as a doublet of different glycosylated forms, since a single band was formed after PNGase F treatment (data not shown), and EDEM has five possible N-glycosylation sites (Figure 1A). Molecular mass standards (kDa) are shown on the left. (C) Inhibition of NHK degradation by proteasome inhibitors. Cells were pulse-labelled for 15 min (P) and chased for 90 min (C), in the presence or absence of the proteasome inhibitors, lactacystin or MG132. MG135, a calpain inhibitor, was used as a control of MG132 treatment (indicated by –). Signals from immunoprecipitated NHK were compared with those of mock-transfected cells without proteasome inhibitor during pulse-labelling. (D) Autoradiogram showing coimmunoprecipitation of NHK and EDEM-HA in cells treated with lactacystin. An arrowhead, arrows, open triangle and dots indicate the same species as in Figure 3B. Open circle denotes another coimmunoprecipitated bands. The right panel (lanes 9–16) shows a 2.5 times longer exposure than the gel shown in the left panel (lanes 1–8).
EDEM lacks α1,2-mannosidase activity
To determine whether EDEM has α1,2-mannosidase activity, we expressed EDEM as a fusion protein secreted by COS-7 cells, in parallel with a fusion protein of mouse Man IB as a positive control. Three independent experiments clearly showed that protein A-EDEM has no α1,2-mannosidase activity, in contrast to protein A-Man IB (data not shown). This lack of enzymatic activity, in spite of its sequence similarity to class I α1,2-mannosidases, is consistent with the fact that EDEM lacks the two Cys residues that are conserved in all enzymatically active class I α1,2-mannosidases, including ER Man I (Figure 1A). The disulfide bond formed from these conserved Cys residues was shown previously to be required for enzymatic activity (Lipari and Herscovics, 1996).
EDEM accelerates the degradation of misfolded α1-antitrypsin
We then examined the possibility that EDEM is involved in ERAD by examining its effect on the degradation of α1-antitrypsin (A1AT) variant null (Hong Kong) (NHK; Sifers et al., 1988). A1AT is a plasma protein belonging to the serine protease inhibitor superfamily. The NHK genetic variant misfolds in the ER and is subsequently degraded by the cytoplasmic proteasome (Liu et al., 1999). Lack of A1AT in the serum is known to cause emphysema (Crystal, 1989). It was shown previously that treatment of cells with α1,2-mannosidase inhibitors prevents NHK degradation (Liu et al., 1999, 1997). When NHK was expressed in human embryonic kidney (HEK) 293 cells, it was degraded with a half-life of ∼2 h. Remarkably, overexpression of EDEM-HA with NHK accelerated the degradation of the A1AT variant, and decreased its half-life to ∼1 h (Figure 3A). A small shift in the electrophoretic mobility of NHK immunoprecipitated during the chase period is most likely due to trimming of the Man9 form to the Man8 form, because this shift was inhibited by the α1,2-mannosidase inhibitor kifunensine (Figure 4A). EDEM-HA consistently coprecipitated with NHK using antibodies to either HA-tag or to A1AT (Figure 3B and D, and see below). Analysis by velocity sedimentation revealed that both EDEM-HA and NHK were recovered in the same fractions of ∼11S, confirming that they were not in the aggregates (data not shown). Treatment of cells with the proteasome inhibitors lactacystin and MG132 (Z-Leu-Leu-Leu-H) (Tanaka, 1998) prevented the degradation of NHK in both the presence and absence of EDEM-HA expression (Figure 3C). These results support the idea that EDEM is directly involved in the ERAD pathway.
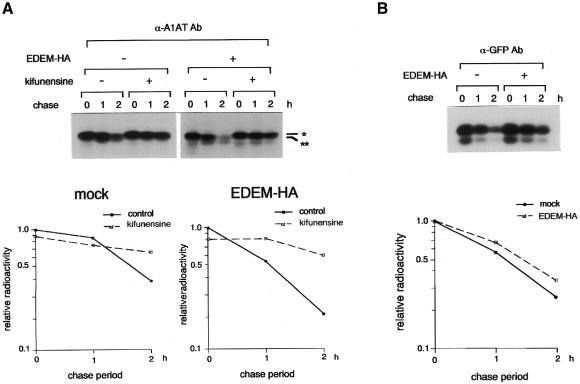
Fig. 4. EDEM accelerates the degradation of misfolded protein in an α1,2-mannosidase-dependent manner. (A) Autoradiogram showing the inhibition of NHK degradation by an α1,2-mannosidase inhibitor kifunensine. Metabolic labelling and immunoprecipitation of transfected HEK 293 cells was performed as described in Figure 3. The position of the predicted Man9 form of the A1AT NHK is shown by *, and that of the Man8 form by **. Intensity of NHK at 0 h chase without kifunensine was arbitrarily set to 1.0, and plotted as in Figure 3A. (B) Degradation of non-glycosylated protein in the ER. pEYFP-ER (Clontech, Palo Alto, CA) was transfected to HEK 293 cells and the intracellular fate of expressed GFP was examined with or without the coexpression of EDEM-HA, as described in A1AT NHK.
Coimmunoprecipitation of NHK using anti-HA antibody was prominent in cells treated with lactacystin during the chase period (Figure 3D, lane 16 versus lane 15). Comparable amounts of labelled NHK in EDEM-HA coexpressing cells were detected in both pulse and chase lanes in the presence of lactacystin (Figure 3D, lanes 7 and 8), but its electrophoretic mobility was different. Following pulse-labelling, NHK is most likely in the Man9 form (Figure 3D, lane7), whereas after the chase it is mostly in the Man8 form (Figure 3D, lane 8). Since the amount of NHK coprecipitated with EDEM-HA was much higher in the latter sample (Figure 3D, lane 16 versus lane 15), EDEM appears to bind preferentially to NHK in the Man8 form. Furthermore, NHK was secreted into the medium in mock-transfected cells treated with lactacystin (Figure 3D, lane 10), but coexpression of EDEM-HA greatly reduced this secretion (Figure 3D, lane 12), providing further evidence that EDEM-HA binds preferentially to the Man8 form of NHK within the ER. Essentially the same coprecipitation and secretion profiles were observed in cells treated with MG132 (data not shown).
EDEM accelerates the degradation of misfolded A1AT depending on the extent of mannose trimming from the N-linked oligosaccharides
Next, we asked whether accelerated degradation of misfolded glycoprotein by EDEM depends on trimming of the N-linked oligosaccharides. We added kifunensine to inhibit trimming of α1,2-mannose residues from N-glycans (Gonzalez et al., 1999; Liu et al., 1999; Tremblay and Herscovics, 1999). Under these conditions, it was shown previously that misfolded A1AT NHK accumulates in the ER as a Man9-containing form, and that its subsequent degradation is inhibited (Liu et al., 1999). Overexpression of EDEM-HA did not accelerate the degradation of NHK in the presence of kifunensine (Figure 4A), indicating that the EDEM-enhanced degradation of NHK requires mannose trimming by ER Man I. The same result was obtained using the α1,2-mannosidase inhibitor 1-deoxymannojirimycin (Fuhrmann et al., 1984) (data not shown).
Mannosamine prevents oligosaccharide precursor elongation resulting in the transfer of Man5-7-containing N-glycans to glycoproteins (Van Leeuwen and Kearse, 1997). The degradation of NHK in the presence of mannosamine was not accelerated by coexpression of EDEM-HA (data not shown), indicating further that the effect of EDEM specifically requires trimming of mannose residues from Man9 of the misfolded glycoprotein.
Finally, the involvement of EDEM in the degradation of non-glycosylated substrate was examined (Figure 4B). When a variant of GFP (pEYFP-ER, Clontech) was expressed in the ER of 293 cells, newly synthesized green fluorescent protein (GFP) variant was degraded with a half-life of ∼70 min. Overexpression of EDEM had no effect on the degradation of EYFP-ER, indicating that the effect of EDEM on degradation is specific for misfolded glycoproteins.
DISCUSSION
We have cloned a novel mouse gene encoding a type II ER transmembrane protein, and named it Edem. In spite of its sequence homology with ER mannosidase I, it lacks enzymatic activity as a processing α1,2-mannosidase. EDEM interacts with misfolded A1AT NHK and accelerates its degradation by proteasomes in an α1,2-mannosidase-dependent manner. Thus, EDEM links ER misfolding of glycoproteins to retrotranslocation and proteasomal degradation.
We have identified Edem as an ER stress inducible gene, based on the assumption that the ERAD machinery must be upregulated by ER stress. Further support for this assumption was recently provided by reports showing a close correlation between the unfolded protein response (UPR; Chapman et al., 1998) and ERAD in yeast (Casagrande et al., 2000; Travers et al., 2000). Travers and coworkers reported that a large number of genes involved in ERAD and in the secretory pathway were UPR targets. We exclude the possibility that overexpression of EDEM-HA upregulates the UPR and accelerates the degradation of misfolded protein secondarily, because the synthesis of BiP protein is not upregulated by overexpression of EDEM-HA (data not shown).
The present results indicate that misfolded secretory glycoproteins in the ER interact with EDEM in a manner similar to the binding of immature glycoproteins to calnexin (Trombetta and Helenius, 1998). While interaction of nascent molecules with calnexin leads to productive folding, trapping of misfolded glycoproteins by EDEM appears to be followed by degradation. Although proteasomal degradation of misfolded proteins is reportedly preceded by transport through the Sec61p channel (Plemper et al., 1997; Zhou and Schekman, 1999), the molecular mechanisms remain unknown. Given that EDEM is functionally upstream of Sec61p, analysis of molecules which interact with EDEM may reveal how the ER quality control machinery is linked to proteasomal degradation.
Speculation
EDEM accelerates the degradation of misfolded A1AT depending on the extent of mannose trimming from N-linked glycans. Furthermore, EDEM-HA preferentially binds misfolded A1AT bearing Man8GlcNAc2 oligosaccharides (Figure 3D). Since EDEM has no α1,2-mannosidase enzymatic activity in spite of its sequence homology, it is possible that EDEM is the postulated Man8-binding lectin which recognizes misfolded glycoproteins in the ERAD pathway (Jakob et al., 1998), but additional work will be required to demonstrate directly EDEM lectin activity in vitro.
METHODS
Cloning of Edem cDNA. The suppression subtractive hybridization library of tunicamycin-treated BALB/c 3T3 cells was created using the PCR-select cDNA subtraction kit according to the manufacturer’s recommendations (Clontech). A 2.3 kb Edem cDNA was cloned from a mouse 15.5 day embryo cDNA library (Gibco-BRL, Rockville, MD). Marathon-ReadyTM cDNA from mouse liver (Clontech) was used as template for 3′-RACE to obtain 5.8 kb Edem cDNA, and several overlapping DNA fragments were cloned and sequenced. The program SOSUI (Hirokawa et al., 1998) was used to predict the putative transmembrane region.
Cell culture and drug treatment. BALB/c 3T3, COS-7 and HEK 293 cells were cultured in DMEM supplemented with 10% fetal bovine serum. Cells were treated with 5 µg/ml tunicamycin, 12.5 mM 2-deoxyglucose or 7 µM A23187 in order to induce ER stress. Cells were heat-shocked for 90 min at 42°C, and allowed to recover for 2 h at 37°C, or treated with 100 µM arsenite, in order to induce cytoplasmic stress. Kifunensine (kindly provided by Fujisawa Pharmaceutical Co., Osaka, Japan) was added at a final concentration of 5 µg/ml 4 h prior to pulse-labelling and was present in the medium during the chase period. 1-deoxymannojirimycin (Sigma, St Louis, MO) was added in the medium at a concentration of 1 mM. Lactacystin (Kyowa Medics Co., Tokyo, Japan) and MG132 (Z-Leu-Leu-Leu-H) were added at a final concentration of 20 µM (Tanaka, 1998).
Plasmids and transfection. A C-terminal HA tag was added to Edem cDNA by PCR to produce EDEM-HA in the expression vector pCMV·SPORT2 (Gibco-BRL). The vector pcDNA3.1 (+) (Invitrogen, Carlsbad, CA), which carries the CMV promoter, was used for mock transfection. Human A1AT cDNA was cloned into pREP9, and two nucleotides were deleted using QuickChangeTM site-directed mutagenesis (Stratagene, La Jolla, CA) to create the plasmid encoding the A1AT folding-incompetent variant NHK. FuGENETM 6 transfection reagent (Boehringer Mannheim, Indianapolis, IN) was used for plasmid transfections.
In vitro translation. In vitro translation and translocation of EDEM-HA was carried out using rabbit reticulocyte lysate (Promega, Madison, WI) and canine pancreatic microsomes, and the reaction products were labelled with [35S]methionine (NEN, Boston, MA). For proteinase K digestion, lysates were kept on ice for 30 min with 200 µg/ml of the enzyme.
Metabolic labelling and immunoprecipitation. Cells were labelled with 35S-Promix (Amersham-Pharmacia, Amersham, UK) in medium lacking methionine, and chased in medium containing excess methionine. Cells were lysed in a buffer containing 1% NP-40, 150 mM NaCl, 50 mM Tris–HCl pH 8.0 and protease inhibitors (1 mM N-methyl maleimide, 1 mM phenylmethylsulphonyl fluoride, 1 µg/ml leupeptin and 1 µg/ml pepstatin A) (Satoh et al., 1996). Anti-HA antibody (mouse monoclonal antibody, clone 12CA5, Boehringer Mannheim) and anti-A1AT antibody (goat polyclonal antibody, MBL, Nagoya, Japan) were used for immunoprecipitation. In experiments designed to detect NHK degradation, immunoprecipitated samples were resolved by 10% SDS–PAGE until the Evans Blue dye reached the bottom of a 15 cm gel to distinguish between the Man8 and Man9 forms of NHK.
α1,2-Mannosidase activity. Edem cDNA lacking the N-terminal 29 amino acids (including the putative transmembrane region) was fused in-frame with the IgG binding domain of protein A in pPak, as described previously for mouse α1,2-mannosidase IB (Herscovics et al., 1994). Both EDEM and Man IB-containing plasmids were transfected into COS-7 cells, and secreted fusion proteins were collected with IgG-Sepharose. Duplicate beads were assayed for α1,2-mannosidase activity, as described previously (Herscovics et al., 1994). Expression of comparable amounts of fusion proteins on the beads was confirmed by western blotting using HRP-conjugated IgG.
Supplementary data. Supplementary data are available at EMBO reports Online.
Supplementary Material
Acknowledgments
ACKNOWLEDGEMENTS
We thank Mr B. Sleno for technical assistance. This study was supported by grants from the Ministry of Education, Science, Sports and Culture of Japan (N.H., I.W. and K.N.), and a grant (A.H.) and fellowship (L.O.T.) from the Medical Research Council of Canada.
References
- Bonifacino J.S. and Weissman, A.M. (1998) Ubiquitin and the control of protein fate in the secretory and endocytic pathways. Annu. Rev. Cell. Dev. Biol., 14, 19–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casagrande R., Stern, P., Diehn, M., Shamu, C., Osario, M., Zúñiga, M., Brown, P.O. and Ploegh, H. (2000) Degradation of proteins from the ER of S. cerevisiae requires an intact unfolded protein response pathway. Mol. Cell, 5, 729–735. [DOI] [PubMed] [Google Scholar]
- Chapman R., Sidrauski, C. and Walter, P. (1998) Intracellular signaling from the endoplasmic reticulum to the nucleus. Annu. Rev. Cell. Dev. Biol., 14, 459–485. [DOI] [PubMed] [Google Scholar]
- Crystal R.G. (1989) The α1-antitrypsin gene and its deficiency states. Trends Genet., 5, 411–417. [DOI] [PubMed] [Google Scholar]
- Ellgaard L., Molinari, M. and Helenius, A. (1999) Setting the standards: quality control in the secretory pathway. Science, 286, 1882–1888. [DOI] [PubMed] [Google Scholar]
- Fuhrmann U., Bause, E., Legler, G. and Ploegh, H. (1984) Novel mannosidase inhibitor blocking conversion of high mannose to complex oligosaccharides. Nature, 307, 755–758. [DOI] [PubMed] [Google Scholar]
- Gonzalez D.S., Karaveg, K., Vandersall-Nairn, A.S., Lal, A. and Moremen, K.W. (1999) Identification, expression, and characterization of a cDNA encoding human endoplasmic reticulum mannosidase I, the enzyme that catalyzes the first mannose trimming step in mammalian Asn-linked oligosaccharide biosynthesis. J. Biol. Chem., 274, 21375–21386. [DOI] [PubMed] [Google Scholar]
- Herscovics A., Schneikert, J., Athanassiadis, A. and Moremen, K.W. (1994) Isolation of a mouse Golgi mannosidase cDNA, a member of a gene family conserved from yeast to mammals. J. Biol. Chem., 269, 9864–9871. [PubMed] [Google Scholar]
- Hirokawa T., Boon-Chieng, S. and Mitaku, S. (1998) SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics, 14, 378–379. [DOI] [PubMed] [Google Scholar]
- Jakob C.A., Burda, P., Roth, J. and Aebi, M. (1998) Degradation of misfolded endoplasmic reticulum glycoproteins in Saccharomyces cerevisiae is determined by a specific oligosaccharide structure. J. Cell Biol., 142, 1223–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopito R.R. (1997) ER quality control: the cytoplasmic connection. Cell, 88, 427–430. [DOI] [PubMed] [Google Scholar]
- Kutay U., Ahnert-Hilger, G., Hartmann, E., Wiedenmann, B. and Rapoport, T.A. (1995) Transport route for synaptobrevin via a novel pathway of insertion into the endoplasmic reticulum membrane. EMBO J., 14, 217–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipari F. and Herscovics, A. (1996) Role of the cysteine residues in the α1, 2-mannosidase involved in N-glycan biosynthesis in Saccharomyces cerevisiae. The conserved Cys340 and Cys385 residues form an essential disulfide bond. J. Biol. Chem., 271, 27615–27622. [DOI] [PubMed] [Google Scholar]
- Lipari F. and Herscovics, A. (1999) Calcium binding to the class I α1, 2-mannosidase from Saccharomyces cerevisiae occurs outside the EF hand motif. Biochemistry, 38, 1111–1118. [DOI] [PubMed] [Google Scholar]
- Liu Y., Choudhury, P., Cabral, C.M. and Sifers, R.N. (1997) Intracellular disposal of incompletely folded human α1-antitrypsin involves release from calnexin and post-translational trimming of asparagine-linked oligosaccharides. J. Biol. Chem., 272, 7946–7951. [DOI] [PubMed] [Google Scholar]
- Liu Y., Choudhury, P., Cabral, C.M. and Sifers, R.N. (1999) Oligosaccharide modification in the early secretory pathway directs the selection of a misfolded glycoprotein for degradation by the proteasome. J. Biol. Chem., 274, 5861–5867. [DOI] [PubMed] [Google Scholar]
- Nagase T. et al. (1996) Prediction of the coding sequences of unidentified human genes. VI. The coding sequences of 80 new genes (KIAA0201–KIAA0280) deduced by analysis of cDNA clones from cell line KG-1 and brain. DNA Res., 3, 321–329. [DOI] [PubMed] [Google Scholar]
- Plemper R.K. and Wolf, D.H. (1999) Retrograde protein translocation: ERADication of secretory proteins in health and disease. Trends Biochem. Sci., 24, 266–270. [DOI] [PubMed] [Google Scholar]
- Plemper R.K., Böhmler, S., Bordallo, J., Sommer, T. and Wolf, D.H. (1997) Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature, 388, 891–895. [DOI] [PubMed] [Google Scholar]
- Satoh M., Hirayoshi, K., Yokota, S.-i., Hosokawa, N. and Nagata, K. (1996) Intracellular interaction of collagen-specific stress protein HSP47 with newly synthesized procollagen. J. Cell Biol., 133, 469–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sifers R.N., Brashears-Macatee, S., Kidd, V.J., Muensch, H. and Woo, S.L.C. (1988) A frameshift mutation result in a truncated α1-antitrypsin that is retained within the rough endoplasmic reticulum. J. Biol. Chem., 263, 7330–7335. [PubMed] [Google Scholar]
- Tanaka K. (1998) Proteasomes: structure and biology. J. Biochem., 123, 195–204. [DOI] [PubMed] [Google Scholar]
- Travers K.J., Patil, C.K., Wodicka, L., Lockhart, D.J., Weissman, J.S. and Walter, P. (2000) Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell, 101, 249–258. [DOI] [PubMed] [Google Scholar]
- Tremblay L.O. and Herscovics, A. (1999) Cloning and expression of a specific human α1, 2-mannosidase that trims Man9GlcNAc2 to Man8GlcNAc2 isomer B during N-glycan biosynthesis. Glycobiology, 9, 1073–1078. [DOI] [PubMed] [Google Scholar]
- Trombetta E.S. and Helenius, A. (1998) Lectins as chaperones in glycoprotein folding. Curr. Opin. Struct. Biol., 8, 587–592. [DOI] [PubMed] [Google Scholar]
- Vallée F., Karaveg, K., Herscovics, A., Moremen, K.W. and Howell, P.L. (2000) Structural basis for catalysis and inhibition of N-glycan processing class I α1, 2-mannosidases. J. Biol. Chem., 275, 41287–41298. [DOI] [PubMed] [Google Scholar]
- Van Leeuwen J.E.M. and Kearse, K.P. (1997) Reglucosylation of N-linked glycans is critical for calnexin assembly with T cell receptor (TCR) α proteins but not TCRβ proteins. J. Biol. Chem., 272, 4179–4186. [DOI] [PubMed] [Google Scholar]
- Zhou M. and Schekman, R. (1999) The engagement of Sec61p in the ER dislocation process. Mol. Cell, 4, 925–934. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.